1. Introduction
In the past few decades, evolutionary algorithms (EAs) [
1] have been commonly adopted for tackling multi-objective optimization problems (MOPs) [
2] with multiple contradictory objectives to pursue, earning the name multi-objective evolutionary algorithms (MOEAs) [
3]. To date, large numbers of MOEAs have already been proposed and can be briefly divided into the following categories: the dominance-based framework that uses the nondominated sorting method for environmental selection, such as the most classic dominance-based framework NSGA-II [
4], its improved version based on reference points [
5], a strengthened dominance-based algorithm for many-objective optimization [
6], and one with explicit variable space diversity management [
7]; the decomposition-based framework that decomposes an MOP into single-objective problems based on weight vectors, such as the most classic decomposition-based framework MOEA/D [
8], its improved version based on differential evolution [
9] or stable matching [
10], and one based on symbiotic organism search [
11]; the indicator-based framework that utilizes specific performance indicators for truncation, such as the most classic indicator-based framework based on the hypervolume indicator [
12], one based on boundary protection [
13], one based on analyzing dominance move [
14], and a bi-indicator-driven indicator-based framework [
15]. There are in addition many other excellent MOEA frameworks, such as the surrogate-based framework [
16], the classifier-based framework [
17], a three-stage MOEA [
18], and the multitasking MOEA [
19].
With the help of population-based searching abilities and no need for domain knowledge, MOEAs have been widely utilized to solve real-life optimization problems, such as network construction [
20], task offloading [
21], community detection [
22], and the bi-objective feature selection problem [
23] that has been specially focused on in this work. More specifically, feature selection [
24] is a data preprocessing technique, selecting only a subset of useful features for classification, especially in high-dimensional datasets [
25]. Aiming at minimizing the ratio of selected features, i.e., the first objective, and the ratio of classification errors, the second objective, feature selection then becomes a multi-objective optimization problem, formally presented as follows [
26].
where
D represents the full number of features that can be selected in the decision space, and
M denotes the number of objectives to be optimized, which is set to two in this work.
is the objective vector of solution
, and
denotes the corresponding objective value in the direction of the
ith objective. Moreover,
represents the decision vector of a certain solution, where
indicates that the
ith feature is selected and
indicates not being selected.
To be more specific, the first objective function
, which denotes the ratio of selected features in this work, is defined as follows:
the value of which discretely ranges from 0 to 1 (i.e.,
). In addition, given the results of
(True Positive),
(True Negative),
(False Positive), and
(False Negative), the second objective function
, denoting the ratio of classification errors related to the above selected features, can be further defined as follows:
However, despite of their various applications in the field of optimization, traditional MOEAs may still face the curse of dimensionality in tackling the bi-objective feature selection problem, when the number of features increases to high dimensionality, causing a lack of search abilities in finding nondominated solutions. Many existing research works have already attempted to solve this issue [
27], but many of these either have complicated algorithm frameworks or need to preset a large number of fixed parameters. Furthermore, it is also difficult to balance well algorithm performance between relatively lower-dimensional datasets and higher-dimensional ones, in terms of both diversity and convergence.
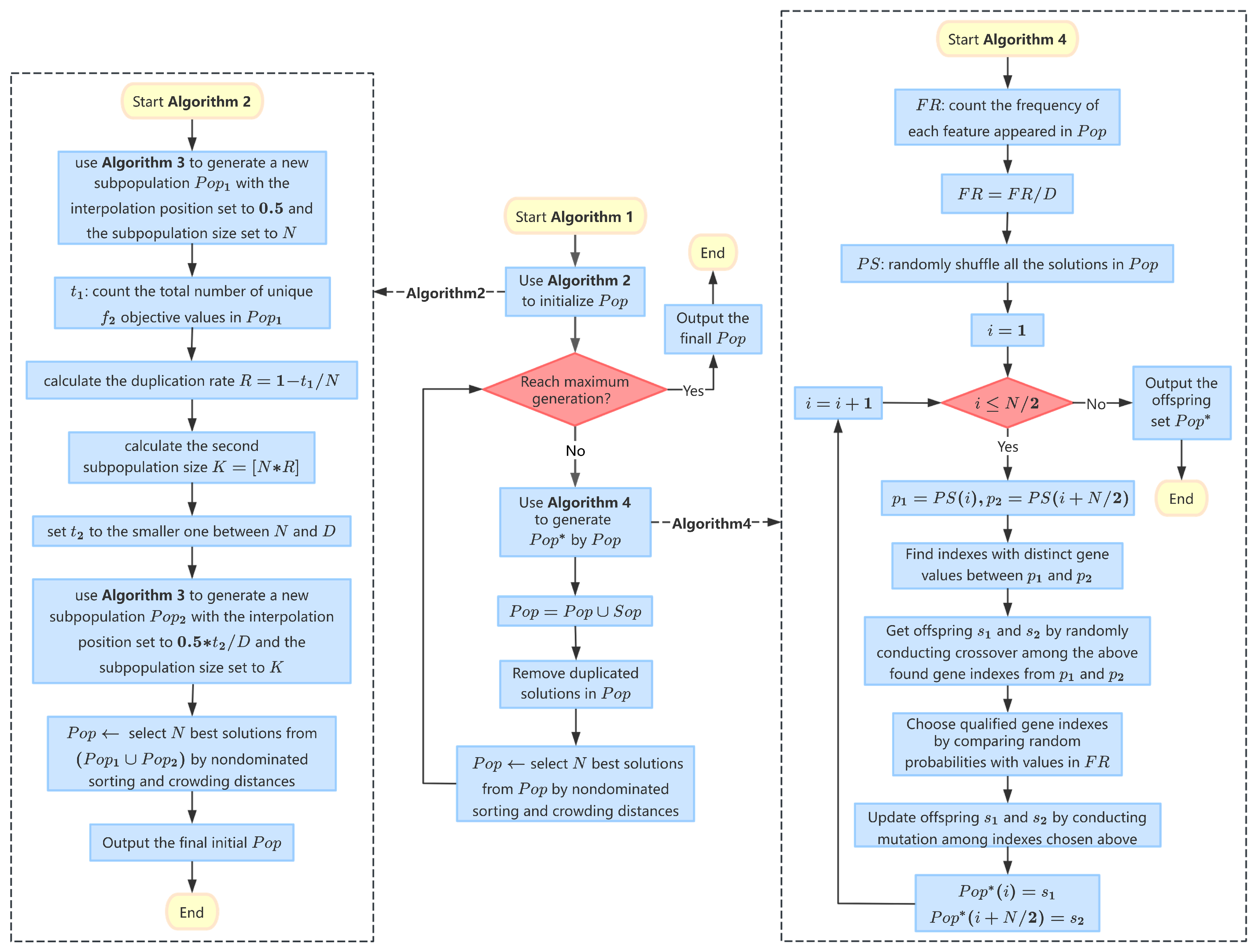
Therefore, in this paper, a simple and effective MOEA framework based on the specially designed adaptive initialization and reproduction mechanisms (abbreviated as AIR) is proposed for addressing the bi-objective feature selection problem in classification datasets. In the proposed AIR algorithm, an adaptive initialization mechanism is designed to provide a promising start for the later evolution and to pursue faster convergence, while an adaptive reproduction method is also adopted to generate more diverse offspring and to avoid potential pre-maturity in convergence. Furthermore, combining the proposed adaptive initialization and reproduction methods together not only increases the search abilities of AIR in finding nondominated solutions, but also helps to maintain a better balance between population diversity and algorithm convergence across the whole evolution. In this way, AIR is then able to deal with more universal optimization environments, across various feature dimensions, from a lower dimensionality to a higher one.
Moreover, it should also be noted that the primary focus of this paper is not on parameter tuning but rather on introducing an innovative adaptive initialization and reproduction strategy for MOEAs in the context of bi-objective feature selection. The key parameters utilized in the adaptive initialization and reproduction processes of AIR are not arbitrarily set but are instead derived from an analysis of the solution distribution in the objective space and the feature frequency within the current population. This adaptive mechanism allows AIR to dynamically balance the exploration and exploitation of the solution space, maintaining a healthy diversity while gradually converging toward promising regions. This is particularly advantageous when dealing with high-dimensional datasets, where the complexity of the feature space and the trade-offs between the two objectives (feature reduction and classification accuracy) are most pronounced. By adaptively adjusting key parameters in response to the evolving characteristics of the population, AIR can effectively navigate the challenges of high-dimensional bi-objective feature selection, which is a significant contribution to the field of MOEAs.
The remainder of this paper is organized as follows. First, related works about multi-objective evolutionary feature selection are discussed in
Section 2. Then, the proposed AIR algorithm and its two important components (AI and AR) are illustrated in
Section 3. Furthermore, the essential experimental setups are given in
Section 4, while all the empirical results are analyzed in
Section 5. Finally, the conclusions and future work are both summarized in
Section 6.
2. Related Works
The field of multi-objective evolutionary feature selection adopts MOEAs for tackling feature selection problems, which can be roughly categorized into wrapper-based approaches or filter-based ones [
28]. In general, wrapper-based methods [
29,
30] normally adopt certain classification models, such as SVM (Support Vector Machine) or KNN (k-Nearest Neighbor), to evaluate the corresponding classification accuracy as the result of the currently selected feature subsets. Conversely, filter-based methods [
31,
32] do not rely on any classifier, but directly analyze the explicit or implicit relationship between features and the corresponding classes in the classification datasets, without verifying the classification result for any selected feature subset. Thus, a wrapper based approach is normally more accurate but could consume considerable computational cost due to the real-time classification process during evolution.
Therefore, this paper focuses on studying wrapper-based approaches for evolutionary bi-objective feature selection, attempting to increase the search abilities by improving the initialization and reproduction processes, and to decrease the computational cost by selecting fewer features for classification. Many existing works have already studied similar topics and proposed many inspiring MOEAs specifically for addressing the bi-objective feature selection problem, in recent years.
For instance, in 2020, Xu et al. [
33] proposed a segmented initialization method and an offspring modification method, named SIOM; however, its key parameter settings are all fixed and cannot be adaptively altered for different classification datasets, which is not suitable for tackling high-dimensional feature selection. Tian et al. in 2020 [
34] proposed a sparse MOEA for tackling bi-objective feature selection, named SparseEA, based on the analyses of each single feature performance before the start of the main evolution; however, the analyses may require a large computational cost as the dimensionality of features increases. Subsequently, Zhang et al. further [
35] proposed an improved version of the previous Sparse, named SparseEA2, which has a modified two-layer encoding scheme assisted by variable grouping techniques but still incurs considerable computational cost for solving high-dimensional feature selection problems. In 2021, Xu et al. [
36] proposed a duplication analysis-based evolutionary algorithm, which is combined with an efficient reproduction method; however, its performance has not yet been tested in cases of high-dimensional feature selection. In 2022, Cheng et al. [
25] proposed a steering matrix-based MOEA specifically for tackling high-dimensional feature selection; however, its performance on relatively lower-dimensional datasets is not validated, which might restrict its universality. Later, inspired by the aforementioned work [
36], Jiao et al. in 2024 [
37] designed a problem reformulation mechanism to further improve the process of handling solution duplication, named PRDH (which we use as a comparison algorithm in our experiments), whose applicability remains unconfirmed in different MOEA frameworks. In the same year, Xu et al. [
38] proposed a bi-search mechanism-based evolutionary algorithm, named BSEA (which we use as a comparison algorithm in our experiments), which was specifically designed to overcome the challenge of search abilities for high-dimensional feature selection problems; however, this relies on a rather complicated multi-task MOEA framework consisting of two dynamic subpopulations. Most recently, Hang et al. [
26] designed a probe population-based initialization method to improve the search ability in high-dimensional feature space; however, its iterative exploration of probe populations may incur considerable computational cost. More related works about evolutionary multi-objective feature selection in classification can be found in the most recent survey [
27].
Thus, in this work, the design of a simple and effective MOEA framework is attempted, with the aim of improving the initialization and reproduction processes, with adaptively set parameters, to create a more universal algorithm suitable for handling both lower-dimensional classification datasets and higher-dimensional ones.
6. Conclusions and Future Work
In this work, an MOEA based on adaptive initialization and reproduction mechanisms termed AIR is specifically designed for tackling bi-objective feature selection, especially for the cases of relatively higher-dimensional classification datasets. In AIR, an adaptive initialization method is designed in order to provide a promising and hybrid initial population, not only covering the middle region in the objective space but also adaptively exploring the relatively front region of interest with much smaller sizes of selected feature subsets. Moreover, an adaptive reproduction method is also designed in order to provide more offspring diversity and to maintain a delicate balance between convergence and diversity, avoiding the potential pre-maturity of evolution. It should also be noted that the general framework of AIR is rather simple and effective, making it capable of adapting different kinds of optimization environment from lower to higher feature dimensionality. All the key parameters in the proposed AIR algorithm are not fixed but are adaptively adjusted based on the evolving characteristics of the population, which allows AIR to dynamically balance exploration and exploitation, maintaining diversity while gradually converging towards promising regions. The adaptive settings for those key parameters are the major contribution and innovation of this work in solving complex bi-objective feature selection problems, especially in high-dimensional datasets where static parameter settings may not be effective.
The promising performance and potential advantages of AIR have been comprehensively analyzed and verified in the experiments, by comparing with five state-of-the-art MOEAs on multiple performance indicators and a series of 20 real-life classification datasets. In general, the proposed AIR performs overall the best, with promising search abilities in terms of both optimization and classification.
In the future work, the applicability of AIR to more kinds of discrete multi-objective optimization problems, such as community node detection and neural network construction, will be further researched. Moreover, it is also planned to incorporate more extensive comparisons with methods from different fields to provide a more comprehensive evaluation of the proposed approach.