Abstract
Higher temperature conditions during the final stages of rice seed development (seed filling and maturation) are known to cause damage to both rice yield and rice kernel quality. The western and central parts of Japan especially have seen record high temperatures during the past decade, resulting in the decrease of rice kernel quality. In this study, we looked at the rice harvested from a town in the central Kanto-plains (Japan) in 2010. The daytime temperatures were above the critical limits ranging from 34 to 38 °C at the final stages of seed development and maturity allowing us to investigate high-temperature effects in the actual field condition. Three sets of dry mature rice seeds (commercial), each with specific quality standards, were obtained from Japan Agriculture (JA Zen-Noh) branch in Ami-town of Ibaraki Prefecture in September 2010: grade 1 (top quality, labeled as Y1), grade 2 (medium quality, labeled as Y2), and grade 3 (out-of-grade or low quality, labeled as Y3). The research objective was to examine particular alterations in genome-wide gene expression in grade 2 (Y2) and grade 3 (Y3) seeds compared to grade 1 (Y1). We followed the high-temperature spike using a high-throughput omics-approach DNA microarray (Agilent 4 × 44 K rice oligo DNA chip) in conjunction with MapMan bioinformatics analysis. As expected, rice seed quality analysis revealed low quality in Y3 > Y2 over Y1 in taste, amylose, protein, and fatty acid degree, but not in water content. Differentially expressed gene (DEG) analysis from the transcriptomic profiling data revealed that there are more than one hundred upregulated (124 and 373) and downregulated (106 and 129) genes in Y2 (grade 2 rice seed) and Y3 (grade 3 rice seed), respectively. Bioinformatic analysis of DEGs selected as highly regulated differentially expressed (HRDE) genes revealed changes in function of genes related to metabolism, defense/stress response, fatty acid biosynthesis, and hormones. This research provides, for the first time, the seed transcriptome profile for the classified low grades (grade 2, and out-of-grade; i.e., grade 3) of rice under high-temperature stress condition.
1. Introduction
With the rise in mean global temperatures, the earth’s biosphere is warming up gradually. According to the statistics of the North American Space Agency’s (NASA) earth observatory data, the average global temperature increased above 1 °C since the year 1880 [1]. In the next 100 years, the surface temperatures are expected to rise between 2 °C to 6 °C if greenhouse gas emissions continue. This is a serious threat for our future generations as it directly influences the habitable conditions—both flora and fauna. A rise in temperature induces heat stress and seriously affects plants, which play a key role in providing food, oxygen, and shelter to several species of fauna including humans. Moreover, its effects on the productivity of food crops will result in a food crisis for the growing population [2]. Recently, Zhao et al. (2017) [3] estimated that for every one-degree rise in global temperature, yields of food crops like wheat, rice, maize, and soybean will be reduced by 6%, 3.2%, 7.4%, and 3.1%, respectively [3]. Therefore, there is a grave need to understand the effects of increasing temperatures and the biological responses induced in food crops, to address challenges in developing next-generation crops [2,3].
Heat stress damages both physiological and molecular level mechanisms in plants [4,5,6,7]. Major damages include scorching and abscission of leaves, shoot and stems, fruit/seed damage, reduced photosynthesis, seed germination, increase in reactive oxygen species (ROS), and osmotic stress [5,6,8,9,10,11]. Furthermore, the ROS accumulated during heat stress damages molecular components by inducing oxidative stress [12,13]. For example, the formation of hydroxyl radicle could induce irreversible DNA single-strand breaks, peroxidation of lipids, protein proteolysis, and damage of photosynthesis system II [14,15,16]. In addition, hydrogen peroxide (H2O2) formed during heat stress could alter the balance between starch biosynthesis and degradation mechanisms by upregulation of starch degradation and downregulation of the biosynthesis genes. Additionally, plants also develop heat-tolerance by responding to heat stress by altering their gene expression and synthesizing specific heat shock proteins (HSPs), transporters, and enzymes.
Rice (Oryza sativa), being one of the top three food crops produced and consumed by nearly 3.5 billion people around the world, has been well studied for its yield and quality with respect to changes in global temperatures. According to the Food and Agriculture Organization (FAO) of the United Nations, Asia is the largest producer of rice in the world (nearly 90%). The average rice production statistics between 1994–2018 show that China and India stand as the top two producers (193 and 140 million tons respectively), while Japan is ranked 10th (11 million tons) in the world. Japan is also the 13th largest consumer of rice in the world as of 2018. In Japan, as rice is a primary staple food crop with high socio-economic importance, any reduction in yield and damage to the quality of the grain is a serious issue. The decrease in rice productivity is generally due to reduced pollen germination and in turn, affects the spikelet fertility and yield [17,18,19,20,21]. Moreover, spikelet sterility occurs when air temperature reaches a threshold limit of 35 °C during flowering time [22]. For example, in 2007, the Kanto and Tokaido regions of Japan faced a marked rise in temperatures to 40 °C during the summer months. The samples collected in this region showed high rates of spikelet fertility damages [19]. In addition to high-temperature injury in grain productivity, it also affects the quality of rice. For example, if rice crops are exposed to high temperatures during the first two weeks after an early emergence, the rice grain turns into immature kernels with white portions [23,24,25,26]. A decade back, high proportions of first-grade rice kernel were produced in the warmer regions (west Japan) like Kyushu, while production was lower in northern regions of Japan (like Tohoku and Hokkaido), due to cold damage. However, in the past years, the rice crop quality from North to West Japan has been completely reversed due to climatic changes [18]. In the year 2010, the average temperature across Japan approached 28 °C to 29 °C, as compared to normal day temperatures (26 °C). That year recorded the hottest summer (June-September 2010) ever experienced with day temperatures ranging between 35 °C to 38 °C in East, South, Central, and West regions of Japan. On average, the daytime temperatures were nearly 1.8 and 2 °C above normal in August. An exposure to such anomalies in temperature for a couple of hours is enough to induce spikelet sterility and reduce crop productivity in rice. As such these conditions during 2010 resulted in producing milky white kernels in the first-grade rice kernel crop, and, therefore reduced their production throughout Japan (Figure 1). This grain chalkiness is due to the induced production of starch hydrolyzing enzymes (α-amylases) as a result of high temperatures [27,28,29,30].
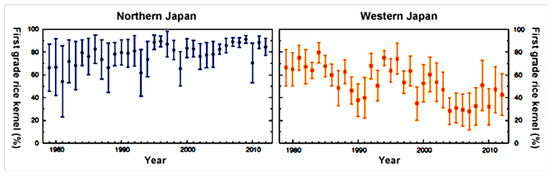
Figure 1.
Temporal change in the proportion of first grade rice kernel in Northern Japan and Western Japan. This figure, created based on Ministry of Agriculture, Forestry and Fisheries data, has been obtained with permission from Prof. Shunji Ohta, Faculty of Human Sciences, Waseda University-Recent Impacts of Climatic Extreme Events on Everyday Food in Japan: The Need for an Adaptation Strategy for Climate Change [31].
It was observed that the productivity and quality of rice also depend on its variety along with the temperature. However, no studies were reported on how different varieties of the same cultivar respond to heat stress in the open field conditions. Especially in Japan, Koshihikari is a highly grown rice cultivar/variety for the past few decades, and also is the most widely affected variety due to climate changes in recent years. Understanding the importance of this cultivar in Japan, the current study focuses on exploring transcriptome level differences under heat stress between grades 1, 2, and 3 (labeled as Y1, Y2, and Y3 for the purpose of the experiment) of the Koshihikari rice variety. Each grade of rice is categorized by Japan’s National Food Agency (NFA) based on the grain’s physiological characteristics like weight per volume, moisture content, appearance, region in which it was grown, etc., which determine its quality [32]. For this open field sample, sampling was done from a region in East Japan (Kanto-plains) during the high-temperature season of the year 2010, and included three different grades of rice. The seed samples were analyzed for genome-wide gene expression (DNA microarray technique) and compared using bioinformatic techniques for identifying differentially expressed genes (DEG).
2. Results and Discussion
2.1. Koshihikari Rice Seed Quality in Grades 1 to 3
The seed quality was tested based on quality criteria like taste value, percentage of amylase, protein, water content, and fatty acid degree in the rice seed (Table 1). Grade 1 (Y1) Koshihikari rice is identified to have a high taste value of 86. Compared to Y1, grade 2 (Y2) and grade 3 (Y3) are relatively less tasty by 5 to 3 points as determined by professional analysis. In addition, the Y2 and Y3 grains have a high percentage of amylose and water content in comparison to Y1, indicating sticky and chalky rice. In general, lower amylose content means lower chalkiness of the grains, resulting in harder rice. Low water content indicates lower moisture levels which in turn produce less sticky rice after cooking [33,34]. Quality of rice also depends on its aroma and flavor. The surface lipids of grains form free fatty acids as they undergo hydrolysis during storage of the grain. The free fatty acids formed in this way are susceptible to oxidation and eventual formation of hydrocarbons such as aldehydes and ketones, which give a foul odor to the seeds [35,36]. Y1 was found to have a lesser degree (15.5) of unsaturated fatty acids in comparison to Y2 (17) and Y3 (20), indicating the presence of good aroma with grade 1. The overall grain quality analysis clearly indicated that under extreme temperatures, the grain quality is affected more (to negative values) in Y3 followed by Y2, and is least affected in Y1 (Grade 1).

Table 1.
Rice grain (cv. Koshihikari * seed) quality analyses **.
2.2. Investigation of the Koshihikari Rice Seed Transcriptomes in Grades 2 and 3
This work looks at differences in transcripts accumulated in the dried endosperm after the maturation process. From the analysis of DEGs in Y3 and Y2 in comparison to Y1, a greater number of DEGs were observed in Y3 (502 genes) than Y2 (230 genes), as shown in Figure 2. Individually, a total of 373 upregulated and 129 downregulated genes were observed in Y3 while Y2 resulted in 124 and 106 up and downregulated genes, respectively. Among all the DEGs in grades Y3 and Y2, similar expression patterns were also observed for a few common genes present in both grades. In this category, there are nearly 59 upregulated genes and 33 downregulated genes.
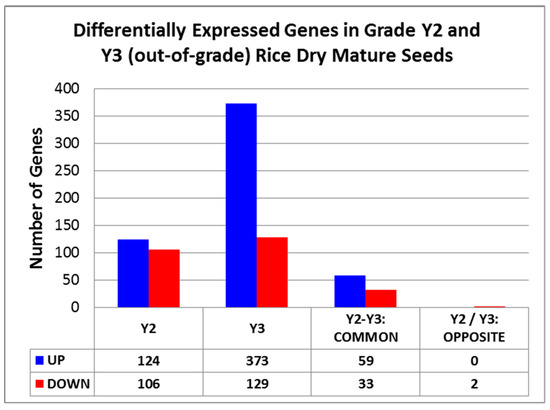
Figure 2.
Figure indicating the differentially expressed genes (DEGs) in Y2 and Y3.
2.3. MapMan Analysis of Koshihikari Rice Seed Differentially Expressed Genes in Grades 2 and 3
In order to visualize the DEGs that are involved in key pathways, a total of 42,560 Rice Japonica genes were plotted against the pathway maps of the Mapman tool. Among all the genes, DEGs with high fold changes were selected for the analysis, as discussed in methods for Mapman analysis. From the fold change cut-off threshold, a total of 161 and 490 highly regulated differentially expressed (HRDE) genes for each Y2 and Y3 were identified. Among these, 92 genes are common for both grades. A total of 68 and 398 genes are unique for Y2 and Y3, respectively resulting in a total of 560 HRDEs for both. An overview of these HRDEs in Y1-Y2 and Y1-Y3, and the related 36 pathway bins, are shown in (Figure 3A,B). Results clearly indicate that the highest fraction of the gene regulation occurred in 16, 17, 20, 26, 27, 28, 29, 30, 33, and 34 Mapman bins. The number of genes associated with each bin, specific to each pathway, is listed in supplementary Table S1. These bins are related to secondary metabolism, hormone metabolism, stress response, miscellaneous enzyme families, RNA, DNA, protein, signaling, development, and transport. In addition, bin 35 has the most differentially expressed genes; however, these genes are without any functional annotation.

Figure 3.
Mapman Bins for functional categorization of high temperature-responsive genes in (A) Y2: grade 2 Koshihikari rice seed and (B) Y3: grade 3 Koshihikari rice seed. Non-redundant 640 genes which expression are changed over 2-fold in the seeds of both grades 2 (Y2) and 3 (Y3) were functionally categorized into MapMan bins as described in Materials and Methods. The heat map with grid boxes shows the genes (blue, upregulated; red, downregulated) in each BIN for each grade seeds. Number of genes associated to each bin is listed in supplementary Table S1. The 36 BINS abbreviations: PS, photosynthesis; maCHO, major carbohydrate metabolism; miCHO, minor carbohydrate metabolism; G, glycolysis; FM, fermentation; GL/GC, gluconeogenese/glyoxlate cycle; OPP, oxidative pentose phosphate; TCA/OT, tricarboxylic acid/organic acid transformations; MET/ATPs, mitochondrial electron transport/adenosine triphosphate; CW, cell wall; L, lipid metabolism; N-, nitrogen metabolism; AA, amino acid metabolism; S-A, sulfur assimilation; MH, metal handling; S, secondary metabolism; H, hormone metabolism; Co-F/V, co-factor and vitamin metabolism; TS, tetrapyrrole synthesis; ST, stress; RR, redox regulation; P, polyamine metabolism; N, nucleotide metabolism; BioDX, biodegradation of xenobiotics; C1, C1-metabolism; MISC, miscellaneous; RNA, ribonucleic acid; DNA, deoxyribonucleic acid; PR, protein; SIG, signaling; C, cell; mRNA, messenger RNA; D, development; T, transport; NA, not assigned; MN, mineral nutrition.
The overall expression profile indicates that stress response genes (bin 20) are strongly regulated in both Y2 and Y3. Y3 exhibited a larger number of strongly upregulated genes in comparison to Y2. Further, the differential gene expression patterns of Y2 and Y3 in three major processes, namely cell function, metabolism, and abiotic-biotic stress (Figure 4, Figure 5 and Figure 6) were explored.
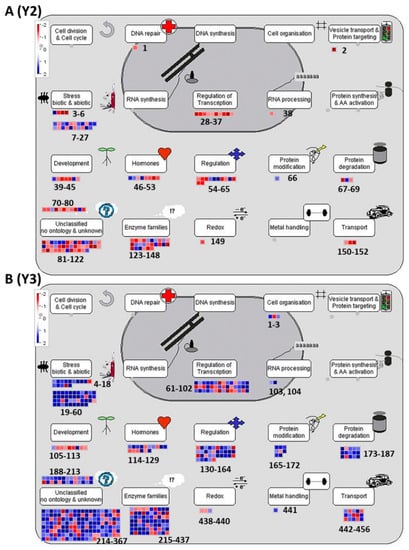
Figure 4.
A cell function map of highly regulated differentially expressed (HRDE) genes that are categorized based on various functions, generated using Mapman. (A) Map for HRDE genes related to Koshihikari rice grade Y2. (B) Map for HRDE genes related to Koshihikari rice grade Y3. Blue and red colored data points indicated highly up or downregulated genes. Numbering for each data point on the figure was given in left to right order to identify relevant gene information from supplementary Table S2. These numbers represent serial numbers for the genes in the supplementary table accordingly.
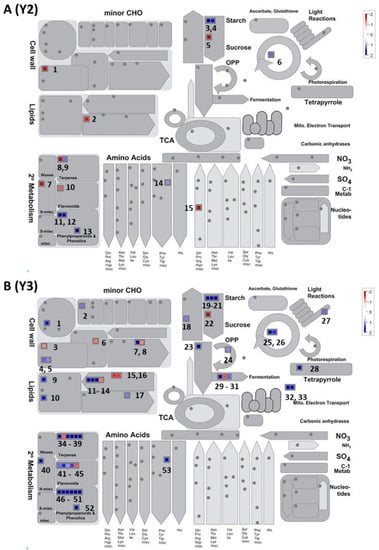
Figure 5.
A metabolism overview map of highly regulated differentially expressed (HRDE) genes that are categorized into various metabolic pathways generated using Mapman. (A) Map for HRDE genes related to Koshihikari rice grade Y2. (B) Map for HRDE genes related to Koshihikari rice grade Y3. Numbering for each data point on the figure was given in left to right order to identify relevant gene information from supplementary Table S3. These numbers represent serial numbers for the genes in the supplementary table accordingly. Expression of specific genes of interest like PME, cellulose synthases, pectin lyases, XET, raffinose synthase, TAG lipases, DGK, Omega-6-desaturases, SPT2, amylase and Susy family genes can be observed in Y2(1, 3), Y3(6), Y3(7, 8), Y3(4), Y3(2), Y2(2), Y3(9), Y3(12, 13, 14), Y3 (10), respectively.
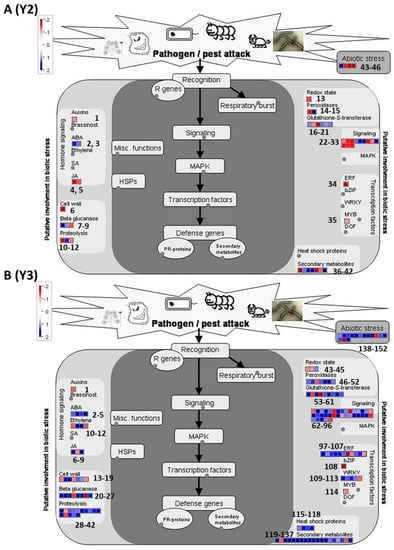
Figure 6.
An abiotic-biotic map of differentially expressed genes that are categorized into abiotic and biotic stress-responsive pathways generated using Mapman. (A) Map for HRDE genes related to Koshihikari rice grade Y2. (B) Map for HRDE genes related to Koshihikari rice grade Y3. Numbering for each data point on the figure was given in left to right order to identify relevant gene information from supplementary Table S4. These numbers represent serial numbers for the genes in the supplementary table accordingly. Expression of specific genes of interest like sHsp, MYB4, DREB1/CBF, ERF, OsWRKY, GGPS, anthocyanins, and flavonoid biosynthesis, COMT, laccasses, OsSAUR, FIP1, lipoxygenase2 and OPR family genes can be observed in Y3(115/138), Y2/Y3(35/114), Y3(101, 102), Y3(98), Y3(110-112), Y2(41),Y3(133-135), Y3(178,179), Y2/Y3(47/186), Y2/Y3(1), Y2/Y3(2,3/2,3,4,5), Y2/Y3 (4/10) and Y2/Y2 (5/11,12) grid boxes, respectively. Y2/Y3 represents similar genes detected in both Y2 and Y3.
For each of the above pathway maps, a total of 15, 151 and 46 HRDE genes of Y2 and 53, 457 and 152 HRDE genes of Y3, were mapped as data points. All these genes are listed in supplementary Tables S2–S4, along with their bin-numbers and functions. For the cell function, metabolism-overview and biotic-abiotic stress pathway maps (Figure 4, Figure 5 and Figure 6), the numbering for each data point was given in left to right order to identify relevant gene information from supplementary Tables S2–S4. Serial numbers for the genes in the supplementary tables were numbered accordingly. Overall mapping results indicate that around 650 genes related to various cellular functions were differentially regulated in both Y2 and Y3, respectively (Figure 4, supplementary Table S2). Y2 appears to respond specifically by down regulation of genes in biotic-abiotic stress, transcription, RNA processing, protein degradation, hormones and regulation. While Y3 exhibited quite opposite response by upregulation of genes in respective mechanisms, with few downregulated genes. Genes related to protein modification are downregulated in both Y2 and Y3.
Specifically, from the metabolic-overview map (Figure 5), results indicate that highly up-, or downregulated genes are mainly related to cell wall, lipid, starch, and secondary metabolic pathways. In Y2, very few genes were differentially regulated (7 downregulated and 8 upregulated). Among these, genes related to cell wall remodeling proteins (pectin methyl esterase’s- PMEs), sucrose synthase, triacylglycerol (TAG) lipase, phosphotase synthase and a few secondary metabolism-related genes (tyrosine decarboxylate, terpene cyclase, isoflavonoid reductase) were highly downregulated. On the other hand, Y3 data showed upregulation of cell wall modification proteins like transglucosylases (XET), diacylglycerol kinases, omega-6 fatty acid desaturases, male sterility proteins, α-amylase isozymes, rubisco interacting proteins, mitochondrial electron-transport proteins (transposons) and many proteins related to secondary metabolites (terpenes, flavonoids, and phenolics) (Supplementary Table S3). Out of both Y2 and Y3, commonly upregulated metabolic pathways were related to starch, chitinase, phenylpropanoids and phenolics, and aromatic amino acid synthases. While the commonly downregulated genes are related to PME and sucrose synthase.
On the other hand, among the cellular functions, the biotic and abiotic stress responses are the major events during heat stress. Y2 exhibited downregulation of genes related to auxins, jasmonic acid (JA), lipoxygense, PME, proteases, peroxidases, protein kinases, ATP binding proteins, signaling G-proteins, calcium ion binding proteins and abiotic stress-related germin like proteins (Figure 6). On the other hand, Y3 showed more upregulated genes especially related to glucanase, proteolysis, peroxidases, glutathione-S-transferases, signaling, transcription factors (TFs), HSPs, secondary metabolites, and abiotic stress responses. Despite heat stress, in Y2 there were no genes related to ethylene, HSP, and TFs like b-zip that crossed the threshold set for detection of HRDE genes, in our two-color dye-swap DNA microarray strategy. There are genes with a 1-fold upregulation which are not considered as the threshold is set to 2. So, there is a minimal level of gene expression in Y2 indicating that it is not as sensitive as Y3 to heat stress.
2.4. High Temperature-Triggered Regulatory Events in Koshihikari Rice Seeds of Grades 2 and 3
Overall results indicated that the effect of high temperature on Koshihikari rice of grades 2 and 3 is quite different. Most of the genes were downregulated in Y2 type, while Y3 exhibited upregulation.
2.4.1. Cell Wall Damage Repair
Initial response to heat shock is observed in cell wall remodeling proteins like PMEs and XETs. PMEs are involved in regulating cell wall plasticity, porosity, and modulation of Ca2+ ion channels [37], while XETs are involved in secondary cell wall strengthening [38,39]. The regulation of XETs and PMEs under heat shock is previously observed in rice, Arabidopsis, and many plant varieties [38,40,41]. During heat stress, Y2 and Y3 exhibited down regulation of PMEs genes Os09g37360.1 and Os04g46740.1, respectively. In addition, Y3 alone shows downregulation of genes related to cellulose synthesis (Os02g09930.1) and pectin lyases (Os10g26940.1 and Os02g15690.1), while upregulation of XET gene (Os06g22919.2) indicating cell wall response in Y3 is more sensitive to heat shock. An upregulation of the raffinose synthase gene (Os08g38710.1) in Y3, which prevents plants from oxidative stress damage [42,43,44], was also observed. Besides, the pectinase activity upregulation of cellulases like endo-glucosidase, prominently in Y3 can be seen (supplementary Table S3, Metabolism overview; and Figure 5).
2.4.2. Lipid Remodelling
The lipid signaling is another key process involved in abiotic stress responses by the plants. In Y2, there is no more than one gene related to lipid mechanism which was identified as the HRDE gene. This gene (Os08g04800.1) codes for a TAG lipase protein and is related to lipid degradation. TAG lipase was found to be strongly downregulated (−1.63). TAG lipases de-esterify fatty acids from TAG (accumulated in lipid bodies) or plastglobular lipids and are generally accumulated during seed germination [45]. TAG lipases are observed to be accumulated under heat stress in Arabidopsis [45,46]; however, the current data shows the opposite result. It is to be noted that rice and Arabidopsis are different, and therefore, the same gene could have unique functions in different plants and tissues. On the other hand, in Y3 there were no genes related to TAG lipases identified as HRDEs. However, the same gene was found in Y3 with very minimal (three times less than Y2) downregulation (not HRDE). This indicated that in this case, strong downregulation of TAG lipases could be common in Koshihikari as this is a DEG analysis in comparison to Y1 also. So, the result in Y3 indicates that Y3 might have started the accumulation of this gene under heat stress.
Furthermore, the genes related to diacyl-glycerol kinases (DGK), phospholipid synthesis, sphingolipid transferases, and fatty acid omega 6 desaturases were strongly upregulated in Y3 (supplementary Table S3, Metabolism overview; and Figure 5). In general, DGKs are involved in phosphatidic acid (PA) synthesis as well as sphingo-lipid synthesis. The PA produced by the mediation of DGKs has a positive regulatory impact on sphingo-lipid kinases and a negative effect on abscisic acid synthesis pathways [47]. PA is also involved in the growth and development of plant root hair and also functions as membrane-localized signal to recruit specific proteins and change their activity [48,49,50]. Therefore, by upregulation of this DGK specific gene (Os04g54200.1), Y3 is preparing for abiotic stress due to increased temperatures.
The fatty acid desaturases, on the other hand, are involved in maintaining membrane fluidity and are usually upregulated under low-temperature stress. However, studies also identified that high temperatures also enhance desaturases involved in the eukaryotic pathway [51]. In the current work upregulation of omega-6-desturases (Os07g23410.2, Os07g23430.1, Os02g48560.5) FAD2 and FAD3 related genes was observed. This clearly indicates a remodeling of fatty acids in endoplasmic reticulum [52]. In addition, the FAD2 gene was also found to be downregulated. This could be possibly due to a stop in the FA 18:1 synthesis from plastids [53,54].
Sphingo-lipids, on the other hand, also play an essential role in preventing plants from heat stress. Therefore, the data were screened for any changes in Sphingo-lipid genes and resulted in the identification of an upregulated of serine palmitoyl transferase 2 (SPT2) (Os01g70370.1) in Y3. This enzyme is involved in de novo synthesis of sphiganine and di-hydro shpinganine to form sphingo-lipids and ceramides. The ceramides further induce at least one heat shock protein-like αβ-crystallin upon heat stress [55]. In accordance with this, an upregulation of “α-crystallin-Hsps IbpA,” a small Hsp (sHsp) related gene (Os02g48140.1) was also observed (Supplementary Table S4, Aiotic-Biotic stress; Figure 6). The SPT2 enzyme is known to play a key role in the male gametophyte development in Arabidopsis [56]. Upregulation of SPT2 might be a heat shock recovery mechanism in order to prevent gametophyte damage.
2.4.3. Transcription Factor Activation
Besides differential expression of major genes involved in the metabolic process, several changes in the TF genes related to abiotic-stress responses were also observed. Specifically, in both Y2 and Y3 the MYB and bZIP TF’s were downregulated while upregulation of AP2/ERF and OsWRKY was observed only in Y3 (Supplementary Table S4, Aiotic-Biotic stress; Figure 6). MYB4 TF-related gene (Os09g36730.1) was downregulated in both Y2 and Y3. Recent studies in Arabidopsis have indicated that upregulation of MYB4 TFs induce the biosynthesis of secondary metabolites (hydroxycinnamate esters) that in turn increase UV-B hypersensitivity in plants [57,58]. Downregulation of these MYB4 TFs in Y2 and Y3 indicate increased tolerance to UV-B radiation. AP2/ERF TFs were upregulated in Y3 alone. The AP2/ERF (APETALA2/ethylene response factor) TF’s are one of the most important groups in plants that help in the stress defense mechanism [59]. These TF’s induce a set of abiotic stress-related genes. In this study, AP2/ERF family proteins like dehydration-responsive element-binding protein (DREB1/CBF) (Os09g35020.1 and Os09g35010.1) and ethylene-responsive element-binding factor (ERF) (Os04g46220) were upregulated in Y3. These genes were known to play a key role in ABA independent stress tolerance, especially related to osmotic stress [60,61]. The other major class of TFs identified in this study is the OsWRKY TFs. The WRKY TF’s are well known for their roles in the regulation of plant growth, development and apoptosis, and responses to biotic and abiotic stresses [62]. There are 74 and 109 WRKY members in each of Arabidopsis and rice genome, respectively [63,64]. In the current analysis, especially in Y3, an upregulation of OsWRKY genes 24, 28, 45, and 49 (Os01g61080.1, Os06g44010.1, Os05g25770.1, Os05g49100.1), and downregulation of OsWRKY24 (Os07g02060.1) was observed. However, no such specific responses were observed in Y2. Among these, the OsWRKY28 and OsWRKY45 were well studied. OsWRKY28 known to be highly responsive to As(V) and regulation of As(V)/Pi uptake or tolerance in rice [65]. In addition, Cai et al. (2014) showed that OsWRKY28 rice mutants resulted in various disorders like downregulation of JA biosynthesis genes in the shoots, irregular spikelet development, altered flower closing, and anther dehiscence and eventually resulting in lower fertility [66]. In the current study, JA gene expression was observed to decrease in Y2 while OsWRKY28 is not differentially regulated. On the other hand, in Y3, upregulation of OsWRKY28 along with JA biosynthesis genes is seen. In addition, JA is also known to regulate OsWRKY45 gene expression [67]. The OsWRKY45 family is also known to participate in ABA and salt stress signaling [68]. Therefore, it is obvious from our study that the expression of OsWRKY28, JA, and OsWRKY45 are linked to each other. This clearly indicates that Y3 might be moving towards heat stress damage recovery by overexpressing these TFs and in turn producing stress-responsive hormones.
2.4.4. Secondary Metabolites
Secondary metabolites are produced by plants through several metabolic pathways to support their survival in the environment. Some of the most commonly studied secondary metabolites are alkaloids, flavonoids, and terpenoids. Most plants regulate synthesis of these compounds for their survival based on the environmental conditions. Among most environmental factors, temperature is known to influence plant secondary metabolite production significantly [69]. In the current study, upregulation of some genes related to ent-kaurene synthase, anthocyanins, flavonoid biosynthesis, lignin biosynthesis, and laccases was observed. Three genes related to ent-kaurene synthase were found to be strongly upregulated in Y3 (Os04909900.1, Os02936140, and Os02936210), while only one gene (Os01914630) was mildly upregulated in Y2. These enzymes are known for their role in gibberellin phytohormone biosynthesis and serve as intermediates in specific di-terpenoid metabolism [70]. Additionally, specific genes belonging to the anthocyanins and flavonoid biosynthesis pathway in Y3 were found to be upregulated. Some of these are chalcone synthase (Os11g32650.1), leucoanthocyanidin reductase (Os06g44170.1) and dihydroflavonol-4-reductase (DFR) (Os03g08624.1). On the contrary, in Y2, these genes were not found. However, the isoflavonone reductase related gene with moderate expression was found to be downregulated in both Y2 (Os01g01660.1) and Y3 (Os02g56460.1). This class of flavonoids was identified to exhibit antioxidative functions in plants like tomato [71]. These are also responsible for fruit and flower color in several plants. In Arabidopsis, the mutants with DFR deficient genes resulted in decreased levels of proanthocyanidin brown tannins on their seed coats [72,73]. Another important metabolite that is very essential for plants is lignin. It provides structural support for the plants to grow and also protect the plant from pathogens. Its accumulation could be considered as a repair mechanism to prevent cell wall damage during heat stress. Genes related (Os04g01470.1, Os04g09680.1, and Os11g42200.1) to lignin biosynthesis were also found to be upregulated in both Y2 and Y3. Os04g01470.1 and Os04g09680.1 are the caffeic acid 3-O-methyltransferase (COMT) genes. These are involved in converting caffeic acid to ferulic acid and 5-hydroxyferulic acid to sinapic acid [74]. Both the genes were observed to be upregulated in Y3, while the former was only observed in Y2. In addition, upregulation of laccases (Os11g42200.1) was observed in both Y2 and Y3. Higher laccase activity indicates induced catalysis of oxidative coupling between phenylpropane units to form lignin [75]. Heat stress in plants could also lead to osmotic stress due to disruption of osmotic hemostasis. In order to prevent osmotic stress plants produce carotenoids [74]. Here in Y2, a gene (Os01g14630.1) geranyl diphosphate synthase (GPPs) related to osmotic stress and carotenoid synthesis was strongly downregulated compared to Y3. Upregulation of GGPS during stress conditions leading to enhanced osmotic stress tolerance was found in Arabidopsis thaliana [76,77]. The current result is quite opposite to previous reports. It could be due to abundance of GPPs in leaves compared to seed and differential expression of specific genes in specific tissues.
2.4.5. Starch Metabolism
Starch metabolism is a key process in regulating rice quality and germination [23,33]. Two key enzymatic processes involved in this pathway are related to starch hydrolysis and sucrose synthesis. Here, in both Y2 and Y3, few amylase genes (Os08g36910.2 and Os09g28400.1) were found to be commonly upregulated (supplementary Table S3, Metabolism overview; and Figure 5). In addition, one more gene related to amylase (Amy1) (Os02g52700.1) was also found to upregulated in Y3 alone. It was previously found that upregulation of Amy1 related genes produced chalky grains by degrading the starch reserves in the ripening grains [24,25,26]. It appears to be more in Y3 as multiple copies of such genes are upregulated. On the other hand, cleavage of sucrose is very important for accumulation of starch in the seeds. This is mediated by sucrose synthase (Susy) genes which converts sucrose to UDP glucose. The effect of heat stress could suppress these genes and was observed here. A gene related to the Susy family (SUS4, Os07g42490.1) was downregulated in both Y2 and Y3. Downregulation of this gene indicates a reduction in starch content and indirectly contributing to grain chalkiness along with amylases.
2.4.6. Hormone Regulated Gene Expression
Hormones play a major role in regulating stress-responsive mechanisms in plants. Several hormones are involved in the activation or inactivation of specific genes related to stress tolerance. In this experiment few hormone-regulated genes were found to be differentially expressed in Y2 and Y3. Two different SAUR class proteins OsSAUR18 (Os04g43740.1) and OsSAUR23 (Os04g56690.1) were observed in both Y2 and Y3, respectively (Supplementary Table S4, Abiotic-Biotic stress; and Figure 6). In general SAUR class genes were induced in the presence of auxins. The reduced expression of OsSAUR genes 18 and 23 in Y2 and Y3 could indicate low levels of endogenous auxins. A decrease in endogenous auxins and SAUR expression was previously observed in barley and Arabidopsis [78,79]. This could be due to the disruption of the auxin metabolism caused by heat stress. Previously, such effect due to heat stress was identified in rice under heat stress which could lead to inhibition of pollen tube elongation resulting in decreased spikelet fertility [80]. Further, ABA-induced GRAM domain-containing proteins like FIP1 were also found to be upregulated, indicating possible accumulation of ABA. ABA is known to regulate starch biosynthesis genes and plays a key role in grain filling under high temperatures [81]. The induced expression of the FIP1 genes could therefore suggest a heat-responsive mechanism in rice. Two F1P1 genes (os10g34730.1 and Os04g44500.1) were found to be similarly regulated in both Y2 and Y3. Additionally, genes os02g42430.1 and Os04g44510.1 were observed to be upregulated in Y3 alone indicating a stronger response to high temperatures. In addition to the above findings, unique changes in genes related to jasmonate synthesis were detected. Expression of genes related to the JA biosynthesis pathway, like lipoxygenase 2 (Os03g52860.1), 12-Oxo-PDA-reductase family genes OPR (Os06g11200.1) and OsOPR5 (Os06g11210.1) is also observed in our study. Between Y2 and Y3 the expression of these genes is opposite. Lipoxygenase-2 and OPR were downregulated in Y2, while in Y3 the lipoxygenase-2 and OsOPR5 are upregulated and OPR is not. Previously JA levels and these genes were identified to be upregulated under drought stress and downregulated under heat stress in rice [82]. Accordingly, from the current results, it can be understood that Y2 is experiencing heat stress. Whereas, Y3 might be experiencing both drought and heat stress, as downregulation of the OPR gene was also observed here.
2.4.7. Concluding Remarks
This is the first such study targeting a field situation where an experiment was designed after observing the heat, and talking to the farmers in Tsukuba city. In summary, the rice being eaten by the people has been found to have certain characteristics of heat stress-like response, and current work clarified the genome-wide changes in genes, under the indicated experimental conditions and the environment from where they were harvested. The present study also provides a unique database for the readers and scientific community to further utilize and research upon.
3. Materials and Methods
3.1. Plant Material
The experiment used japonica-type rice cultivar (O. sativa L. cv. Koshihikari) seeds. Dry mature seeds of cv. Koshihikari were the commercially available rice and obtained from Japan Agriculture (JA Zen-Noh, Japan) branch in Ami-town of Ibaraki prefecture (Kanto region), Japan, in September 2010. Three sets of seeds were obtained as grade 1 (labeled Y1), grade 2 (labeled Y2), and out-of-grade (Grade 3, labeled as Y3), as defined by JA Zen-Noh.
3.2. Seed Quality Analysis
Dry mature seeds (grain) were analyzed for its quality using a commercial rice grain analyzer service (Satake Corporation, Tokyo). The analysis consisted of the following: taste value, amylose, protein, moisture, and fatty acid degree.
3.3. Rice Whole Genome DNA Microarray Analysis
Dry mature seeds (12 of each grade of Y1-Y3) were used for preparing fine powders in liquid nitrogen. Briefly, the seeds were placed in a pre-chilled mortar and pestle containing liquid nitrogen, ground completely to a very fine powder with the chilled pestle in liquid nitrogen and stored at −80 °C till used for RNA extraction. For total RNA extraction, the stored sample powder (~100 mg) was transferred to a 2 mL sterile microfuge tube, followed by addition of 0.9 mL of CTAB buffer [a 10 mL volume of buffer contains 0.5 mL (50 mM) of 1 M stock Tris–HCl solution (pH 8.0), 1.0 mL (5 mM) of 500 mM ethylenediaminetetraacetic acid (EDTA, pH 8.0), 0.2 g (2%, w/v) of CTAB, 1.68 mL (0.84 M) of 5 M NaCl, and 0.1 M β-mercaptoethanol, which is added just before use of the solution]. The contents were mixed by vortexing for 30 s and incubated for 5 min at RT. After an addition of 0.8 mL of phenol-chloroform-isoamylalcohol (PCIA; 25:24:1), the homogenate was mixed well (by gentle shaking) for 5 min at RT. After centrifugation at 15,000 × g for 5 min at 4 °C, an aliquot (0.6 to 0.7 mL) of the upper phase was transferred to a 1.5-mL sterile microfuge tube, followed by addition of 1 volume of chloroform, and the mixture was centrifuged at 15,000 × g for 5 min at 4 °C. The resulting supernatant was transferred to another 1.5 mL microfuge tube and 0.033 volume of 3 M sodium acetate, pH 5.5 and 1 volume of 2-isopropanol were added. The mixture was incubated for 15 min on ice and then centrifuged at 15,000 × g for 5 to 10 min at 4 °C to collect the RNA. The supernatant was completely removed and the pellet was dissolved in 0.1 mL of RNase-free water (SDW; double sterilized distilled water) followed by using the RNeasy mini protocol for RNA cleanup exactly as described by the manufacturer (QIAGEN, Gaithersburg, MD, USA). To verify the quality of this RNA, the yield and purity were determined spectrophotometrically (NanoDrop, Wilmington, DE, USA) and visually confirmed using formaldehyde-agarose gel electrophoresis.
After extraction of high-quality total RNA from dehusked seeds using a modified CTAB extraction protocol, a rice 4 × 44K custom (eARRAY, AMAdid-017845) oligo DNA microarray chip (G2514F: Agilent Technologies, Palo Alto, CA, USA) was used for genome-wide gene profiling as described previously [83]. The flip labeling (dye-swap or reverse labeling with Cy3 and Cy5 dyes) procedure was used to nullify the dye bias associated with unequal incorporation of the two Cy dyes into cRNA [84,85,86]. The dye-swap approach which is well established in our laboratories and research provides a more stringent selection condition for profiling differentially expressed genes rather than simply doing only 2 or 3 replicates, which overlook the dye bias [86,87,88,89,90,91,92]. The experimental design for the DNA microarray analysis of rice seed transcriptome using a two-color dye-swap approach (Figure 7A) is shown along with the extracted seed total RNA quality (Figure 7B).
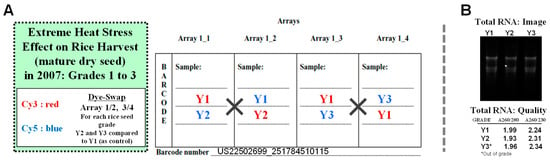
Figure 7.
Experimental design for the DNA microarray analysis of high temperature affected mature dry rice (cv. Koshihikari) seed grades 2 and 3 transcriptome. A two-color dye-swap approach was used (A) and followed by checking the quality of extracted seed total RNA (B). Three sets of dry mature rice seeds (commercial) were obtained Japan Agriculture (JA Zen-Noh) branch in Ami-town of Ibaraki prefecture in September 2010, as grade 1 (labeled as Y1), grade 2 (labeled as Y2), and grade 3 (out-of-grade, labeled as Y3). Gene expressions genome-wide in grade 2 (Y2) and grade 3 (Y3) seeds over the grade 1 (Y1) was carried out using an Agilent 4 × 44K rice oligo DNA chip.
Total RNA (800 ng) for each Y1, Y2, and Y3 sample were labeled with either Cy3 or Cy5 dye using an Agilent Low RNA Input Fluorescent Linear Amplification Kit. Hybridization and wash processes were performed according to the manufacturer’s instructions. Hybridized microarray chips were scanned using the Agilent Microarray Scanner G2565BA. To detect differentially expressed significant genes between control and treated samples, each slide image was processed by Agilent Feature Extraction software (version 9.5.3.1). Normalization of Cy3 and Cy5 signals was performed by LOWESS (locally weighted linear regression), which calculates the log ratio of dye-normalized Cy3- and Cy5-signals. The significance (P) value is based on the propagate error and universal error models. In this analysis, the threshold of significantly expressed differential genes was set to <0.01 (for the confidence that the feature was not differentially expressed). Lists of differentially expressed gene [up- (≥2.0 fold) and down- (≤0.5 fold) regulated genes] were generated and annotated using the Agilent GeneSpring version GX 10.
The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (GEO) and are accessible through GEO series accession number GSE79405.
3.4. MapMan Analysis
To analyze the gene expression changes, rice genes were mapped onto metabolic pathways using MapMan tool (version 3.5.1, Max Plant Institute of Molecular Plant Physiology, Germany) [93]. To map genes onto their respective pathways MapMan uses information from its datasets (Mappings), where all annotated genes of the respective organism were classified into BINS based on their function [94]. In this work, a rice mapping file (Rice_japonica_automatic_mapping08) from MapMan database was used as a template. Out of 43,494 genes detected from our microarray data, 41,446 genes were present in the mapping file and the remaining 2048 (having ID_ TIGRv4S1) were not annotated. The annotated gene expression data was used here and pre-processed it using a PERL script to meet the locus_name identifier criteria similar to that available in the mapping file. Once this is done the expression data file with modified gene ID’s were successfully classified into various BINS (Table S1-Mapman Bins) by MapMan. For the final mapping, the upregulated and downregulated genes were selected, and their fold change transformed into Log2 (fold) data was used. A total of 41,446 differentially expressed genes were sorted to select up (≥2.0 fold) and down (≤0.5 fold) regulated genes were generated and annotated using the Agilent GeneSpring version GX 10. This resulted in 161 and 490 HRDE-genes for each Y1-Y2 and Y1-Y3, respectively. This sorted gene expression data was used to map onto Mapman pathways. The genes which are differentially expressed are indicated either as highly up- or downregulated are represented in blue and red-colored data points (grid boxes), respectively. A gradient in blue or red indicate genes with medium up or downregulation.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4433/11/5/528/s1, Table S1: Rice Japonica genes associated with Mapman BINS; Table S2: Highly expressed differentially regulated (HRDE) Genes mapped on Cell Function map; Table S3: Highly expressed differentially regulated (HRDE) Genes mapped on Metabolism Overview map; Table S4: Highly expressed differentially regulated (HRDE) Genes mapped on Abiotic and Bioic stress map.
Author Contributions
R.K.B. wrote the paper, and R.R. edited the paper along with A.S. and G.K.A., and R.R., A.S. and G.K.A. discussed the initial idea for the experiment towards an experimental approach; R.R. designed the experiment and performed the experiment with J.S.; K.C. supported with the MapMan analyses along with R.K.B., and S.K. provided the DNA microarray chip probe design; M.Y. provided the seeds and supported the rice seed quality analysis; S.S. provided the facilities and support for the DNA microarray analysis. All authors approved the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Available online: https://earthobservatory.nasa.gov/world-of-change/global-temperatures (accessed on 1 April 2020).
- Lobell, D.B.; Gourdji, S.M. The influence of climate change on global crop productivity. Plant Physiol. 2012, 160, 1686–1697. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Liu, B.; Piao, S.; Wang, X.; Lobell, D.B.; Huang, Y.; Huang, M.; Yao, Y.; Bassu, S.; Ciais, P.; et al. Temperature increase reduces global yields of major crops in four independent estimates. Proc. Natl. Acad. Sci. USA 2017, 114, 9326–9331. [Google Scholar] [CrossRef] [PubMed]
- Bita, C.E.; Tom Gerats, T. Plant tolerance to high temperature in a changing environment: Scientific fundamentals and production of heat stress-tolerant crops. Front. Plant Sci. 2013, 4, 273. [Google Scholar] [CrossRef]
- Qu, A.L.; Ding, Y.F.; Jiang, Q.; Zhu, C. Molecular mechanisms of the plant heat stress response. Biochem. Biophys. Res. Commun. 2013, 432, 203–207. [Google Scholar] [CrossRef] [PubMed]
- Hatfield, J.L.; Prueger, J.H. Agroecology: Implications for Plant Response to Climate Change. In Crop Adaptation to Climate Change; Yadav, S.S., Redden, R.J., Hatfield, J.L., Lotze-Campen, H., Hall, A.E., Eds.; Wiley-Blackwell: West Sussex, UK, 2011; pp. 27–43. [Google Scholar]
- Ohama, N.; Sato, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Transcriptional regulatory network of plant heat stress response. Trends Plant Sci. 2017, 22, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Wahid, A.; Gelani, S.; Ashraf, M.; Foolad, M. Heat tolerance in plants: An overview. Environ. Exp. Bot. 2007, 61, 199–223. [Google Scholar] [CrossRef]
- Hua, J. From freezing to scorching, transcriptional responses to temperature variations in plants. Curr. Opin. Plant Biol. 2009, 12, 568–573. [Google Scholar] [CrossRef]
- Kim, E.H.; Kim, Y.S.; Park, S.H.; Koo, Y.J.; Do Choi, Y.; Chung, Y.Y.; Lee, I.J.; Kim, J.K. Methyl jasmonate reduces grain yield by mediating stress signals to alter spikelet development in rice. Plant Physiol. 2009, 149, 1751–1760. [Google Scholar] [CrossRef]
- Wang, D.; Heckathorn, S.A.; Kumar Mainali, K.; Tripathee, R. Timing effects of heat-stress on plant ecophysiological characteristics and growth. Front. Plant Sci. 2016, 7, 1629. [Google Scholar] [CrossRef]
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef]
- Andreeva, V.A. Involvement of Peroxidase in the Protective Plant Mechanism. In Biochemical, Molecular and Physiological Aspects of Plant Peroxidases; Lobarzewski, J., Greppin, H., Penel, C., Gaspar, T., Eds.; University of Geneva: Geneva, Switzerland, 1991; pp. 433–442. [Google Scholar]
- Hiramoto, K.; Ojima, N.; Sako, K.; Kikugawa, K. Effect of plant phenolics on the formation of the spin-adduct of hydroxyl radical and the DNA strand breaking by hydroxyl radical. Biol. Pharm. Bull. 1996, 19, 558–563. [Google Scholar] [CrossRef] [PubMed]
- Moller, I.M.; Jensen, P.E.; Hansson, A. Oxidative modifications to cellular components in plants. Ann. Rev. Plant Biol. 2007, 58, 459–481. [Google Scholar] [CrossRef] [PubMed]
- Camejo, D.; Jiménez, A.; Alarcón, J.J.; Torres, W.; Gómez, J.M.; Sevilla, F. Changes in photosynthetic parameters and antioxidant activities following heat–shock treatment in tomato plants. Func. Plant Biol. 2006, 33, 177–187. [Google Scholar] [CrossRef]
- Yoshida, S. Effects of temperature on growth of the rice plant (Oryza sativa L.) in a controlled environment. Soil Sci Plant Nutr. 1973, 19, 299–310. [Google Scholar] [CrossRef]
- Jagadish, S.V.; Muthurajan, R.; Oane, R.; Wheeler, T.R.; Heuer, S.; Bennett, J.; Craufurd, P.Q. Physiological and proteomic approaches to address heat tolerance during anthesis in rice (Oryza sativa L.). J. Exp. Bot. 2010, 61, 143–156. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, T.; Ishimaru, T.; Kondo, M.; Kuwagata, T.; Yoshimoto, M.; Fukuoka, M. Spikelet sterility of rice observed in the record hot summer of 2007 and the factors associated with its variation. J. Agric. Meteorol. 2011, 67, 225–232. [Google Scholar] [CrossRef]
- Shrivastava, P.; Saxena, R.R.; Xalxo, M.S.; Verulkar, S.B. Effect of high temperature at different growth stages on rice yield and grain quality traits. J. Rice Res. 2012, 5, 29–42. [Google Scholar]
- Fu, G.; Feng, B.; Zhang, C.; Yang, Y.; Yang, X.; Chen, T.; Zhao, X.; Jin, Q.; Tao, L. Heat stress is more damaging to superior spikelets than inferiors of rice (Oryza sativa L.) due to their different organ temperatures. Front. Plant Sci. 2016, 7, 163. [Google Scholar] [CrossRef]
- Satake, T.; Yoshida, S. High temperature-induced sterility in indica rices at flowering. Jpn. J. Crop Sci. 1978, 47, 6–17. [Google Scholar] [CrossRef]
- Sharma, K.P.; Sharma, N. Influence of high temperature on sucrose metabolism in chalky and translucent rice genotypes. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 2018, 88, 1275–1284. [Google Scholar] [CrossRef]
- Tashiro, T.; Wardlaw, I. The effect of high-temperature on kernel dimensions and the type and occurrence of kernel damage in rice. Aust. J. Agric. Res. 1991, 42, 485–496. [Google Scholar] [CrossRef]
- Zakaria, S.; Matsuda, T.; Tajima, S.; Nitta, Y. Effect of high temperature at ripening stage on the reserve accumulation in seed in some rice cultivars. Plant Prod Sci. 2002, 5, 160–168. [Google Scholar]
- Nakata, M.; Fukamatsu, Y.; Miyashita, T.; Hakata, M.; Kimura, R.; Nakata, Y.; Kuroda, M.; Yamaguchi, T.; Yamakawa, H. High temperature-induced expression of rice α-amylases in developing endosperm produces chalky grains. Front. Plant Sci. 2017, 8, 2089. [Google Scholar] [PubMed]
- Morita, S.; Wada, H.; Matsue, Y. Countermeasures for heat damage in rice grain quality under climate change. Plant Prod. Sci. 2016, 19, 1–11. [Google Scholar] [CrossRef]
- Hakata, M.; Kuroda, M.; Miyashita, T.; Yamaguchi, T.; Kojima, M.; Sakakibara, H.; Mitsui, T.; Yamakawa, H. Suppression of α-amylase genes improves quality of rice grain ripened under high temperature. Plant Biotechnol. J. 2012, 10, 1110–1117. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Eixarch, M.; Ellis, R.H. Temporal sensitivity of rice seed development from spikelet fertility to viable mature seed to extreme temperature. Crop Sci. 2015, 55, 354–364. [Google Scholar]
- Yuliawan, T.; Handoko, I. The effect of temperature rise to rice crop yield in Indonesia uses Shierary Rice Model with Geographical Information System (GIS) feature. Procedia Environ. Sci. 2016, 33, 214–220. [Google Scholar] [CrossRef]
- Available online: https://yab.yomiuri.co.jp/adv/wol/dy/opinion/society_131209.html (accessed on 1 April 2020).
- Ohtsubo, K.; Kobayashi, A.; Shimizu, H. Quality evaluation of rice in Japan. Jpn. Agric. Res. Q. 1993, 27, 95–101. [Google Scholar]
- Yamakawa, H.; Hirose, T.; Kuroda, M.; Yamaguchi, T. Comprehensive expression profiling of rice grain filling-related genes under high temperature using DNA microarray. Plant Physiol. 2007, 144, 258–277. [Google Scholar] [CrossRef]
- Yamakawa, H.; Hakata, M. Atlas of rice grain filling-related metabolism under high temperature: Joint analysis of metabolome and transcriptome demonstrated inhibition of starch accumulation and induction of amino acid accumulation. Plant Cell Physiol. 2010, 51, 795–809. [Google Scholar]
- Yasumatsu, K.; Moritaka, S. Fatty acid compositions of rice lipid and their changes during storage. Agric. Biol. Chem. 1964, 28, 257–264. [Google Scholar] [CrossRef]
- Yamamatsu, K.; Moritaka, S.; Wada, S. Stale flavor of stored rice. Agric. Biol. Chem. 1966, 30, 483–486. [Google Scholar]
- Wu, H.C.; Bulgakov, V.P.; Jinn, T.L. Pectin Methyl esterase’s: Cell wall remodeling proteins are required for plant response to heat stress. Front. Plant Sci. 2018, 9, 1612. [Google Scholar] [CrossRef] [PubMed]
- Rienth, M.; Torregrosa, L.; Luchaire, N.; Chatbanyong, R.; Lecourieux, D.; Kelly, M.T.; Romieu, C. Day and night heat stress trigger different transcriptomic responses in green and ripening grapevine (vitis vinifera) fruit. BMC Plant Biol. 2014, 14, 108. [Google Scholar] [CrossRef] [PubMed]
- Tenhaken, R. Cell wall remodeling under abiotic stress. Front. Plant Sci. 2015, 5, 771. [Google Scholar] [CrossRef] [PubMed]
- Gall, L.H.; Philippe, F.; Domon, J.M.; Gillet, F.; Pelloux, J.; Rayon, C. Cell wall metabolism in response to abiotic Stress. Plants 2015, 4, 112–166. [Google Scholar] [CrossRef]
- Huang, Y.C.; Wu, H.C.; Wang, Y.D.; Liu, C.H.; Lin, C.C.; Luo, D.L.; Jinn, T.L. Pectin methyl esterase 34 contributes to Hhat tolerance through its role in promoting stomatal movement. Plant Physiol. 2017, 174, 748–763. [Google Scholar] [CrossRef]
- Nishizawa, A.; Yabuta, Y.; Shigeoka, S. Galactinol and raffinose constitute a novel function to protect plants from oxidative damage. Plant Physiol. 2008, 147, 1251–1263. [Google Scholar] [CrossRef]
- Egert, A.; Keller, F.; Peters, S. Abiotic stress-induced accumulation of raffinose in Arabidopsis leaves is mediated by a single raffinose synthase (RS5, At5g40390). BMC Plant Biol. 2013, 13, 218. [Google Scholar] [CrossRef]
- Sengupta, S.; Mukherjee, S.; Basak, P.; Majumder, A.L. Significance of galactinol and raffinose family oligosaccharide synthesis in plants. Front. Plant Sci. 2015, 6, 656. [Google Scholar] [CrossRef]
- Padham, A.K.; Hopkins, M.T.; Wang, T.W.; McNamara, L.M.; Lo, M.; Richardson, L.G.; Smith, M.D.; Taylor, C.A.; Thompson, J.E. Characterization of a plastid triacylglycerol lipase from Arabidopsis. Plant Physiol. 2007, 143, 1372–1384. [Google Scholar] [CrossRef] [PubMed]
- Higashi, Y.; Okazaki, Y.; Takano, K.; Myouga, F.; Shinozaki, K.; Knoch, E.; Fukushima, A.; Saito, K. Heat inducible lipase remodels chloroplastic monogalactosyl diacylglycerol by Liberating α-Linolenic Acid in Arabidopsis Leaves under Heat Stress. Plant Cell 2018, 30, 1887–1905. [Google Scholar] [CrossRef]
- Hou, Q.; Ufer, G.; Bartels, D. Lipid signaling in plant responses to abiotic stress. Plant Cell Environ. 2016, 39, 1029–1048. [Google Scholar] [CrossRef] [PubMed]
- Mishkind, M.; Vermeer, J.E.; Darwish, E.; Munnik, T. Heat stress activates phospholipase D and triggers PIP accumulation at the plasma membrane and nucleus. Plant J. 2009, 60, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Xu, W.; Burke, J.J.; Xin, Z. Role of phosphatidic acid in high temperature tolerance in Maize. Crop Sci. 2010, 50, 2506–2515. [Google Scholar] [CrossRef]
- Escobar-Sepúlveda, H.F.; Trejo-Téllez, L.I.; Pérez-Rodríguez, P.; Hidalgo-Contreras, J.V.; Gómez-Merino, F.C. Diacylglycerol kinases are widespread in higher plants and display inducible gene expression in response to beneficial elements, metal, and metalloid ions. Front. Plant Sci. 2017, 8, 129. [Google Scholar] [CrossRef]
- Li, Q.; Zheng, Q.; Shen, W.; Cram, D.; Fowler, D.B.; Wei, Y.; Zou, J. Understanding the biochemical basis of temperature-induced lipid pathway adjustments in plants. Plant Cell 2015, 227, 86–103. [Google Scholar] [CrossRef]
- Dar, A.A.; Choudhury, A.R.; Kancharla, P.K.; Arumugam, N. The FAD2 Gene in Plants: Occurrence, regulation, and role. Front. Plant Sci. 2017, 8, 1789. [Google Scholar] [CrossRef]
- Byfield, G.E.; Upchurch, R.G. Effect of temperature on delta-9 stearoyl-ACP and microsomal omega-6 desaturase gene expression and fatty acid content in developing soybean seeds. Crop Sci. 2007, 47, 1698. [Google Scholar] [CrossRef]
- Altunoglu, Y.C.; Unel, N.M.; Baloglu, M.C.; Ulu, F.; Can, T.H.; Cetinkaya, R. Comparative identification and evolutionary relationship of fatty acid desaturase (FAD) genes in some oil crops: The sunflower model for evaluation of gene expression pattern under drought stress. Biotechnol. Biotechnol. Equip. 2018, 32, 846–857. [Google Scholar] [CrossRef]
- Jenkins, G.M.; Richards, A.; Wahl, T.; Mao, C.; Obeid, L.; Hannun, Y. Involvement of yeast sphingolipids in the heat stress response of Saccharomyces cerevisiae. J. Biol. Chem. 1997, 272, 32566–32572. [Google Scholar] [CrossRef] [PubMed]
- Teng, C.; Dong, H.; Shi, L.; Deng, Y.; Mu, J.; Zhang, J.; Yang, X.; Zuo, J. Serine palmitoyltransferase, a key enzyme for de novo synthesis of sphingolipids, is essential for male gametophyte development in Arabidopsis. Plant Physiol. 2008, 146, 1322–1332. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Cominelli, E.; Bailey, P.; Parr, A.; Mehrtens, F.; Jones, J.; Tonelli, C.; Weisshaar, B.; Martin, C. Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 2000, 19, 6150–6161. [Google Scholar] [CrossRef]
- Roy, S. Function of MYB domain transcription factors in abiotic stress and epigenetic control of stress response in plant genome. Plant Signal. Behav. 2015, 11, e1117723. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Lv, H.; Li, L.; Liu, J.; Mu, S.; Li, X.; Gao, J. Genome-wide analysis of the AP2/ERF transcription factors family and the expression patterns of DREB genes in Moso Bamboo (Phyllostachys edulis). PLoS ONE 2015, 10, e0126657. [Google Scholar] [CrossRef] [PubMed]
- Lata, C.; Prasad, M. Role of DREBs in regulation of abiotic stress responses in plants. J. Exp. Bot. 2011, 63, 4731–4748. [Google Scholar] [CrossRef] [PubMed]
- Sharoni, A.M.; Nuruzzaman, M.; Rahman, M.A.; Karim, R.; Islam, A.K.M.R.; Hossain, M.M.; Rahman, M.M.; Parvez, M.S.; Haydar, F.M.A.; Nasiruddin, M.; et al. AP2/EREBP transcription factor family genes are differentially expressed in rice seedlings during infections with different viruses. Int. J. Biosci. 2017, 10, 1–14. [Google Scholar]
- Wang, P.; Xu, X.; Tang, Z.; Zhang, W.; Huang, X.Y.; Zhao, F.J. OsWRKY28 regulates phosphate and arsenate accumulation, root system architecture and fertility in rice. Front. Plant Sci. 2018, 9, 1330. [Google Scholar] [CrossRef]
- Ross, C.A.; Liu, Y.; Shen, Q.J. The WRKY Gene Family in Rice (Oryza sativa). J. Integr. Plant Biol. 2007, 49, 827–842. [Google Scholar] [CrossRef]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef]
- Chakrabarty, D.; Trivedi, P.K.; Misra, P.; Tiwari, M.; Shri, M.; Shukla, D.; Kumar, S.; Rai, A.; Pandey, A.; Nigam, D.; et al. Comparative transcriptome analysis of arsenate and arsenite stresses in rice seedlings. Chemosphere 2009, 74, 688–702. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Yuan, Z.; Chen, M.; Yin, C.; Luo, Z.; Zhao, X.; Liang, W.; Hu, J.; Zhang, D. Jasmonic acid regulates spikelet development in rice. Nat. Commun. 2014, 5, 3476. [Google Scholar] [CrossRef] [PubMed]
- Huangfu, J.; Li, J.; Li, R.; Ye, M.; Kuai, P.; Zhang, T.; Lou, Y. The Transcription Factor OsWRKY45 Negatively Modulates the Resistance of Rice to the Brown Planthopper Nilaparvata lugens. Int. J. Mol. Sci. 2016, 17, 697. [Google Scholar] [CrossRef] [PubMed]
- Tao, Z.; Kou, Y.; Liu, H.; Li, X.; Xiao, J.; Wang, S. OsWRKY45 alleles play different roles in abscisic acid signaling and salt stress tolerance but similar roles in drought and cold tolerance in rice. J. Exp. Bot. 2011, 62, 4863–4874. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishna, A.; Ravishankar, G.A. Influence of abiotic stress signals on secondary metabolites in plants. Plant Signal. Behav. 2011, 6, 1720–1731. [Google Scholar] [PubMed]
- Fu, J.; Ren, F.; Lu, X.; Mao, H.; Xu, M.; Degenhardt, J.; Peters, R.J.; Wang, Q. A tandem array of ent-kaurene synthases in maize with roles in gibberellin and more specialized metabolism. Plant Physiol. 2015, 170, 742–751. [Google Scholar] [CrossRef] [PubMed]
- Butelli, E.; Titta, L.; Giorgio, M.; Mock, H.P.; Matros, A.; Peterek, S.; Schiklen, E.G.; Hall, R.D.; Bovy Ag Luo, J.; Martin, C. Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat. Biotechnol. 2008, 26, 1301–1308. [Google Scholar] [CrossRef]
- Shirley, B.W.; Kubasek, W.L.; Storz, G.; Bruggemann, E.; Koornneef, M.; Asubel, F.M.; Goodman, H.M. Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J. 1995, 8, 659–671. [Google Scholar] [CrossRef]
- Wang, H.; Fan, W.; Li, H.; Yang, J.; Huang, J.; Zhang, P. Functional characterization of dihydroflavonol-4-reductase in Anthocyanin Biosynthesis of Purple Sweet Potato Underlies the Direct Evidence of Anthocyanins Function against Abiotic Stresses. PLoS ONE 2013, 8, e78484. [Google Scholar] [CrossRef]
- Davin, L.B.; Lewis, N.G. Phenylpropanoid metabolism: Biosynthesis of monolignols, lignans and neolignans, lignins and suberins. Rec. Adv. Phytochem. 1992, 26, 325–375. [Google Scholar]
- Boudet, A.M.; Lapierre, C.; Grima-Pettenati, J. Biochemistry and molecular biology of lignification. New Phytol. 1995, 129, 203–236. [Google Scholar] [CrossRef]
- Xie, Y.; Xu, D.; Cui, W.; Shen, W. Mutation of Arabidopsis Hy1 causes Uv-C hypersensitivity by impairing carotenoid and flavonoid biosynthesis and the down-regulation of antioxidant defense. J. Exp. Bot. 2012, 63, 3869–3883. [Google Scholar] [CrossRef]
- Chen, W.; He, S.; Liu, D.; Patil, G.B.; Zhai, H.; Wang, F.; Stephenson, T.J.; Wang, Y.; Wang, B.; Valliyodan, B.; et al. A sweetpotato geranylgeranyl pyrophosphate synthase gene, IbGGPS, increases carotenoid content and enhances osmotic stress tolerance in Arabidopsis thaliana. PLoS ONE 2015, 10, e0137623. [Google Scholar] [CrossRef] [PubMed]
- Sakata, T.; Oshino, T.; Miura, S.; Tomabechi, M.; Tsunaga, Y.; Higashitani, N.; Miyazawa, Y.; Takahashi, H.; Watanabe, M.; Higashitani, A. Auxins reverse plant male sterility caused by high temperatures. Proc. Natl. Acad. Sci. USA 2010, 107, 8569–8574. [Google Scholar] [CrossRef] [PubMed]
- Higashitani, A. High temperature injury and auxin biosynthesis in microsporogenesis. Front. Plant Sci. 2013, 4, 47. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Li, G.; Chen, T.; Feng, B.; Fu, W.; Yan, J.; Islam, M.R.; Jin, Q.; Tao, L.; Fu, G. Heat stress induces spikelet sterility in rice at anthesis through inhibition of pollen tube elongation interfering with auxin homeostasis in pollinated pistils. Rice (N. Y.) 2018, 11, 14. [Google Scholar] [CrossRef]
- Mauri, N.; Fernández-Marcos, M.; Costas, C.; Desvoyes, B.; Pichel, A.; Caro, E.; Gutierrez, C. GEM, a member of the GRAM domain family of proteins, is part of the ABA signaling pathway. Sci. Rep. 2016, 6, 22660. [Google Scholar] [CrossRef]
- Umesh, D.K.; Pal, M. Differential role of jasmonic acid under drought and heat stress in rice (Oryza sativa). J. Pharmacogn. Phytochem. 2018, 7, 2626–2631. [Google Scholar]
- Cho, K.; Shibato, J.; Agrawal, G.K.; Jung, Y.H.; Kubo, A.; Jwa, N.S.; Tamogami, S.; Satoh, K.; Kikuchi, S.; Higashi, T.; et al. Integrated transcriptomics, proteomics, and metabolomics analyses to survey ozone responses in the leaves of rice seedling. J Proteome Res. 2008, 7, 2980–2998. [Google Scholar] [CrossRef]
- Cho, K.; Shibato, J.; Kubo, A.; Kohno, Y.; Satoh, K.; Kikuchi, S.; Sarkar, A.; Agrawal, G.K.; Rakwal, R. Comparative analysis of seed transcriptomes of ambient ozone-fumigated 2 different rice cultivars. Plant Signal. Behav. 2013, 8, e26300. [Google Scholar] [CrossRef]
- Altman, N. Replication, variation and normalization in microarray experiments. Appl. Bioinform. 2005, 4, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Martin-Magniette, M.L.; Aubert, J.; Cabannes, E.; Daudin, J.J. Evaluation of the gene-specific dye bias in cDNA microarray experiments. Bioinformatics 2005, 21, 1995–2000. [Google Scholar] [CrossRef] [PubMed]
- Rosenzweig, B.A.; Pine, P.S.; Domon, O.E.; Morris, S.M.; Chen, J.J.; Sistare, F.D. Dye bias correction in dual-labeled cDNA microarray gene expression measurements. Environ. Health Perspect. 2004, 112, 480–487. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.; Kubo, A.; Shibato, J.; Agrawal, G.K.; Saji, H.; Rakwal, R. Global identification of potential gene biomarkers associated with ozone-induced foliar injury in rice seedling leaves by correlating their symptom severity with transcriptome profiling. Int. J. Life Sci. 2012, 6, 1–13. [Google Scholar] [CrossRef]
- Hirano, M.; Rakwal, R.; Shibato, J.; Sawa, H.; Nagashima, K.; Ogawa, Y.; Yoshida, Y.; Iwahashi, H.; Niki, E.; Masuo, Y. Proteomics and transcriptomics-based screening of differentially expressed proteins and genes in brain of Wig rat: A model for attention deficit hyperactivity disorder (ADHD) research. J. Proteome Res. 2008, 7, 2471–2489. [Google Scholar] [CrossRef] [PubMed]
- Tano, K.; Mizuno, R.; Okada, T.; Rakwal, R.; Shibato, J.; Masuo, Y.; Ijiri, K.; Akimitsu, N. MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Lett. 2010, 584, 4575–4580. [Google Scholar] [CrossRef]
- Ogawa, T.; Rakwal, R.; Shibato, J.; Sawa, C.; Saito, T.; Murayama, A.; Kuwagata, M.; Kageyama, H.; Yagi, M.; Satoh, K.; et al. Seeking gene candidates responsible for developmental origins of health and disease (DOHaD). Congenit. Anom. 2011, 51, 110–125. [Google Scholar] [CrossRef]
- Hori, M.; Nakamachi, T.; Rakwal, R.; Shibato, J.; Nakamura, K.; Wada, Y.; Tsuchikawa, D.; Yoshikawa, A.; Tamaki, K.; Shioda, S. Unraveling the ischemic brain transcriptome in a permanent middle cerebral artery occlusion model by DNA microarray analysis. Dis. Models Mech. 2011, 5, 270–283. [Google Scholar] [CrossRef]
- Usadel, B.; Poree, F.; Nagel, A.; Lohse, M.; Czedik-Eysenberg, A.; Stitt, M. A guide to using MapMan to visualize and compare Omics data in plants: A case study in the crop species, Maize. Plant Cell Environ. 2009, 32, 1211–1229. [Google Scholar] [CrossRef]
- Thimm, O.; Blaesing, O.; Gibon, Y.; Nagel, A.; Meyer, S.; Krüger, P.; Selbig, J.; Müller, L.A.; Rhee, S.Y.; Stitt, M. MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004, 37, 914–939. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).