Abstract
Pseudomonas aeruginosa is an opportunistic pathogen of public health concern. This study aimed to investigate the prevalence of P. aeruginosa, some virulence factors, and antimicrobial resistance patterns and highlight the potential pathways of horizontal blaSHV-resistant gene transfer from diverse sources. A total of 220 samples were collected from fish (n = 90), water (n = 30), poultry (n = 50), and humans (n = 50). All samples were isolated, confirmed by the Vitek 2 system, and tested against antimicrobial agents. Some virulence and resistance genes were examined by PCR and sequenced for the blaSHV-resistant gene from four selected isolates from each source. SPSS v26, with chi-squared tests and Pearson correlations (p < 0.05), was implemented for statistical investigation. P. aeruginosa was isolated at 33.3%, 20%, 14%, and 24% from fish, water, poultry, and humans, respectively. Using the diffusion disk method, extensively drug-resistant (XDR) and multidrug-resistant (MDR) strains were detected. All strains harbored the oprL and toxA genes, while the lasB gene was present in 40% of fish samples but not present in human samples. All strains lacked the exoS gene. The tetA, sul1, blaSHV, and blaTEM resistance genes were detected at different percentages. The blaSHV genes from fish and water isolates were closely related to each other and showed similarity to those of the human isolates. The poultry isolates formed a separate phylogenetic lineage. The emergence of XDR and MDR P. aeruginosa highlights a possible public health threat. Based on the gene similarity between fish and water isolates, our results suggest that these isolates have a common origin. The similarity between the human isolates and environmental isolates (fish and water) raises concerns about possible transmission to humans.
1. Introduction
Pseudomonas aeruginosa (P. aeruginosa) is an opportunistic bacterium that poses a significant concern in both human and veterinary medicine due to its occurrence in several environments, including aquatic life, poultry, and water sources. In fish farming, it produces septicemia and ulceration in fish, leading to serious complications and financial losses [1]. In poultry, it can cause respiratory illnesses, septicemia, and reduced productivity, particularly in intensive farms. Polluted water is a critical source and dissemination vector for this durable pathogen [2]. In humans, P. aeruginosa is the primary cause of nosocomial infection and is frequently resilient to numerous medications, making treatment difficult [3]. Under the One Health approach, its importance is highlighted by its existence in the human, animal, and environmental sectors. Its ecology and patterns of transmission must be understood in order to implement integrated control procedures [4].
Fish constitute a rich source of good-quality protein and nutritional elements such as omega-3 fatty acids, selenium, vitamin D, and iodine [5]. The consumption of fish has recently seen a 3.2% annual increase in comparison to meat products, particularly in developing countries [6]. There are over 200 species in the genus Pseudomonas, such as P. aeruginosa, P. anguilliseptica, P. lundensis, P. putida, P. fragi, and P. fluorescens [7]. Both freshwater and marine water have an abundance of these bacteria [6]. In the poultry industry, P. aeruginosa is a major pathogen that primarily affects impaired hens. It can cause serious infections such septicemia, yolk sac infections, and respiratory diseases, and it is commonly isolated from hatcheries, water supply systems, litter, and poultry enclosure air [8]. The pathogen’s capacity to create biofilms increases its resistance and survival, making antimicrobial treatment and disinfection difficult. Infections can result in increased mortality, slow development rates, and financial losses in the production of broilers [9].
Numerous virulence factors are produced by P. aeruginosa, such as exotoxins (exotoxins S, T, U, and A), pigments (pyomelanin, pyoverdine, and pyocyanine), proteases (which cause tissue destruction), flagella, lectins, siderophores, and different secretion systems (the Type III “injectasome” in particular) [10]. The toxA gene encodes the extracellular enzyme exotoxin A (ETA), which prevents host cells from synthesizing proteins [11]. The type III secretion system’s (T3SS) effector exotoxins—ExoT, ExoS, ExoY, and ExoU—are believed to be a major contributor to P. aeruginosa virulence [12]. LasB elastase breaks down the structural proteins of cells, including elastin, collagen, and non-collagen proteins [13]. P. aeruginosa’s outer membrane proteins (OprI and OprL) are essential for the bacterium’s interactions with its environment and its natural resistance to antibiotics; their presence has been associated with efflux transport systems that affect cell permeability [14]. Because these proteins are specific to this organism, they could be a reliable indicator for the rapid detection of P. aeruginosa in clinical samples [15].
Multidrug resistance (MDR) in bacteria has emerged as one of the 21st century’s most formidable problems because of the rising incidence of illnesses that are difficult to cure and the dearth of appropriate therapeutic options [12]. Due to P. aeruginosa’s innate resistance to several antibiotic classes and its capacity to develop resistance to almost all potent antimicrobial agents, MDR strains can arise [16]. Among the most prevalent genes for antibiotic resistance, blaSHV aids bacteria such as P. aeruginosa in their resistance to β-lactam drugs. This gene has been detected in humans, fish, poultry, and water, among other sources. According to new research on poultry, P. aeruginosa from broiler farms carries the blaSHV gene, frequently in combination with other resistance genes [17]. blaSHV-positive bacteria are also found in farm and natural water, most likely as a result of pollution and animal husbandry waste. Laboratory tests have revealed a substantial frequency of blaSHV in humans, especially in hospital-associated infections [18].
A straightforward, integrated framework is used in the One Health strategy to emphasize the connections between aquatic, animal, and human health. To understand and lessen the risks of disease transmission between species, this point of view is crucial. Recent studies have pointed out the value of such a strategy in combating resistant bacteria and newly developing transmissible diseases. Uddin et al. (2021) [12] suggested the need for continuous detection and control strategies to combat multidrug-resistant P. aeruginosa. Additionally, ref. [4] focused on the necessity of a One Health framework to establish the prevalence of zoonotic threats. By adopting this strategy, this study aimed to assess P. aeruginosa’s prevalence, virulence factors, and resistance patterns and highlight the potential pathways involved in the horizontal gene transfer and environmental dissemination of the blaSHV gene among P. aeruginosa isolates from diverse sources.
2. Materials and Methods
2.1. Animal Ethics
Fish and poultry handling, human sample collection, and other experimental methods were approved by Suez Canal University’s Animal Ethics Review Committee, Egypt, with ethics number SCU-VET-AREC-R-2025018.
2.2. Sampling
Samples were collected from different types of sources in the Ismailia Governorate of Egypt for the isolation of P. aeruginosa. From three freshwater fish farms, 90 moribund fish samples (30 Oreochromis niloticus, 30 Tilapia zillii, and 30 Clarias gariepinus) were gathered. Each fish’s gills, liver, spleen, and kidneys were removed aseptically and placed in aerated plastic bags in accordance with Wamala et al. [19]. Thirty water samples were collected from the three corresponding freshwater fish farms. According to [20], water samples were handled and stored in an icebox. Lung, liver, and spleen samples were collected from 50 broiler chicks suffering from respiratory manifestations. The samples were pooled and handled as one. Fifty swabs were collected from human sputum, wounds, and urine at Suez Canal University Hospital in the Ismailia Governorate. Suez Canal University’s Faculty of Veterinary Medicine’s Bacteriology Laboratory received all samples for bacteriological examination.
2.3. Isolation, Vitek 2 Identification, and PCR Confirmation of Recovered Isolates
Samples were directly streaked onto cetrimide agar (Himedia, Maharashtra, India) and MacConkey’s agar (Oxoid, Cheshire, UK) and then incubated aerobically for 24 h at 37 °C. Yellowish green fluorescent pigment production is commonly associated with Pseudomonads [21]. Recovered colonies were purified to determine their biochemical and phenotypic characteristics. In short, Gram’s stain was used for the morphological identification of each isolate, and the Vitek 2 system (bioMérieux, Marcy-l’Étoile, France) was used for biochemical identification. Additionally, the detected isolates were confirmed using a species-specific set of primers that targeted the P. aeruginosa 16SrRNA gene (PaF: 5′-GGGGGATCTTCGGACCTCA-3′; PaR: 5′TCCTTAGAGTGCCCACCCG-3′), as outlined by Spilker et al. [22].
2.4. Antimicrobial Susceptibility Testing
The obtained isolates were subjected to antimicrobial susceptibility testing utilizing 11 antimicrobial agents (Oxoid, UK) from seven antibiotic classes: the tetracycline class (tetracycline (TE, 30 µg), oxytetracycline (OT, 30 µg)); the aminoglycoside class (amikacin (AK, 30 µg), tobramycin (TOB, 10 µg)); the cephalosporin III class (cefotaxime (CTX, 30 µg), ceftriaxone (CRO, 30 µg)); the β-lactam–β-lactamase inhibitor combination class (amoxicillin clavulanic acid (AMC, 30 µg), ampicillin/sulbactam (SAM, 20 µg)); the folate pathway inhibitor class (sulfamethoxazole/trimethoprim (SXT, 25 µg)); the fluoroquinolone class (ciprofloxacin (CIP, 5 µg)); and the polymyxin class (colistin (CT, 10 µg)). The test was carried out on Muller–Hinton agar plates (Oxoid, UK), which were incubated at 37 °C for 24 h. The test findings were interpreted in light of the CLSI [23].
2.5. Molecular Typing of Isolated P. aeruginosa Strains’ Virulence and Antimicrobial Resistance Genes
A silica-based membrane QIAamp DNA Mini Kit (catalog no. 51304, Hilden, Germany) was used to extract the DNA of purified strains in accordance with the manufacturer’s instructions. Briefly, QIAGEN protease was pipetted into a microcentrifuge tube; then, the sample was added; then, buffer AL was added, and the sample was mixed, incubated, and centrifuged; ethanol (96%) was added and it was mixed and then centrifuged again. Afterwards, the mixture was applied to the QIAamp mini spin column and centrifugated. The QIAamp mini spin column was placed in a clean 2 mL collection tube, and the tube containing the filtrate was discarded. The QIAamp mini spin column was then opened, buffer AW1 was added, and it was centrifuged. The QIAamp mini spin column was next placed in a clean 2 mL collection tube, and the tube containing the filtrate was discarded. Then, the QIAamp mini spin column was opened, buffer AW2 was added, and it was centrifuged. The QIAamp mini spin column was then placed in a new 2 mL collection tube and centrifuged. Following this, the QIAamp mini spin column was placed in a clean microcentrifuge; then, buffer AE was added, and it was incubated and then centrifuged. Genomic DNA templates were quantified using Nanodrop (Nanodrop 1000, Thermo Scientific, London, UK) and afterwards kept at −20 °C until they were required for PCR. Virulence genes (oprL, toxA, lasB, and exoS) and antimicrobial resistance genes (tetA, sul1, blaSHV, and blaTEM) were detected in the P. aeruginosa strains. The retrieved bacteria’s resistance to commercially available antibiotics was confirmed by the selection of antimicrobial-resistant genes. All primers were provided by the Metabion Company in Germany; Table 1 lists the oligonucleotide sequences and PCR settings for each primer. In a T100 gradient thermocycler (Biometra, Jena, Germany), PCR reactions (25 μL) were amplified using the EmeraldAmp GT PCR Master Mix (Code No. RR310A, Takara, Japan). The positive control was a virulent reference strain of P. aeruginosa that was generously supplied by the Animal Health Research Institute in Dokki, Cairo, Egypt. This strain was multidrug-resistant to tetracycline, sulfamethoxazole/trimethoprim, ceftriaxone, and cefotaxime. Meanwhile, a reaction without template DNA functioned as a negative control. After screening the products using 1.5% (w/v) agarose gel electrophoresis (AppliChem GmbH, Darmstadt, Germany), they were photographed.

Table 1.
PCR conditions for testing of P. aeruginosa encoding genes.
2.6. Sequence Analysis of blaSHV-Resistant Gene
The blaSHV gene was sequenced for four P. aeruginosa strains that encoded the blaSHV-resistant gene (one strain from fish, water, poultry, and humans). On an Applied Biosystems 3130 automated DNA sequencer (Applied Biosystems, Thermo Fisher Scientific, Carlsbad, CA, USA), a purified PCR product was sequenced in both the forward and reverse directions using a ready reaction Bigdye Terminator V3.1 cycle sequencing kit (Perkin-Elmer/Applied Biosystems, Foster City, CA, USA), with Cat. No. 4336817. According to Altschul et al. [24], a Basic Local Alignment Search Tool (BLAST) analysis was first conducted to determine sequence identity to GenBank accessions. Mesquite v.3.40, equipped with Clustal v.2.1, was utilized to align and trim the acquired sequences with the existing sequences from GenBank. The jModelTest v.2.1.6 implementation of the Akaike information criterion was used to select the evolution models (TIM3 + F). Using the maximum likelihood approach, a phylogenetic tree was constructed using IQ-Tree v.1.6.10.
2.7. Statistical Analysis
SPSS version 26 (IBM Corp, Armonk, NY, USA) was used to analyze the data. The chi-squared test was used to analyze the categorical data, including the differences in the prevalence of P. aeruginosa isolates from different sources, and to assess the differences in the antimicrobial resistance patterns of the recovered isolates from various sources and the existence of different tested genes among the investigated isolates of different origins. The correlations between the tested variables were estimated by Pearson’s correlations (r). Pearson’s correlations (r) were estimated and visualized using the Hmisc package [25] in the R software, version 4.3.3 (https://www.r-project.org/, accessed on 10 July 2025). Before correlation analyses, variables were tested for normality using a Q-Q plot. The oprL, toxA, and exoS gene variables were excluded from the analyses as they were identical among all isolates under study. The p-values were considered statistically significant if they were less than 0.05. All graphs were generated by the R software, version 4.3.3 [26], using ggplot [27], the Hmisc package [25], and the pheatmap package [28] to generate a radar chart, heatmap, hierarchical clustering heatmap, box plot, stacked bar plot, and correlation plots.
3. Results
3.1. Phenotypic Characteristics of Isolated P. aeruginosa from Different Sources
All recovered isolates were arranged microscopically in double or short chains of Gram-negative bacilli. Typically, P. aeruginosa isolates obtained on cetrimide agar at 37 °C for 24 h had a yellowish green fluorescent pigment and exhibited irregular, large colonies with a fruity odor. On MacConkey’s agar, the bacteria proliferated and appeared as fat, smooth, non-lactose-fermenting colonies with regular edges and an alligator skin appearance from the top view. The Vitek 2 results revealed that the probability of P. aeruginosa identification reached 99%. Additionally, all isolates showed positive results in the PCR amplification of the species-specific 16S rRNA gene.
3.2. Prevalence of P. aeruginosa from Different Sources
Among the examined fish, the overall prevalence of P. aeruginosa was 33.3% (30/90) (Table 2 and Figure 1). The highest prevalence was recorded in Clarias gariepinus (12/30, 40%), followed by Oreochromis niloticus (11/30, 36.6%) and then Tilapia zillii (7/30, 23.3%) (Table 3 and Figure 1). There was no statistically significant difference in the prevalence of P. aeruginosa among various fish species (p > 0.05). Concerning P. aeruginosa’s prevalence in various infected organs, the most common infected organ was the liver, followed by the kidney, spleen, and gills (Table 3 and Figure 2). Statistically, the prevalence of P. aeruginosa among different organs in the examined fish species showed no significant difference (p > 0.05). The overall P. aeruginosa prevalence in water, poultry, and humans was 20%, 14%, and 24%, respectively (Table 2 and Figure 1). The P. aeruginosa prevalence across different human samples was 50% (6/12) in wounds, 33.3% (4/12) in sputum, and 16.7% (2/12) in urine. There was no significant difference in the dissemination of P. aeruginosa among the examined samples from different sources (p = 0.064).

Table 2.
Prevalence of P. aeruginosa isolated from different sources.
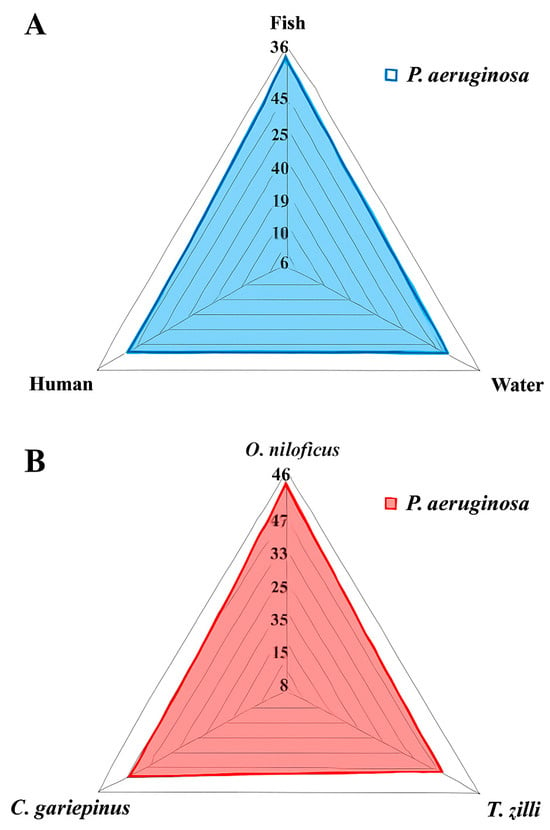
Figure 1.
Prevalence of P. aeruginosa isolated from different locations (A) and fish species (B).

Table 3.
Prevalence of P. aeruginosa isolated from different fish organs.
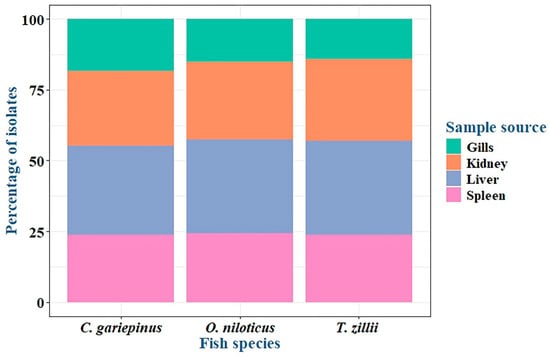
Figure 2.
P. aeruginosa prevalence in organs of infected fish species.
3.3. P. aeruginosa Antimicrobial Susceptibility Testing
The antimicrobial susceptibility of the retrieved P. aeruginosa isolates from fish consisted of remarkable resistance to tetracycline (100%), sulfamethoxazole/trimethoprim (100%), oxytetracycline (93%), amikacin (83%), tobramycin (83%), amoxicillin clavulanic acid (83%), ampicillin/sulbactam (83%), ceftriaxone (80%), and cefotaxime (73%). Moreover, 100% of isolates were sensitive to colistin. P. aeruginosa isolates from water showed 100% resistance to oxytetracycline, amikacin, tobramycin, amoxicillin clavulanic acid, ampicillin/sulbactam, and ciprofloxacin and 83% resistance to tetracycline, cefotaxime, and sulfamethoxazole/trimethoprim. Additionally, they showed 67% resistance to ceftriaxone but were 100% sensitive to colistin. P. aeruginosa isolates from humans showed 100% resistance to tobramycin, cefotaxime, ceftriaxone, amoxicillin, clavulanic acid, and ampicillin/sulbactam; 83% resistance to tetracycline, oxytetracycline, amikacin, and sulfamethoxazole/trimethoprim; and 67% resistance to ciprofloxacin. However, all human P. aeruginosa isolates were sensitive to colistin, reaching 100%. P. aeruginosa isolates from poultry showed 100% resistance to tetracycline, oxytetracycline, ampicillin/sulbactam, amoxicillin, clavulanic acid, cefotaxime, sulfamethoxazole/trimethoprim, amikacin, and tobramycin, while 71.4% of isolates were sensitive to colistin, as shown in Table 4 and Figure 3. Statistical analysis revealed no significant variations (p > 0.05) in the antimicrobial resistance patterns of the tested P. aeruginosa isolates from various sources to all tested antimicrobials.

Table 4.
The antimicrobial resistance patterns of the isolated P. aeruginosa from different sources.
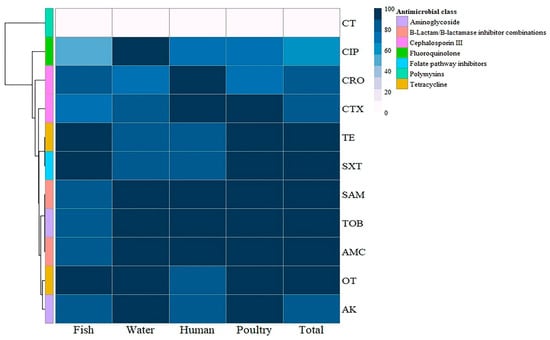
Figure 3.
Hierarchical clustering heatmap showing the antibiogram of the isolated P. aeruginosa from different sources. The color code on the right of the heatmap refers to the resistance percentage to the tested antimicrobial agent. TE: Tetracycline, OT: Oxytetracycline, AK: Amikacin, TOB: Tobramycin, CTX: Cefotaxime, CRO: Ceftriaxone, AMC: Amoxycillin/Clavulanic Acid, SAM: Ampicillin/Sulbactam, SXT: Sulfamethoxazole/Trimethoprim, CIP: Ciprofloxacin, CT: Colistin.
3.4. Virulence and Antimicrobial Resistance Genes of P. aeruginosa Strains
Concerning virulence genes, the recovered P. aeruginosa strains isolated from fish commonly harbored the virulence genes oprL (100%) and toxA (100%), followed by lasB (40%), while the exoS gene was not detected (0%). P. aeruginosa strains isolated from water and poultry commonly harbored the virulence genes oprL (100%), toxA (100%), and lasB (100%), while the exoS gene was not detected (0%). P. aeruginosa strains isolated from humans commonly harbored the virulence genes oprL (100%) and toxA (100%), while the lasB and exoS genes were not detected (0%) (Table 5 and Figure 4 and Figure 5).

Table 5.
Prevalence of virulence and antimicrobial resistance genes among P. aeruginosa strains from different sources by PCR.
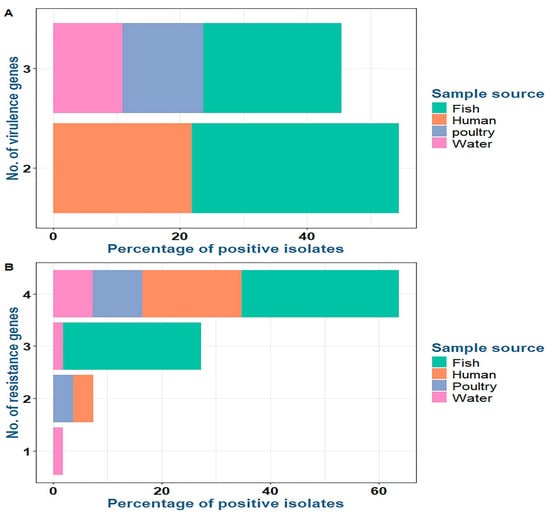
Figure 4.
Distribution of virulence (A) and resistance (B) genes among P. aeruginosa isolates from various sources.
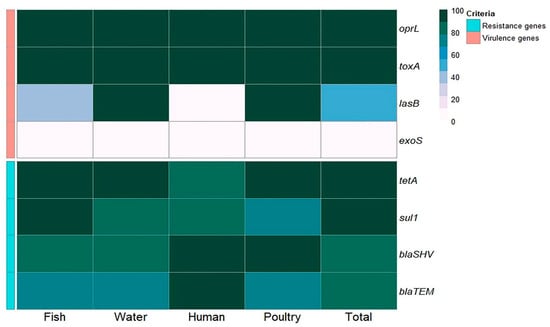
Figure 5.
Frequency of antibiotic resistance and virulence genes in P. aeruginosa isolates obtained from various sources.
Concerning antimicrobial resistance genes, the recovered P. aeruginosa strains isolated from fish commonly harbored tetA (tetracycline resistance) (100%), sul1 (sulfonamide resistance) (100%), and blaSHV (β-lactam resistance) (80%), followed by blaTEM (β-lactam resistance) (73.3%). P. aeruginosa strains isolated from water harbored tetA (100%), sul1 (83.3%), and blaSHV (83.3%), followed by blaTEM (66.7%). P. aeruginosa strains isolated from poultry harbored tetA, blaSHV, sul1, and blaTEM at 100%, 100%, 71.4%, and 71.4%, respectively. P. aeruginosa strains isolated from humans harbored blaSHV, blaTEM, tetA, and sul1 at 100%, 100%, 83.3%, and 83.3%, respectively (Table 5 and Figure 4 and Figure 5). There was a statistically significant difference in the prevalence of the lasB and sul1 genes among P. aeruginosa isolates from different sources (p < 0.001). There was no statistically significant difference in the distributions of tetA, blaSHV, and blaTEM resistance genes among P. aeruginosa isolates from different sources (p > 0.05). There were statistically significant differences in the distributions of two and three resistance genes (p < 0.001) and two and three resistance genes (p = 0.021, 0.004) among P. aeruginosa isolated from fish, water, poultry, and human samples. There were no statistically significant differences in the distributions of one and four virulence genes among P. aeruginosa isolated from fish, water, poultry, and human samples (p = 0.109 and 0.322, respectively) (Table 5).
3.5. Patterns of Multidrug Resistance in Recovered P. aeruginosa Strains
The study’s findings indicated that 53.3% (16/30) of the retrieved P. aeruginosa isolates obtained from fish were XDR to six antimicrobial classes (tetracyclines: TE and OT; folate pathway inhibitors: SXT; aminoglycosides: AK and TOB; β-lactam–β-lactamase inhibitor combinations: SAM and AMC; cephalosporin III: CTX and CRO; and fluoroquinolone: CIP) and harbored the tetA, sul1, blaTEM, and blaSHV resistance genes. Moreover, 16.7% (5/30) of the isolated P. aeruginosa strains were MDR to five different classes (tetracyclines: TE and OT; folate pathway inhibitors: SXT; β-lactam–β-lactamase inhibitor combinations: SAM and AMC; aminoglycosides: AK and TOB; and cephalosporin III: CRO) and harbored the tetA, sul1, and blaSHV resistance genes. In addition, 13.3% (4/30) of the isolated P. aeruginosa strains were MDR to five different classes (tetracyclines: TE and OT; folate pathway inhibitors: SXT; aminoglycosides: AK and TOB; β-lactam–β-lactamase inhibitor combinations: SAM and AMC; and cephalosporin III: CTX) and harbored the tetA, sul1, and blaTEM resistance genes. Moreover, 10% (3/30) of the isolated P. aeruginosa strains were MDR to three classes (tetracyclines: TE and OT; folate pathway inhibitors: SXT; and cephalosporin III: CTX) and harbored the tetA, sul1, and blaSHV resistance genes. In addition, 6.7% (2/30) of the isolated P. aeruginosa strains were MDR to three classes (tetracyclines: TE; folate pathway inhibitors: SXT; and cephalosporin III: CTX) and harbored the tetA, sul1, and blaTEM resistance genes (Table 6).

Table 6.
Multidrug resistance patterns and antimicrobial resistance gene distribution among P. aeruginosa strains (n = 30 for fish, n = 6 for water, n = 7 for poultry, and n = 12 for humans).
Concerning P. aeruginosa isolates obtained from water, 66.6% (4/6) were XDR to six antimicrobial classes (tetracyclines: TE and OT; β-lactam–β-lactamase inhibitor combinations: SAM and AMC; cephalosporin III: CTX and CRO; folate pathway inhibitors: SXT; fluoroquinolone: CIP; and aminoglycosides: TOB and AK) and harbored the tetA, sul1, blaSHV, and blaTEM resistance genes. In addition, 16.7% (1/6) were XDR to six antimicrobial classes (tetracyclines: TE and OT; β-lactam–β-lactamase inhibitor combinations: SAM and AMC; cephalosporin III: CTX; folate pathway inhibitors: SXT; fluoroquinolone: CIP; and aminoglycosides: TOB and AK) and harbored the tetA, sul1, and blaSHV resistance genes. Moreover, 16.7% (1/6) were MDR to four antimicrobial classes (tetracyclines: OT; β-lactam–β-lactamase inhibitor combinations: SAM and AMC; fluoroquinolone: CIP; and aminoglycosides: TOB and AK) and harbored the tetA and blaSHV resistance genes (Table 6).
Concerning P. aeruginosa isolates obtained from poultry, 71.4% (5/7) were XDR to six antimicrobial classes (tetracyclines: TE and OT; β-lactam–β-lactamase inhibitor combinations: SAM and AMC; cephalosporin III: CTX and CRO; folate pathway inhibitors: SXT; fluoroquinolone: CIP; and aminoglycosides: TOB and AK) and harbored the tetA, sul1, blaSHV, and blaTEM resistance genes. In addition, 28.6% (2/7) were MDR to five antimicrobial classes (tetracyclines: TE and OT; β-lactam–β-lactamase inhibitor combinations: SAM and AMC; cephalosporin III: CTX; folate pathway inhibitors: SXT; and aminoglycosides: TOB and AK) and harbored the tetA and blaSHV resistance genes (Table 6).
Regarding P. aeruginosa isolates obtained from humans, 66.7% (8/12) were XDR to six antimicrobial classes (aminoglycosides: TOB and AK; tetracyclines: TE and OT; cephalosporin III: CTX and CRO; β-lactam–β-lactamase inhibitor combinations: SAM and AMC; fluoroquinolone: CIP; and folate pathway inhibitors: SXT) and harbored the tetA, sul1, blaSHV, and blaTEM resistance genes. In addition, 16.7% (2/12) were MDR to five antimicrobial classes (tetracyclines: TE and OT; cephalosporin III: CTX and CRO; β-lactam–β-lactamase inhibitor combinations: SAM and AMC; aminoglycosides: TOB; and folate pathway inhibitors: SXT) and harbored the tetA, sul1, blaSHV, and blaTEM resistance genes. Moreover, 16.7% (2/12) were MDR to three antimicrobial classes (aminoglycosides: TOB and AK; cephalosporin III: CTX and CRO; and β-lactam–β-lactamase inhibitor combinations: SAM and AMC) and harbored the tetA, sul1, blaSHV, and blaTEM resistance genes (Table 6). The conducted analysis showed that, throughout all examined sources, XDR isolates were more prevalent than MDR isolates. Sixteen fish isolates, five water isolates, five poultry isolates, and eight human isolates were found to be XDR, while 14, 1, 2, and 4 isolates from the same sources were found to be MDR. The higher incidence of XDR over MDR poses challenges in providing proper treatment and emphasizes the urgent need for integrated One Health programs and the improved oversight of antibiotics to prevent subsequent transmission.
The clustering patterns of P. aeruginosa isolates based on the phenotypic antimicrobial resistance genes and virulence genes displayed medium diversity and polyclonality. Of the 55 examined isolates, only 12 isolates belonged to various lineages. Moreover, three main branches (I, II, and III) and eight clusters were observed in our results, and close relatedness was determined between P. aeruginosa isolates from human and fish samples (Figure 6).
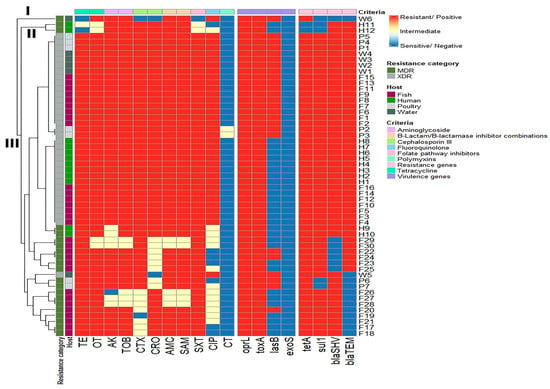
Figure 6.
Hierarchical clustering heatmap depicting the overall distribution of the P. aeruginosa isolates depending on their phenotypic antimicrobial resistance patterns, resistance genes, and virulence genes. Different sample sources, resistance categories, and antimicrobial classes are color-coded on the right side of the heatmap. TE: Tetracycline, OT: Oxytetracycline, AK: Amikacin, TOB: Tobramycin, CTX: Cefotaxime, CRO: Ceftriaxone, AMC: Amoxycillin/Clavulanic Acid, SAM: Ampicillin/Sulbactam, SXT: Sulfamethoxazole/Trimethoprim, CIP: Ciprofloxacin, CT: Colistin.
The relatedness of the investigated phenotypic antimicrobial resistance genes and virulence genes exhibited low diversity, with two branches (A and B) and one cluster. The SAM, AMC, and TOB antimicrobials were clustered together. Additionally, we identified a highly positive, significant correlation between the TE and OT phenotypes and the tetA (r = 0.69 each, p < 0.001) and sul1 (r = 0.73 and 0.4, respectively, p < 0.01) genes, as well as a significant (p < 0.0001) positive correlation between the tetA and sul1 genes (r = 0.61). We determined a significant (p < 0.05) positive correlation between the lasB gene and the SAM, AMC, TOB, and AK phenotypes (r = 0.29, 0.29, 0.29, and 0.33, respectively). Moreover, the blaSHV gene correlated significantly and positively with the OT (r = 0.31, p-value < 0.05), CIP (r = 0.43, p-value < 0.01), and CRO (r = 0.72, p-value < 0.0001) phenotypes, and there was a significant positive correlation between the blaTEM gene and the CTX, CIP (r = 0.78 and 0.44, respectively, p-value < 0.0001), CRO (r = 0.3, p < 0.05), TOB, AMC, and SAM phenotypes, as well as the sul1 gene (r = 0.29 each, p-value < 0.05) (Figure 7).
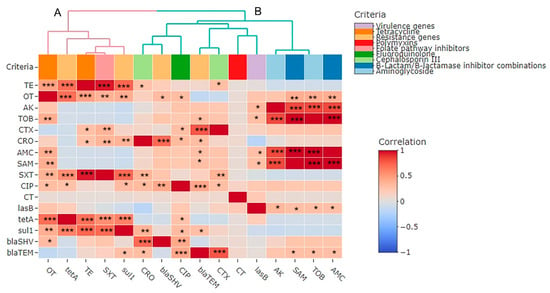
Figure 7.
Pairwise correlations (r) between phenotypic antimicrobial resistance genes and virulence genes of P. aeruginosa isolates exhibited with two branches (A,B). The scale on the right of the figure refers to the correlation coefficient (r). Stars refer to a significant correlation: * p < 0.05, ** p < 0.01, *** p < 0.001. Variables that are identical among all strains are excluded and thus not shown in this figure. TE: Tetracycline, OT: Oxytetracycline, AK: Amikacin, TOB: Tobramycin, CTX: Cefotaxime, CRO: Ceftriaxone, AMC: Amoxycillin/Clavulanic Acid, SAM: Ampicillin/Sulbactam, SXT: Sulfamethoxazole/Trimethoprim, CIP: Ciprofloxacin, CT: Colistin.
P. aeruginosa strains recovered from fish, water, poultry, and humans had MAR index values ranging from 0.27 to 0.91 (Table 7 and Figure 8). There were no statistically significant differences in the prevalence of MAR indices and resistance to antimicrobial classes (p > 0.05) among P. aeruginosa isolates obtained from fish, water, poultry, and human samples (Table 7).

Table 7.
Frequency of resistance to various antimicrobial agents in P. aeruginosa isolates belonging to various sources.
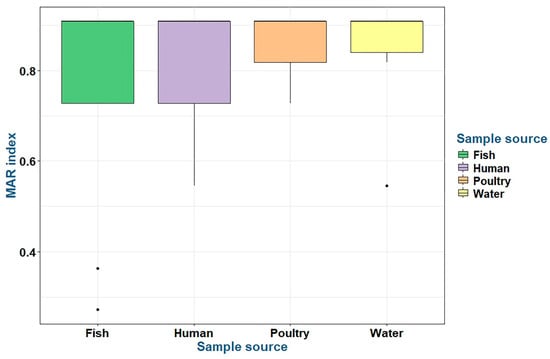
Figure 8.
The multiple antibiotic resistance (MAR) indices of the tested P. aeruginosa isolates belonging to various sources.
3.6. Sequence Analysis of blaSHV Gene in P. aeruginosa Strains from Fish, Water, and Humans
Four P. aeruginosa isolates—originating from fish, water, poultry, and human sources—were selected for phylogenetic analysis based on their phenotypic resistance to β-lactam antibiotics and the presence of the blaSHV gene, as confirmed by molecular screening. Sequencing of the blaSHV gene was performed for each isolate. The sequences obtained from the fish and water isolates were submitted to GenBank under the accession numbers MZ700497 and MZ700498 (Figure 9), respectively. Comparative sequence analysis revealed a high degree of similarity between these two environmental isolates, suggesting a close genetic relationship or potentially a common origin.
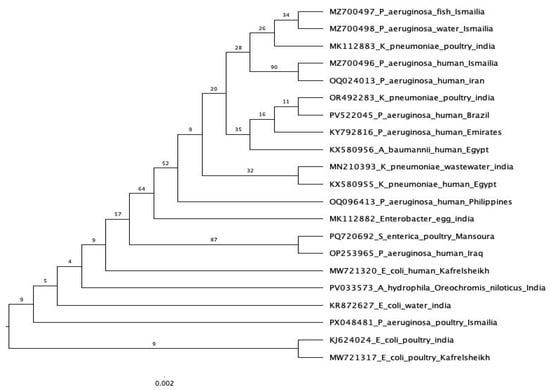
Figure 9.
Phylogenetic analysis of the blaSHV gene sequence from different sources. All other sequences are derived from the NCBI GenBank database.
The blaSHV gene sequence from the human isolate, submitted under GenBank accession number MZ700496 (Figure 9), exhibited high similarity to blaSHV-1 sequences previously reported from clinical P. aeruginosa strains in Iran. Interestingly, this human-derived sequence also showed greater similarity to the sequences from the fish and water isolates in the Ismailia Governorate than to the poultry-derived isolate.
The blaSHV gene sequence from the poultry isolate was submitted under GenBank accession number PX048481 (Figure 9). Phylogenetic analysis placed the poultry-derived sequence in a distinct clade, separate from the other sources analyzed in this study. Notably, this sequence exhibited greater similarity to a blaSHV gene from E. coli isolated from water in India, rather than to any of the local isolates examined.
4. Discussion
Pseudomonas aeruginosa is recognized as a significant pathogen responsible for ulcerative infections in fish populations. These infections have elevated mortality rates, caused substantial economic losses, and reduced efficiency in fish farming [16], in addition to having zoonotic implications [29]. In the current study, P. aeruginosa’s overall prevalence among the analyzed fish was 33.3%. The highest incidence was seen in Clarias gariepinus (40%), followed by Oreochromis niloticus (36.6%) and then Tilapia zillii (23.3%). The liver was the most significantly affected organ. The results aligned with [18,30], which found that the P. aeruginosa prevalence in fish was 31.57% and 31.5%, respectively. Meanwhile, ref. [31] found a higher incidence of P. aeruginosa in Clarias gariepinus than in Oreochromis niloticus. Moreover, ref. [1] regularly isolated P. aeruginosa from liver samples.
The studies [7,19,31] revealed a decrease in the P. aeruginosa incidence rates in fish of 12%, 4.2%, and 13.8%, respectively. Geographical dispersion, environmental factors, host vulnerability, and the sample collection season may all contribute to variations in prevalence. P. aeruginosa was discovered in 20% of the water samples, which differed from [32], where the authors isolated P. aeruginosa from various water sources, such as lakes, ponds, and rivers, with a percentage of 61.5%. The status of the water in which the fish were hunted is considered an indicator of the bacterial load; hence, fish could acquire harmful microorganisms from their natural aquatic habitats [33].
It is notable that P. aeruginosa was isolated from poultry at a percentage of 14%. In large-scale chicken farms, where even a small percentage of P. aeruginosa might affect a large number of birds, this percentage may appear insignificant, yet it represents a notable concern. Our results were nearly identical to those in [34], which stated that the isolation rate of P. aeruginosa in broiler chickens was 18% in Beni-Suef and Fayoum, Egypt. In contrast, ref. [35] recorded a high isolation rate of P. aeruginosa from broiler chickens in Fayoum, Egypt, reaching 77.5%. These findings are within the ranges reported in other countries. This percentage is significant from a clinical and financial standpoint because P. aeruginosa is not a typical component of the poultry flora and is frequently linked to illness outbreaks. P. aeruginosa infections can result in low weight gain, high chick mortality, and a greater need for antibiotics. Furthermore, it may serve as a covert source of infection or antibiotic resistance in birds that appear to be in good health [36]. As a result, even a 14% rate would necessitate regular screening, improved sanitation, and conscientious antibiotic use in chicken farms.
The results revealed a high isolation rate of P. aeruginosa from the pus of infected wounds and burns in human samples (50%), followed by sputum (33.3%) and then urine samples (16.7%). This finding is almost in line with the study [37] regarding the isolation rate of P. aeruginosa from pus (50%), followed by sputum (25%) and then urine samples (16.67%). In contrast, ref. [38] found that the organism was responsible for 16% of nosocomial pneumonia cases, 11% of hospital-acquired urinary tract infections, and 8% of surgical wound infections. Differences across studies may arise from differing hospital conditions and sample sizes.
The long-term use of antimicrobial drugs to treat Pseudomonas infections leads to the emergence of multidrug-resistant strains in aquatic environments via R-plasmid transfer [39]. According to [40], P. aeruginosa is resistant to several antibiotics, including aminoglycosides, quinolones, and β-lactams. Regarding antimicrobial sensitivity tests, all isolates recovered from different sources did not show resistance to colistin, a result confirmed in [18], which found P. aeruginosa to be 100% sensitive to colistin. In contrast, ref. [41] described the 100% resistance of P. aeruginosa isolates to colistin. Furthermore, all isolates showed resistance to other tested antimicrobial agents, with different percentages. These findings were consistent with the data in [42], where the authors determined the high resistance of P. aeruginosa to ampicillin/sulbactam and amoxicillin/clavulanic acid (68%).
The current data highlight XDR P. aeruginosa resistance to six antimicrobial classes, including the tetA, sul1, blaTEM, and blaSHV resistance genes. These findings are congruent with those in [18,43]. The indiscriminate use of antibiotics and evolving antibiotic resistance genes may lead to the emergence of MDR strains [44]. The MAR value in this investigation was above 0.2, echoing the results of [43]. The survival ability of P. aeruginosa strains in a polluted environment is indicated by an MAR value of more than 0.2, and the high rate of deterioration in the samples under investigation may have been caused by the presence of several antibiotic residues [45]. The evolution of numerous and widespread drug-resistant P. aeruginosa strains with public health implications is highlighted in this study; as a result, preventing food poisoning requires proper handling, storage, and transportation procedures, as well as antimicrobial drug surveillance systems [46]. Furthermore, MDR P. aeruginosa strains were found in three or five antimicrobial classes and had the tetA, sul1, blaTEM, or blaSHV resistance genes. These findings align with [47], where the authors detected a high rate of resistance for tetracyclines and low resistance for colistin, and 24.3% of isolates were MDR. Meanwhile, ref. [48] revealed 97.5% resistance to sulfamethoxazole/trimethoprim and [49] detected colistin sensitivity amounting to 85%, amikacin resistance of 64%, and ceftriaxone and cefotaxime resistance of 56.52%, with 32.8% of P. aeruginosa isolates from humans. In contrast, in [49,50], the authors discovered the lowest resistance to amikacin. The difference in bacterial resistance prevalence rates among studies may arise from many factors, including the type of clinical specimen investigated, community hygiene, and antimicrobial agent exposure [48]. P. aeruginosa is broadly resistant to numerous medications and becomes progressively resistant to most existing antibiotics, making it a major concern in hospital settings all over the world [51].
According to the current study’s PCR results, every tested strain was positive for the oprL and toxA genes, which is in line with [35]. P. aeruginosa’s outer membrane proteins, known as L-lipoproteins, give the bacterium resistance to antiseptics and other antimicrobial agents. Because they are limited to Pseudomonad species, they may be a valuable target for identifying and assessing the virulence of Pseudomonads in clinical specimens [52]. The toxA gene on the virulent P. aeruginosa chromosome encodes exotoxin A, an extracellular component of the bacterium. It functions similarly to the diphtheria toxin in that it prevents the host cell from producing proteins [53]. The lasB gene was absent in human isolates, while it was found in 40%, 100%, and 100% of fish, poultry, and water isolates, respectively. Moreover, ref. [54] detected lasB at 41.2%. LasB hydrolyzes various host proteins, destroying host tissues and the immune response and promoting inflammation [55]. In this research, the exoS gene was absent in all strains, and this is almost aligned with [55], where the authors detected a minor prevalence of the exoS gene (5%). Antimicrobial resistance genes were distributed in different percentages in all isolates, which is in line with [18,56]. These resistant and highly virulent bacteria can colonize new environments and cause disease more efficiently [57].
The phylogenetic relationships observed in this study highlight potential pathways of horizontal gene transfer and environmental dissemination of the blaSHV gene among P. aeruginosa isolates from diverse sources. The close genetic similarity between the fish and water isolates suggests a shared environmental reservoir or direct contamination linkage, likely facilitated by aquatic ecosystems. The similarity between the human isolates and environmental isolates (fish and water) raises concerns about possible transmission from environmental sources to humans, especially in regions with inadequate wastewater management. In contrast, the poultry isolates formed a separate phylogenetic lineage, indicating a distinct origin or the acquisition of the blaSHV gene, potentially influenced by agricultural antimicrobial usage. The similarity between the poultry isolates and an E. coli strain from India may reflect the international dissemination of resistance genes through the food chain or animal trade. These findings underscore the importance of integrated One Health surveillance approaches to monitor and control the spread of antibiotic resistance genes across the environmental, animal, and human health sectors. Our results follow [58], where the authors stated that bacteria are widespread in the aquatic habitat and can cause significant mortality in wild and cultivated fish because of illnesses caused by numerous bacterial species. Most bacterial pathogens are a part of the natural microflora prevalent in aquatic environments; however, they can also act as opportunistic or secondary infections. Many patients acquire infections linked to P. aeruginosa, posing a public health concern [18]. Nowadays, Pseudomonas species cause foodborne diseases that humans can acquire by eating contaminated food and prepared goods, handling contaminated seafood [59], or by eating raw fish and its byproducts [60]. The potential for antimicrobial-resistant gene expression in P. aeruginosa to circulate among individuals, poultry, fish, and water supplies constitutes one of the most notable public health implications highlighted by our results. Tracking and combating these infections on a global scale is vital, since the high incidence and resistance patterns found in our isolates indicate that these sources may serve as repositories for resilient strains. Our results support the urgent need for comprehensive antimicrobial oversight along with consolidated One Health policies to prevent the spread of resistance and maintain human and animal health.
5. Conclusions
Our study revealed the concerning prevalence of XDR and MDR P. aeruginosa isolates from fish, water, poultry, and humans in Egypt. Remarkably, all strains harbored the oprL and toxA virulence genes. The tetA, sul1, blaSHV, and blaTEM resistance genes were also detected at high prevalences in all isolates. The close genetic similarity between the fish and water isolates suggests a shared environmental reservoir or direct contamination linkage, likely facilitated by aquatic ecosystems. The similarity between the human isolates and environmental isolates (fish and water) raises concerns about possible transmission to humans. These findings underscore the urgent need for stringent control measures to mitigate the spread of MDR and XDR P. aeruginosa and to address the misuse of antimicrobials in fish farming, in line with the One Health perspective.
Author Contributions
Conceptualization, A.W. and M.E.; methodology, A.E. and M.A.E.-A.; software, W.A.H. and M.A.E.-A.; validation, A.M.A.M. and S.M.A.-R.; formal analysis, S.I.A.-S. and S.A.-S.; investigation, S.A.-S. and F.A.; resources, S.I.A.-S. and A.A.; data curation, W.E.-D. and E.M.A.-A.; writing—original draft preparation, A.W., A.E. and M.E.A.H.; writing—review and editing, A.W. and M.E.A.H.; visualization, S.I.A.-S. and S.A.-S.; supervision, A.W. and M.E.A.H.; project administration, S.M.A.-R.; funding acquisition, A.M.A.M. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Deanship of Scientific Research, Vice Presidency for Graduate Studies and Scientific Research, King Faisal University, Saudi Arabia (Grant No. KFU 253318).
Institutional Review Board Statement
Fish and poultry handling, human sample collection, and other experimental methods were approved by Suez Canal University’s Animal Ethics Review Committee, Egypt (SCU-VET-AREC-R-2025018, approved on 1 July 2025).
Informed Consent Statement
Not applicable.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.
Acknowledgments
This work was supported by the Deanship of Scientific Research, Vice Presidency for Graduate Studies and Scientific Research, King Faisal University, Saudi Arabia (Grant No. KFU 253318).
Conflicts of Interest
The authors declare that they have no competing interests.
References
- Abou Elez, R.M.M.; Zahra, E.M.F.; Gharieb, R.M.A.; Mohamed, M.E.M.; Samir, M.; Saad, A.M.; Merwad, A.M.A. Resistance patterns, virulence determinants, and biofilm genes of multidrug-resistant P. aeruginosa isolated from fish and fish handlers. Sci. Rep. 2024, 14, 24063. [Google Scholar] [CrossRef]
- Marouf, S.; Li, X.; Salem, H.M.; Ahmed, Z.S.; Nader, S.M.; Shaalan, M.; Awad, F.H.; Zhou, H.; Cheang, T. Molecular detection of multidrug-resistant P. aeruginosa of different avian sources with pathogenicity testing and in vitro evaluation of antibacterial efficacy of silver nanoparticles against multidrug-resistant P. aeruginosa. Poult. Sci. 2023, 102, 102995. [Google Scholar] [CrossRef]
- Reynolds, D.; Kollef, M. The Epidemiology and Pathogenesis and Treatment of P. aeruginosa Infections: An Update. Drugs 2021, 81, 2117–2131. [Google Scholar] [CrossRef]
- World Health Organization. WHO Bacterial Priority Pathogens List, 2024: Bacterial Pathogens of Public Health Importance, to Guide Research, Development, and Strategies to Prevent and Control Antimicrobial Resistance; World Health Organization: Geneva, Switzerland, 2024. [Google Scholar]
- Morshdy, A.E.M.; Hussein, M.A.; Mohamed, M.A.A.; Hamed, E.; El-Murr, A.E.; Darwish, W.S. Tetracycline residues in tilapia and catfish tissue and the effect of different cooking methods on oxytetracycline and doxycycline residues. J. Consum. Prot. Food Saf. 2022, 17, 387–393. [Google Scholar] [CrossRef]
- Ben Mhenni, N.; Alberghini, G.; Giaccone, V.; Truant, A.; Catellani, P. Prevalence and antibiotic resistance phenotypes of Pseudomonas spp. in fresh fish fillets. Foods 2023, 12, 950. [Google Scholar] [CrossRef] [PubMed]
- Abd El Tawab, A.A.; Maarouf, A.A.; Ahmed, N.M. Detection of Virulence factors of Pseudomonas species isolated from fresh water fish by PCR. Benha Vet. Med. J. 2016, 30, 199–207. [Google Scholar] [CrossRef]
- Rizk, A.M.; Elsayed, M.M.; Abd El Tawab, A.A.; Elhofy, F.I.; Soliman, E.A.; Kozytska, T.; Brangsch, H.; Sprague, L.D.; Neubauer, H.; Wareth, G. Phenotypic and genotypic characterization of resistance and virulence in P. aeruginosa isolated from poultry farms in Egypt using whole genome sequencing. Vet. Microbiol. 2024, 292, 110063. [Google Scholar] [CrossRef]
- Odoi, H.; Boamah, V.E.; Boakye, Y.D.; Agyare, C. Prevalence and Phenotypic and Genotypic Resistance Mechanisms of Multidrug-Resistant P. aeruginosa Strains Isolated from Clinical, Environmental, and Poultry Litter Samples from the Ashanti Region of Ghana. J. Environ. Public Health 2021, 2021, 9976064. [Google Scholar] [CrossRef]
- Behzadi, P.; Baráth, Z.; Gajdács, M. It’s not easy being green: A narrative review on the microbiology, virulence and therapeutic prospects of multidrug-resistant P. aeruginosa. Antibiotics 2021, 10, 42. [Google Scholar] [CrossRef]
- Horii, T.; Muramatsu, H.; Monji, A.; Miyagishima, D. Release of exotoxin A, peptidoglycan and endotoxin after exposure of clinical P. aeruginosa isolates to carbapenems in vitro. Chemotherapy 2005, 51, 324–331. [Google Scholar] [CrossRef] [PubMed]
- Uddin, T.M.; Chakraborty, A.J.; Khusro, A.; Zidan, B.R.M.; Mitra, S.; Emran, T.B.; Dhama, K.; Ripon, M.K.H.; Gajdács, M.; Sahibzada, M.U.K. Antibiotic resistance in microbes: History, mechanisms, therapeutic strategies and future prospects. J. Infect. Public Health 2021, 14, 1750–1766. [Google Scholar] [CrossRef]
- Chadha, J.; Harjai, K.; Chhibber, S. Revisiting the virulence hallmarks of P. aeruginosa: A chronicle through the perspective of quorum sensing. Environ. Microbiol. 2022, 24, 2630–2656. [Google Scholar] [CrossRef]
- Sabzehali, F.; Rahimi, H.; Goudarzi, H.; Goudarzi, M.; Izad, M.H.Y.; Chirani, A.S.; Jalali, S.A.; Faghihloo, E. Functional engineering of OprF-OprI-PopB as a chimeric immunogen and its cross-protective evaluation with GM-CSF against P. aeruginosa: A comprehensive immune informatics evaluation. Inform. Med. Unlocked 2021, 25, 100673. [Google Scholar] [CrossRef]
- Zhao, Y.; Xu, H.; Wang, H.; Wang, P.; Chen, S. Multidrug resistance in P. aeruginosa: Genetic control mechanisms and therapeutic advances. Mol Biomed 2024, 5, 62. [Google Scholar] [CrossRef]
- Shahrokhi, G.R.; Rahimi, E.; Shakerian, A. The prevalence rate, pattern of antibiotic resistance, and frequency of virulence factors of P. aeruginosa strains isolated from fish in Iran. J. Food Qual. 2022, 2022, 8990912. [Google Scholar] [CrossRef]
- Islam, R.; Ferdous, F.B.; Hoque, M.N.; Asif, N.A.; Rana, M.L.; Siddique, M.P.; Rahman, M.T. Characterization of beta-lactamase and virulence genes in P. aeruginosa isolated from clinical, environmental and poultry sources in Bangladesh. PLoS ONE 2024, 19, e0296542. [Google Scholar]
- Algammal, A.M.; Mabrok, M.; Sivaramasamy, E.; Youssef, F.M.; Atwa, M.H.; El-Kholy, A.W.; Hetta, H.F.; Hozzein, W.N. Emerging MDR- P. aeruginosa in fish commonly harbor opr L and tox A virulence genes and bla TEM, bla CTX-M, and tet A antibiotic-resistance genes. Sci. Rep. 2020, 10, 15961. [Google Scholar] [CrossRef]
- Wamala, S.P.; Mugimba, K.K.; Mutoloki, S.; Evensen, Ø.; Mdegela, R.; Byarugaba, D.K.; Sørum, H. Occurrence and antibiotic susceptibility of fish bacteria isolated from Oreochromis niloticus (Nile tilapia) and Clarias gariepinus (African catfish) in Uganda. Fish. Aquat. Sci. 2018, 21, 6. [Google Scholar] [CrossRef]
- Yisa, J.; Tijani, J.O. Analytical studies on water quality index of river Landzu. Am. J. Appl. Sci. 2010, 7, 453–458. [Google Scholar] [CrossRef]
- Lamont, I.L.; Martin, L.W. Identification and characterization of novel pyoverdine synthesis genes in P. aeruginosa. Microbiology 2003, 149, 833–842. [Google Scholar] [CrossRef]
- Spilker, T.; Coenye, T.; Vandamme, P.; LiPuma, J.J. PCR-based assay for differentiation of P. aeruginosa from other Pseudomonas species recovered from cystic fibrosis patients. J. Clin. Microbiol. 2004, 42, 2074–2079. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing; Clinical Lab Standards Institute: Malvern, PA, USA, 2016; Volume 35, pp. 16–38. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Harrell, F.E., Jr. Hmisc: Harrell Miscellaneous. CRAN Contrib. Packag. 2003. Available online: https://cran.r-project.org/web/packages/Hmisc/index.html (accessed on 6 January 2023).
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2024. [Google Scholar]
- Wickham, H.; Chang, W.; Henry, L.; Pedersen, T.L.; Takahashi, K.; Wilke, C.; Woo, K.; Yutani, H.; Dunnington, D.; van den Brand, T. ggplot2: Create Elegant Data Visualisations Using the Grammar of Graphics. CRAN Contrib. Packag. 2007. [Google Scholar] [CrossRef]
- Kolde, R. Pheatmap: Pretty heatmaps. R Package Version 1.0.10. 2012. Available online: https://CRAN.R-project.org/package=pheatmap (accessed on 6 January 2023).
- Abdulhaq, N.; Nawaz, Z.; Zahoor, M.A.; Siddique, A.B. Association of biofilm formation with multi drug resistance in clinical isolates of P. aeruginosa. EXCLI J. 2020, 19, 201. [Google Scholar] [PubMed]
- Sowmya, R.; Chinta, S.S.; Sreedevi, B.; Suresh, Y.; Vinayaka Siddhartha, P.; Madhava Rao, T. Molecular characterization and antibiogram of P. aeruginosa isolated from fish sold in markets of Tirupati, India: Molecular characterization and antibiogram of P. aeruginosa. Fish. Technol. 2024, 61. [Google Scholar] [CrossRef]
- Ali, H.; Awad, A.; Maarouf, A. Molecular Detection of some Virulence Factors of P. aeruginosa Isolated from Freshwater Fishes at Qalubiya Governorate, Egypt. Benha Vet. Med. J. 2023, 43, 80–84. [Google Scholar] [CrossRef]
- Nasreen, M.; Sarker, A.; Malek, M.; Ansaruzzaman, M.; Rahman, M. Prevalence and resistance pattern of P. aeruginosa isolated from surface water. Adv. Microbiol. 2015, 5, 74–81. [Google Scholar] [CrossRef]
- Alawy, A.E.; El-Tras, W.F.; El Raiy, H.R.; Khater, D.F. Impact of industrial wastewater on water and fish quality of Nile River in Kafr El-Zayat, Egypt. Benha Vet. Med. J. 2015, 28, 78–87. [Google Scholar] [CrossRef][Green Version]
- Hassan, W.H.; Ibrahim, A.M.K.; Shany, S.A.S.; Salam, H.S.H. Virulence and resistance determinants in P. aeruginosa isolated from pericarditis in diseased broiler chickens in Egypt. J. Adv. Vet. Anim. Res. 2020, 7, 452–463. [Google Scholar] [CrossRef]
- Morsi, A.I.; Abdallah, M.; Rafequ, A.; Elkhayat, M. Incidence and virulence gene profiling of P. aeruginosa in broiler chickens from Fayoum Governorate, Egypt. Benha Vet. Med. J. 2024, 47, 78–81. [Google Scholar] [CrossRef]
- Abd El-Ghany, W.A. Pseudomonas aeruginosa infection of avian origin: Zoonosis and one health implications. Vet. World 2021, 14, 2155–2159. [Google Scholar] [CrossRef] [PubMed]
- Ismail, M.; El Lamei, M. Studies on P. septicemia in some tilapia in Ismailia. Suez Canal Vet. Med. J. 2017, 22, 107–117. [Google Scholar]
- Ikeno, T.; Fukuda, K.; Ogawa, M.; Honda, M.; Tanabe, T.; Taniguchi, H. Small and rough colony P. aeruginosa with elevated biofilm formation ability isolated in hospitalized patients. Microbiol. Immunol. 2007, 51, 929–938. [Google Scholar] [CrossRef]
- Abd-El-Maogoud, H.A.-E.-N.; Edris, A.B.M.; Mahmoud, A.H.; Maky, M.A. Occurrence and characterization of Pseudomonas species isolated from fish marketed in Sohag Governorate, Egypt. SVU-Int. J. Vet. Sci. 2021, 4, 76–84. [Google Scholar] [CrossRef]
- Hancock, R.E.; Brinkman, F.S. Function of Pseudomonas porins in uptake and efflux. Annu. Rev. Microbiol. 2002, 56, 17–38. [Google Scholar] [CrossRef]
- Akhi, M.T.; Ghotaslou, R.; Beheshtirouy, S.; Asgharzadeh, M.; Pirzadeh, T.; Asghari, B.; Alizadeh, N.; Ostadgavahi, A.T.; Somesaraei, V.S.; Memar, M.Y. Antibiotic susceptibility pattern of aerobic and anaerobic bacteria isolated from surgical site infection of hospitalized patients. Jundishapur J. Microbiol. 2015, 8, e20309. [Google Scholar] [CrossRef]
- Abd El-Baky, R.M.; Masoud, S.M.; Mohamed, D.S.; Waly, N.G.; Shafik, E.A.; Mohareb, D.A.; Elkady, A.; Elbadr, M.M.; Hetta, H.F. Prevalence and some possible mechanisms of colistin resistance among multidrug-resistant and extensively drug-resistant P. aeruginosa. Infect. Drug Resist. 2020, 13, 323–332. [Google Scholar] [CrossRef]
- Shalmashi, H.; Farajnia, S.; Sadeghi, M.; Tanoumand, A.; Veissi, K.; Hamishekar, H.; Gotaslou, R. Detection of ESBLs types blaCTX-M, blaSHV and blaTEM resistance genes among clinical isolates of P. aeruginosa. Gene Rep. 2022, 28, 101637. [Google Scholar] [CrossRef]
- Farhan, S.M.; Ibrahim, R.A.; Mahran, K.M.; Hetta, H.F.; Abd El-Baky, R.M. Antimicrobial resistance pattern and molecular genetic distribution of metallo-β-lactamases producing P. aeruginosa isolated from hospitals in Minia, Egypt. Infect. Drug Resist. 2019, 12, 2125–2133. [Google Scholar] [CrossRef] [PubMed]
- Talukder, A.; Rahman, M.M.; Chowdhury, M.M.H.; Mobashshera, T.A.; Islam, N.N. Plasmid profiling of multiple antibiotic-resistant P. aeruginosa isolated from soil of the industrial area in Chittagong, Bangladesh. Beni-Suef Univ. J. Basic Appl. Sci. 2021, 10, 44. [Google Scholar] [CrossRef]
- Tiamiyu, A.; Soladoye, M.; Adegboyega, T.; Adetona, M. Occurrence and antibiotic sensitivity of bacterial strains isolated from Nile tilapia, Oreochromis niloticus obtained in Ibadan, Southwest Nigeria. J. Biosci. Med. 2015, 3, 19. [Google Scholar] [CrossRef][Green Version]
- Bahador, N.; Shoja, S.; Faridi, F.; Dozandeh-Mobarrez, B.; Qeshmi, F.I.; Javadpour, S.; Mokhtary, S. Molecular detection of virulence factors and biofilm formation in P. aeruginosa obtained from different clinical specimens in Bandar Abbas. Iran. J. Microbiol. 2019, 11, 25. [Google Scholar] [CrossRef]
- Hasan, S.A.; Najati, A.M.; Abass, K.S. Prevalence and antibiotic resistance of P. aeruginosa isolated from clinical samples in Kirkuk City, Iraq. Eurasian J. Biosci. 2020, 14, 1821–1825. [Google Scholar]
- Pokharel, K.; Dawadi, B.R.; Bhatt, C.P.; Gupte, S. Prevalence of P. aeruginosa and its Antibiotic Sensitivity Pattern. J. Nepal Health Res. Counc. 2019, 17, 109–113. [Google Scholar] [CrossRef]
- Deodurg, P.M.; Doddamani, P.K.; Rana, S.; Mir, B.A. Prevalence and Antimicrobial Susceptibility Pattern of P. aeruginosa in a Tertiary Care Hospital. Res. J. Pharm. Technol. 2014, 7, 517. [Google Scholar]
- Buhl, M.; Peter, S.; Willmann, M. Prevalence and risk factors associated with colonization and infection of extensively drug-resistant P. aeruginosa: A systematic review. Expert Rev. Anti-Infect. Ther. 2015, 13, 1159–1170. [Google Scholar] [CrossRef] [PubMed]
- Remans, K.; Vercammen, K.; Bodilis, J.; Cornelis, P. Genome-wide analysis and literature-based survey of lipoproteins in P. aeruginosa. Microbiology 2010, 156, 2597–2607. [Google Scholar] [CrossRef] [PubMed]
- Aljebory, I.S. PCR detection of some virulence genes of P. aeruginosa in Kirkuk city, Iraq. J. Pharm. Sci. Res. 2018, 10, 1068–1071. [Google Scholar]
- Hassuna, N.A.; Mandour, S.A.; Mohamed, E.S. Virulence constitution of multi-drug-resistant P. aeruginosa in upper Egypt. Infect. Drug Resist. 2020, 13, 587–595. [Google Scholar] [CrossRef]
- Gaviard, C.; Cosette, P.; Jouenne, T.; Hardouin, J. LasB and CbpD virulence factors of P. aeruginosa carry multiple post-translational modifications on their lysine residues. J. Proteome Res. 2019, 18, 923–933. [Google Scholar] [CrossRef]
- Hosu, M.C.; Vasaikar, S.D.; Okuthe, G.E.; Apalata, T. Detection of extended spectrum beta-lactamase genes in P. aeruginosa isolated from patients in rural Eastern Cape Province, South Africa. Sci. Rep. 2021, 11, 7110. [Google Scholar] [CrossRef] [PubMed]
- Beceiro, A.; Tomás, M.; Bou, G. Antimicrobial resistance and virulence: A successful or deleterious association in the bacterial world? Clin. Microbiol. Rev. 2013, 26, 185–230. [Google Scholar] [CrossRef] [PubMed]
- Zaheen, Z.; War, A.F.; Ali, S.; Yatoo, A.M.; Ali, M.N.; Ahmad, S.B.; Rehman, M.U.; Paray, B.A. Common bacterial infections affecting freshwater fish fauna and impact of pollution and water quality characteristics on bacterial pathogenicity. In Bacterial fish diseases; Elsevier: Amsterdam, The Netherlands, 2022; pp. 133–154. [Google Scholar]
- Gram, L.; Huss, H.H. Fresh and processed fish and shellfish. In The Microbiological Safety and Quality of Food; Aspen Publishers: Burlington, MA, USA, 2000; pp. 472–506. [Google Scholar]
- Novotny, L.; Dvorska, L.; Lorencova, A.; Beran, V.; Pavlik, I. Fish: A potential source of bacterial pathogens for human beings. Veterinární Medicína 2004, 49, 343–358. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).