Abstract
Syntrophic acetate oxidation operates close to the thermodynamic equilibrium and very little is known about the participating organisms and their metabolism. Clostridium ultunense is one of the most abundant syntrophic acetate-oxidising bacteria (SAOB) that are found in engineered biogas processes operating with high ammonia concentrations. It has been proven to oxidise acetate in cooperation with hydrogenotrophic methanogens. There is evidence that the Wood-Ljungdahl (WL) pathway plays an important role in acetate oxidation. In this study, we analysed the physiological and metabolic capacities of C. ultunense strain Esp and strain BST on genome scale and conducted a comparative study of all the known characterised SAOB, namely Syntrophaceticus schinkii, Thermacetogenium phaeum, Tepidanaerobacter acetatoxydans, and Pseudothermotoga lettingae. The results clearly indicated physiological robustness to be beneficial for anaerobic digestion environments and revealed unexpected metabolic diversity with respect to acetate oxidation and energy conservation systems. Unlike S. schinkii and Th. phaeum, C. ultunense clearly does not employ the oxidative WL pathway for acetate oxidation, as its genome (and that of P. lettingae) lack important key genes. In both of those species, a proton motive force is likely formed by chemical protons involving putative electron-bifurcating [Fe-Fe] hydrogenases rather than proton pumps. No genes encoding a respiratory Ech (energy-converting hydrogenase), as involved in energy conservation in Th. phaeum and S. schinkii, were identified in C. ultunense and P. lettingae. Moreover, two respiratory complexes sharing similarities to the proton-translocating ferredoxin:NAD+ oxidoreductase (Rnf) and the Na+ pumping NADH:quinone hydrogenase (NQR) were predicted. These might form a respiratory chain that is involved in the reduction of electron acceptors rather than protons. However, involvement of these complexes in acetate oxidation in C. ultunense and P. lettingae needs further study. This genome-based comparison provides a solid platform for future meta-proteomics and meta-transcriptomics studies and for metabolic engineering, control, and monitoring of SAOB.
1. Introduction
Syntrophic acetate oxidation is essential for the biomethanisation of organic matter in engineered anaerobic digestion (AD) processes, in particular, those that are characterised by high ammonia levels [1,2,3]. Engineered biogas processes are being increasingly used in Europe and worldwide due to its potential to meet environmental and climate-related targets and to secure future energy supplies by reducing the exploitation of finite fossil resources and recycling nutrients and energy. When using organic wastes produced by agriculture, municipalities, and food industry as substrates, the biogas process enables a sustainable and economical recycling of nutrients and energy between urban and rural areas. This multi-functionality of the process reduces the competition for land between food and energy production. However, use of high-energy biomass materials, such as industrial food waste, is still challenging, as the degradation of proteinaceous materials releases high levels of ammonia, which has a direct impact on the prevailing methane production pathway, with serious consequences for process stability and efficiency [1,2,3].
Effective processing of organic matter requires the cooperative activity of microbial food chains. Fermentative microorganisms partially degrade polysaccharides, proteins, sugars, and amino acids to carbon dioxide, hydrogen, and short-chain fatty acids, of which acetate is by far the most abundant. Acetate is cleaved to methane and carbon dioxide by the activity of aceticlastic methanogens, a mechanism that is well studied and occurring in anoxic environments, such as aquatic sediments and terrestrial subsurfaces [4]. However, AD processes operating at high ammonia levels rely on so-called syntrophic acetate-oxidising bacteria (SAOB) [5,6,7,8,9,10,11], since aceticlastic methanogens become inhibited under high ammonia conditions [1,2]. The acetate-converting activity of SAOB strongly depends on the abundant presence of hydrogenotrophic methanogens, which enable the thermodynamically unfavourable acetate oxidation (reaction 1) by immediately consuming the hydrogen or formate produced (reaction 2) [12,13,14]. The very small overall free energy change of ΔG0′ −35 kJ mol−1 (reaction 3) needs to be shared by two organisms that are doing the work:
CH3COO− +H+ +2H2O → 2CO2 + 4H2 ΔG0′ = +95 kJ per mol rct.
4H2 + CO2 → CH4 + 2H2O ΔG0′ = −131 kJ per mol rct.
CH3COO− + H+ + → CH4 + CO2 ΔG0′ = −35 kJ per mol rct.
Other anaerobic environments, such as rice paddy fields, subtropical lake sediments, oil reservoirs, and nutrient-enriched soils, have also been shown to express syntrophic acetate oxidation (SAO) activity [15,16,17,18,19,20,21]. However, no SAOB have yet been isolated from these environments.
To date, only five SAOB species have been characterised [22,23,24,25,26]. Three of these, Tepidanaerobacter acetatoxydans, Clostridium ultunense, and Syntrophaceticus schinkii, were recovered from mesophilic biogas processes, whereas Thermacetogenium phaeum and Pseudothermotoga lettingae were isolated from thermophilic biogas facilities. Pure culture and enzymatic studies affiliated all but P. lettingae to the physiological group of acetogens, producing acetate as the main end product by employing the reductive Wood-Ljungdahl (WL) pathway when growing heterotrophically [3,13,23,25,26,27]. Whole cell extract-based enzyme activities and messenger RNA (mRNA) expression studies indicated that the SAOB use a reversal of the WL pathway for oxidising acetate to hydrogen and carbon dioxide [13,27,28,29]. The thermophilic P. lettingae does not express acetogenic metabolism [22], and it must therefore use an alternative pathway. T. acetatoxydans might also use another metabolic route for acetate oxidation, as genome-scale analysis has revealed a lack of F0/F1-ATP (Adenosine triphosphate) synthase, which makes use of the oxidative WL pathway for acetate oxidation energetically unfeasible [30]. In contrast, transcriptomic studies of a methane-producing syntrophic co-culture have demonstrated the expression of the WL pathway genes in S. schinkii [28]. Further, genome-based analysis, which has been thoroughly performed for T. acetatoxydans, Th. phaeum, and S. schinkii, indicates different strategies for energy conservation [29,31,32].
Here, we describe genome-based analysis of the mesophilic SAOB C. ultunense strain Esp and strain BST, which have recently been sequenced [33,34]. We also compare and summarise selected metabolic features and energy-conserving systems of all known SAOB (T. acetatoxydans, S. schinkii, Th. phaeum, P. lettingae, and C. ultunense). Genome-guided analyses reported for Th. phaeum [32], T. acetatoxydans [30] and S. schinkii [28] provided the basis for the comparison. In the case of P. lettingae, we analysed a publically available genome draft.
The knowledge gained in this study will form the basis for future metabolic engineering and synthetic biology in order to employ SAOB as microbial cellular factories for hydrogen and methane production on demand in a controlled and monitored manner. In particular, when using complex conditions, such as organic waste streams as fuel for microbial anodes, a deep understanding of the physiological attributes of potential electroactive bacteria, such as the SAOB, is needed, in order to achieve successful deployment under competitive industrial conditions.
2. Experimental Procedures
2.1. Growth Conditions
Since its isolation, C. ultunense strain Esp (JCM accession number JCM 16670) has been kept as liquid culture in our laboratory. For DNA isolation, cells were transferred to fresh basal medium. Stock solutions and basal medium were prepared, as described by Zehnder et al. [35]. In brief, stock solutions for basal medium A (15 mL), B (15 mL), I (5 mL), F (1 mL), and 0.2 g/L yeast extract were mixed with 1100 mL distilled water and cooked down to a final volume of 900 mL. The medium was cooled and aliquoted under flushing with N2/CO2 (80/20, v/v) to 180 mL portions in 500 mL bottles, sealed with butyl rubber stoppers, screw-topped, and autoclaved for 20 min at 121 °C. The cooled bottles were each supplemented with 10 mL of mixture C1 (containing stock solutions E (1mL, trace elements), G (1 mL, vitamin solution), C (12.5 mL) and 34.5 mL distilled water), 10 mL of mixture C2 (containing stock solutions D (49 mL), H (1 mL), and 0.5 g cysteine-HCl) and 2 mL Na-lactate (1 M) as carbon source, by injecting the mixtures and the substrate directly through a sterile filter (0.2 µm) into the bottles. Final pH of the medium was within the neutral range when checked by pH indicator paper. Bottles were inoculated 1:100 with C. ultunese strain Esp and harvested at 5000× g after four weeks of growth at 37 °C without shaking.
2.2. Isolation of DNA
DNA was isolated from 400 mL fresh culture after four weeks of cultivation. The Blood&Tissue Kit (Qiagen, Hilden, Germany) was used for DNA isolation following the standard protocol provided by the manufacturer. The quality was visualised by agarose gel electrophoresis and the quantity determined by fluorometric measurements using Qubit (Thermo Fisher Scientific, Waltham, MA, USA).
2.3. Genome Sequencing and Assembly
C. ultunense strain Esp was sequenced at the SciLife Laboratory (Uppsala, Sweden). The genome was first sequenced using the Ion Torrent PM system (Thermo Fisher Scientific, https://www.thermofisher.com) with a de novo assembly approach, which produced 281 scaffolds with a total size of 6,159,766 base pairs (bp). Sequencing and assembling details are reported in [34]. In order to improve the quality of the genome, we used HiSeq-2500 mate-pair sequencing (Illumina, https://www.illumina.com) with a library size of 3 Kb in mapping assembly.
Read length of 206 bp, a longest read length of 392 bp, and a total of final library reads of 2,631,078 for single end reads were obtained from Ion Torrent sequencing and 82,386,171 from the Illumina mate pair sequencing. General details about the sequencing performed can be found on the SciLifeLab website [36]. The FastQC software package [37] was used for read quality assessment. After pre-assembly quality checking, the filtered reads were fed into MIRA 4.0 [38] assembler for both mapping and de novo assembly, and the same reads data were also provided to the Newbler 2.8 de novo assembler (Roche, http://www.my454.com). Mapping assembly was undertaken against the available genome of C. ultunense strain BS (DSMZ 10521) (accession No. AZSU00000000) [33]. Four different assemblies were produced: (i) de novo assembly of Ion Torrent reads; (ii) de novo assembly of Illumina reads; (iii) hybrid de novo assembly of Ion Torrent reads and Illumina reads; and, (iv) hybrid mapping assembly of Ion Torrent reads and Illumina reads. Resulting assemblies were compared using the Mauve genome alignment tool [39]. However, during this process, we discovered that the culture used for DNA purification was impure. The impurity could be identified as Thermicanus aegyptius, which draft genome sequence is publically available (BioProject: PRJNA81927, RefSeq: NZ_AZU00000000.1, total length: 3.657 Mb). Thus, the hybrid mapping assembly produced from Ion Torrent reads and Illumina reads was selected for further analysis. Selection was based on N50 statistics, number of contigs, and the length of the largest contig. Scaffolding of the selected assembly was performed using SSPACE [40] and the removal of possible gaps present in scaffolds was performed using GapFiller [41]. Homopolymer errors were corrected manually using Consed [42]. The purity of the scaffolds was also checked against the genome of T. aegyptius DSM 12793. Possible miss-assemblies were corrected manually using Tablet, a graphical viewer for the visualisation of assemblies and read mappings [43]. Based on these analyses, the high-quality permanent genome that was obtained was only half the size (3,093,245 bp) of that sequenced by the Ion Torrent PM system [34]. The genome project is deposited in the Genomes On Line Database [44] with GOLD id Gp0161004, and the genome sequence is deposited in the European Nucleotide Archive database with accession number ERS1433212. Genome statistics are summarized in Table S1. Due to the bioinformatics approach that is applied, we do not expect any contamination of the high-quality draft genome sequence however we cannot entirely exclude incompleteness of the genome.
2.4. Genome Annotation and Genome Comparison
Automated gene modelling was performed using the Microbial Genome Annotation & Analysis Platform MaGe [45], a bacterial genome annotation system. Genes were identified using Prodigal [46] and AMIGene [47] as part of the MaGe genome annotation pipeline. The predicted CDSs (coding sequences) were translated and were used to search the National Center for Biotechnology Information (NCBI) non-redundant database and UniProt, TIGRFam, Pfam, PRIAM, Kyoto Encyclopedia of Genes and Genomes (KEGG), Clusters of Orthologous Groups (COG), and InterPro databases using Basic Local Alignment Search Tool Protein (BLASTP) (https://blast.ncbi.nlm.nih.gov/). Predicted coding sequences were subjected to manual analysis using the MaGe web-based platform, which also provides functional information about proteins and was used to assess and correct genes predicted through the automated pipeline. The predicted functions were also further analysed by the MaGe annotation system. Genomic features of C. ultunense strain Esp, C. ultunense strain BST (accession number AZSU00000000), T. acetatoxydans strain Re1 (accession number HF563609), S. schinkii strain Sp3 (accession number CDRZ00000000), Th. phaeum strain PB (accession number CP003732), and P. lettingae strain TMO (accession number CP000812.1) were compared using the MaGe web-based platform. Prophage sequences were identified using the PHASTER web server [48]. Tandem duplicated genomic regions of protein coding genes were calculated on the basis of identity (≥35%), minLRap (≥0.8) and gene separation (maximum of five consecutive genes) by using (MaGe) [45].
2.5. Phylogenetic Placement and Synteny
The 16S ribosomal DNA (rDNA) sequence alignment was carried out using MUSCLE [49] and a phylogenetic tree was constructed using the maximum-likelihood (ML) algorithm [50] with MEGA 6.06 [51,52]. Bootstrap analysis [53] with 100 replicates was performed to assess the support of the clusters. All predicted gene loci from C. ultunense strain Esp were subjected to linear comparisons with C. ultunense strain BST, S. schinkii, T. acetatoxydans, Th. phaeum, and P. lettingae, using the built-in tool in the MaGe platform with synton size ≥ 3 genes.
3. Results
3.1. Phylogenetic Placement
Phylogenetic analysis of the single 16S rDNA affiliates C. ultunense strain Esp to the Clostridia class within the phylum Firmicutes, and therein to the family Tissierellaceae [54] (Ribosomal data base project (RDP Naive Bayesian rRNA Classifier Version 2.11, 2017-12-11). The 16S rDNA gene of the type strain BST (=DSM 10521) is 99% identical to strain Esp (Figure 1). Comparison of the 16S rDNA gene against the latest available databases from GenBank (2018-01-22) using BLAST under default settings shows that Clostridium sp. MT1 isolated from the human gut (LK021112) is the closest current relative, sharing 97% identity. The closest characterised species is Keratinibaculum paraultunense, sharing 95.3% identity [55]. K. paraultunense was isolated from grassy marshland and it has been characterised as a thermophilic, anaerobic, keranolytic bacterium. The model acetogens Acetobacterium woodii and Moorella thermoacetica share 85% and 83% 16S rDNA gene identity, respectively (Figure 1). C. ultunense strain Esp is also only distantly related to the recently characterised mesophilic SAOB S. schinkii (83% identity) and T. acetatoxydans (82% identity), and to the thermophilic SAOB P. lettingae (76%) and Th. phaeum (85%) (Figure 1).
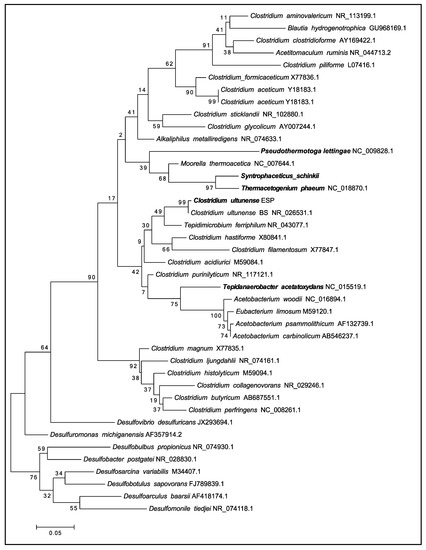
Figure 1.
Maximum likelihood tree of the 16S ribosomal RNA (rRNA) gene showing the phylogenetic position of the syntrophic acetate-oxidising bacteria (SAOB) Clostridium ultunense, Syntrophaceticus schinkii, Tepidanaerobacter acetatoxydans, Pseudothermotoga lettingae, and Thermacetogenium phaeum related to acetogens and sulphate reducers.
Synteny-based analysis revealed that C. ultunense strains BST and Esp have approximately 98% of the total genome size in synteny (Figure S1). Synteny-based analysis with all bacterial genomes that were present in the NCBI database reference sequence (RefSeq) revealed that Alkaliphilus metalliredigens QYMF appeared to be the closest current relative, with approximately 43% of its total genome size in synteny with C. ultunense strain Esp (Figure S2). Likewise, a comparison of all the inferred proteins of C. ultunense strain Esp with all proteins collected in the NCBI RefSeq database revealed the highest number of orthologues with C. ultunense strain BST (3106: 91.87%) and the next highest with A. metalliredigens QYMF (1920: 56.79%).
In general, synteny analysis and comparison of inferred proteins showed a lower relationship of C. ultunense strain Esp to other SAOB than to A. metalliredigens (Table 1, Figure 2).

Table 1.
Summary of synteny analysis (%) and comparison of inferred proteins (number of genes) of C. ultunense strain Esp to C. ultunense strain BST, Alkaliphilus metalliredigens, acetogens (Moorella thermoacetica, Acetobacterium woodii) and syntrophic acetate-oxidising bacteria (SAOB) (S. schinkii, T. acetatoxydans, Th. phaeum, P. lettingae).
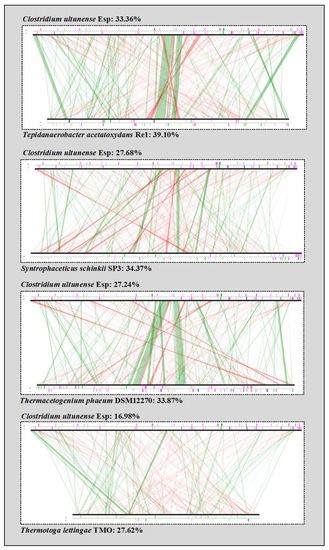
Figure 2.
Synteny comparison of C. ultunense strain Esp and the SAOB S. schinkii, T. acetatoxydans, Th. phaeum, and P. lettingae. The lines indicate syntons between two genomes. Red lines show inversions around the origin of replication. Vertical bars on the border line indicate different elements in genomes, where pink = transposases, or insertion sequences, blue = rRNA and green = transfer RNA (tRNA).
3.2. Genome Properties
The draft genome sequence for C. ultunense strain Esp comprises one scaffold with a total size of 3,093,245 bp and a calculated GC content of 32.40%. The genome showed a protein coding density of 97.97%, with an average intergenic length of 130.04 bp. The genome contains 66 tRNA, four 5S, one 16S, and one 23S rRNA genes (Table S1, Figure S3). No tRNA for selenocysteine incorporation was affiliated automatically by the MaGe pipeline. However, the genome harbours genes for L-selenocysteinyl-tRNA Sec transferase (selA: CUESP_2577, AZSU01_50111), monoselenophosphate synthase (selD: CUESP_2578, AZSU01_50112), and selenocysteinyl-tRNA elongation factor (selB: CUESP_2576, AZSU01_50110), as observed also for S. schinkii [31] and Th. phaeum [32]. For both S. schinkii and Th. phaeum, a selenocysteine-tRNA was annotated. However, selenocysteine-containing proteins are only predicted for C. ultunense Esp/BST and S. schinkii (glycine/betaine reductase complex: CUESP_0484-90, CUESP_02409-11, CUESP_0601-03; AZS01_10478-84, _40044-46, _20010-12 [34]). The genome of C. ultunense strain Esp contains 3303 predicted protein-encoding genes, of which 2198 (66.54%) have been assigned tentative functions. The remaining 1105 ORFs (open reading frames) are hypothetical/unknown proteins. It was possible to allocate 2623 (app. 79.41%) of all the predicted protein-encoding genes to the 23 functional COGs. That number was slightly higher than predicted for C. ultunense strain BST (2594 genes; 77.78%; 21 COGs). In all, 33.7% of the protein-encoding genes fell into the metabolism category, within which they were mainly affiliated to: energy metabolism (5.54%), amino acid transport and metabolism (9.23%), carbohydrate transport and metabolism (5.26%), and inorganic ion transport and metabolism (4.60%) (Table 2). These values are within the range observed for the other SAOB, where carbohydrate metabolism shows the widest range.

Table 2.
Comparison of COG (Clusters of Orthologous Groups) analysis for the SAOB C. ultunense strain Esp, C. ultunense strain BST, T. acetatoxydans, Th. phaeum, P. lettingae, and S. schinkii.
Cultivation experiments indicated that C. ultunense, S. schinkii, and Th. phaeum are very confined in heterotrophic substrate utilisation, in the case of C. ultunense strain Esp encompassing only ethanol, betaine, lactate, cysteine, and raffinose [23,24,25]. P. lettingae instead can use a much broader substrate spectrum [22] and T. acetatoxydans also proved somewhat more versatile [26], both showing higher percentages in this COG category (Table 2).
Both C. ultunense genomes (Esp and BST) were found to harbour several CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) loci. The genomes of strain Esp and BST each harbour one operon for encoding cas proteins (CUESP_2343-2350, AZSU01_30868-30875) and six to nine CRISPR loci were predicted. In T. acetatoxydans, Th. phaeum, and S. schinkii, two operons each have been identified and more than eight CRISPR loci each [28,30,32]. The P. lettingae genome contains one operon encoding CAS proteins (Tlet_0203-0210) and two CRISPR loci were predicted.
Strain Esp and strain BST are both lysogens, as their genomes harbour one prophage each. A smaller number of phage remnants were found in strain Esp. In the case of P. lettingae, only phage remnants were predicted (Table S2). This is within the range that was observed for the other SAOB (Table S2) [28,30,32]. T. acetatoxydans and Th. phaeum are lysogens. The prophage of the latter has been shown to be inducible [32]. All together, the genomes of the SAOBs contain between 1.1% and 3.3% prophage-related genes (Table S2).
3.3. Morphological and Physiological Traits
C. ultunense Esp/BST was shown to be able to form endospores, which is a trait that is common to all SAOB except P. lettingae [22,23,24,25,26]. The master regulator Spo0A needed for sporulation 37 [56] is encoded (CUESP_1786; AZSU01_30330), but the C. ultunense Esp/BST genome lacks genes encoding the phosphorylases (Spo0F, Spo0B), as has been observed in other clostridia 37 [56]. The sporulation-specific sigma factors SigE, SigG, SigF, and SigA were identified (CUESP_1917-18, AZSU01_30452-53; CUESP_1969; AZSU01_30503; CUESP_2058, AZSU01_30586).
The genome encodes both flagellum-related and chemotaxis-related genes (CUESP_1643-1678; AZSU01_30193-30220), which is in accordance with previous observations that C. ultunense Esp/BST possesses a polar flagellum and can perform tumbling movements [24,25]. A similar genotype and phenotype have been described for T. acetatoxydans [30], Th. phaeum [32], and P. lettingae ([22] (Tlet_0622-_0629; 0079-_0081; 1898-_1901; 1819-1826). In contrast, S. schinkii appears to be the only SAOB that lacks the ability to perform chemotactic manoeuvres [28].
C. ultunense Esp/BST can potentially tolerate oxidative stress, as the genome encodes rubrerythrin (AZSU_10385, CUESP_0386), which can reduce hydrogen peroxide (H2O2) to water. Rubrerythrin encoding genes were also reported for S. schinkii, Th. phaeum and T. acetatoxydans [28,30,31,32]. To protect against superoxide radicals (O2−), C. ultunense Esp/BST may express superoxide dismutase (CUESP_0551, _02564; AZSU01_10540, _50098) and superoxide reductase (CUESP_3088; AZSU01_70055). The genome of P. lettingae also encodes rubrerythrin (Tlet_0923) and superoxide reductase (Tlet_1498).
A characteristic of the mesophilic SAOB is their high ammonia tolerance. Potential mechanisms in C. ultunense Esp/BST preventing NH4+/NH3- induced osmotic stress might include putative potassium uptake proteins (CUESP_3204-3205; CUESP_2895-2896), as predicted for S. schinkii [28] and T. acetatoxydans [30]. Homologues have been also predicted for the genomes of P. lettingae (Tlet_0892-0893) and Th. phaeum: (Tph_28930-28960-2, Tph_8850-28840). Potential betaine/glycine transport systems in order to accumulate compatible solute, as found in the genomes of Th. phaeum, S. schinkii [28], and T. acetatoxydans [30], are not predicted for C. ultunense Esp/BST or P. lettingae. However, similarly to T. acetatoxydans [30] and S. schinkii [28], no gene encoding high ammonium affinity glutamine synthetase (GS) was found in the C. ultunense Esp/BST genome. Instead, the low ammonium affinity glutamate dehydrogenase (GDH) pathway appears to play a major assimilatory role (CUESP_1377; ASU01_20769). In contrast, the genomes of the thermophilic SAOB Th. phaeum and P. lettingae harbour homologues to GS (Tph_12990, 08620, 09680, 12980; Tlet_0896, _2058) and GDH (Tph_04750; Tlet_0263). Among the SAOBs that were investigated, putative ammonium transporters were only predicted for Th. phaeum (Tph_08750). V-type ATPases suggested to support T. acetatoxydans in maintaining pH homeostasis [30] were not found in C. ultunense Esp/BST or in the remaining SAOB.
C. ultunense strain Esp harbours about 33 ABC transport systems, of which 11 are predicted to transport oligopeptides and amino acids, which is in agreement with the observed requirement for yeast supplement for growth [24,25]. Five ABC transport systems likely shuffle ribose and guanosine over the membrane, while two transport Fe and Co, respectively (Table S3). Likewise, the majority of the ABC transport systems found encoded by the genomes of P. lettingae (Table S4) and T. acetatoxydans [30] are predicted to transport amino acids and oligopeptides (Table S4). In Th. phaeum and S. schinkii, ABC transport systems are mainly predicted to transport trace elements that are needed as cofactors [28] (Table S5), rather than organic compounds. There are at least four phosphoenolpyruvate (PEP) carbohydrate phosphotransferase systems (PTS) that are encoded in the C. ultunense strain Esp genome. These are predicted to take up bacterial cell wall components, such as N-acetylglucosamine and N-acetylmuramic acids and oligocarbohydrates potentially originating from lichenan or related polymers (Table S6). The general PTS proteins Hpr and EI (CUESP_0086, _0088) and Hpr kinase (CUESP_0224, AZSU_10221) and Hpr-like protein (AZSU_10234, _20320; CUESP_0236, _0914) that are needed for CcpA-dependent regulation of carbon metabolism [57] are encoded elsewhere. Genomic evidence for PTS transport systems and carbon catabolite repression/activation has only been found in T. acetatoxydans [30], and not in S. schinkii [28], Th. phaeum or P. lettingae. One potential TRAP transporter (CUESP_2539-2540) using the ion gradient over the membrane to energise unidirectional transportation of solutes was found encoded in the C. ultunense strain Esp genome. TRAP transporters are only predicted for the genome of T. acetatoxydans [30] and P. lettingae (Tlet_0052-0054), and not for Th. phaeum and S. schinkii [28]. Putative acetate uptake transporters are only predicted for S. schinkii and Th. phaeum (Table S6) [28,32].
3.4. Wood-Ljungdahl Pathway and Acetate Activation
One of the central activities of acetogenic metabolism is mediated by phosphotransacetylase (PTA), which replaces the CoA moiety from acetyl-CoA by an inorganic phosphate molecule to form acetylphosphate. The phosphate moiety is further transferred by the activity of acetate kinase (AckA) to adenosine diphosphate (ADP), producing ATP and the end product acetate. The C. ultunense Esp/BST genome harbours an ackA gene (CUESP_1616, AZSU01_30167), but it lacks a pta gene. Thus, substrate-level phosphorylation through Pta/AckA is strongly impaired. Moreover, only genes encoding proteins of the methyl branch of the WL pathway are present. These include formyl tetrahydrofolate (THF) synthetase (product of CUESP_1461; AZSU01_20850), bifunctional methylene THF dehydrogenase/methenyl THF cyclohydrolase (product of CUESP_1972, AZSU01_30506), and formate dehydrogenase (CUESP_2852-2856; AZSU01_60163-60167). The first two proteins are widely distributed in bacteria, as their activities provide the cells with one-carbon donors at different oxidation levels for biosynthesis. Two potential carbon monoxide dehydrogenases (CODH) were predicted elsewhere: One together in an operon with genes encoding a ferredoxin-binding subunit and an NAD(P)-dependent oxidoreductase (AZSU01_20423-25; CUESP_1023-25); and, the other consisting of two subunits and forming a potential operon with a membrane protein of unknown function (CUESP_2144-47, AZSU01_30669-72). According to synteny maps obtained with genomes of acetogens, neither of those resemble the CODH subunit of the acetyl-CoA/CODH complex as part of the WL pathway. Moreover, the essential CODH-Ni-insertion protein (CooC) is lacking. Genes encoding acetyl CoA synthase subunit, methyltransferase, methylene reductase, and the corrinoid enzyme are absent. Despite this, acetogenic metabolism and acetyl-CoA synthase activities have been reported for the C. ultunense strain BST [13,24]. To achieve this, Pta/AckA activity could potentially be replaced by a predicted ADP-forming Acetyl-CoA synthetase (CUESP_2312, AZSU01_30838). Alternatively, side activities of other phosphate acyltransferases (e.g., CUESP_0095; AZSU_10093; CUESP_1619, AZSU01_30169; CUESP_1223, AZSU01_20621) might substitute for the function of PTA. Enzyme activity measurements that were performed previously [13] are not specific for PTA, because the release of free CoASH from acetyl-CoA is determined [58,59]. Thus, free CoASH can be produced by any acyl-CoA active enzyme, which might explain the observed activity of the cell-free extract [13]. Consequently, the reported weak growth on lactate, pyruvate, ethanol, and L-cysteine must either rely on the abovementioned activities of either ADP-forming Acetyl-CoA synthetase or phosphate acyltransferases. Alternatively, the final product acetyl-CoA is used in anabolic pathways, rather than in energy conservation, and the actual energy source is provided by the added yeast extract. A malate/lactate dehydrogenase-like protein (CUESP_3146), alcohol, and aldehyde dehydrogenase (CUESP_1073-76, AZSU01_20472-74), Pyruvate:ferredoxin/flavodoxin oxidoreductases (CUESP_2474/75; AZSU01_50009/10; CUESP_2527, _0225; AZSU01_50061, _10222), and L-cysteine disulphydrase (CUESP_2922, _2143) are encoded by the C. ultunense Esp/BST genome.
In contrast, the genomes of S. schinkii, T. acetatoxydans, and Th. phaeum harbour the WL pathway genes as well as genes encoding phosphotransacetylase and acetate kinase [28,30,32], which is in accordance with the acetogenic metabolism that is observed in pure cultures. P. lettingae, in which growth has been described as non-acetogenic [22], does not harbour genes that are related to phosphotransacetylase, acetyl-CoA synthetase, ADP-forming acetyl-CoA synthetase, carbon monoxide dehydrogenase, methyltransferase, methylene THF reductase, or format dehydrogenase/lyase. However, genes for acetate kinase (Tlet_0293), formyl THF synthetase (Tlet_1513), and bifunctional methylene THF dehydrogenase/methenyl cyclohydrolase/(Tlet_1514) are encoded.
3.5. Respiratory Complexes
Two potential ion motive electron transport complexes were identified in the genome of C. ultunense strain Esp (Table 3). An operon encodes a protein complex (CUESP_0375-0379; AZSU01_10374-10378), which shows similarity to the Na+ pumping NADH:quinone hydrogenase (Na+-NQR), which is a respiratory complex that is widely distributed in marine bacteria, which couples electron transport to the generation of a sodium gradient [60]. It is composed of six subunits (NQRA-F) pumping out Na+, while electrons travel from reduced nicotinamide adenine dinucleotide (NADH) to quinone via a minimum of five redox carriers [60,61]: one Flavin adenine dinucleotide FAD and one 2Fe-2S cluster in the peripheral NQRF subunit, one flavin mononucleotide (FMN) in the hydrophilic NQRC subunit, and one FMN and riboflavin in the membrane integral NQRB subunit. NQRF is the site for NADH oxidation and NQRA has been identified as the quinol formation site. The complex has also been identified in the genome of the SAOB P. lettingae (Tlet_0865-0869, Table 3) and its close relative Pseudothermotoga thermarum [62]. The genes are arranged in the order NqrBCDEF and share between 36% and 55% identity (Figure 3). However, the genome lacks a NQRA subunit gene that contains the quinone binding site, as has also been reported for P. lettingae and P. thermarum [62]. The genomes of T. acetatoxydans, Th. phaeum, and S. schinkii do not encode any Nqr genes [28,30,32].

Table 3.
Energy-conserving systems and related proteins predicted for the SAOB C. ultunense strain Esp, C. ultunense strain BST, T. acetatoxydans strain Re1, Th. Phaeum strain PB, P. lettingae strain TMO, and S. schinki strain Sp3 [28,30,32].
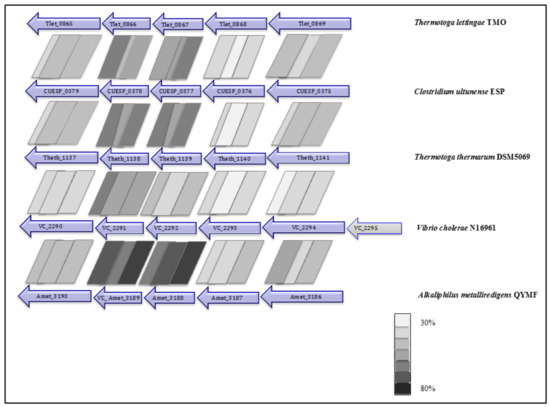
Figure 3.
Organisation of the Nqr genes in C. ultunense, P. lettingae, Pseudothermotoga thermarum, Vibrio cholera, and Alkaliphilus metalliredigens.
As found in T. acetatoxydans [30] and S. schinkii [28], the genomes of C. ultunese Esp/BST (AZSU01_50145-50150; CUESP_2612-2617) and P. lettingae (Tlet_0286-0291), contain a gene cluster each coding for the respiratory Rnf complex. The Rnf complex is a ferredoxin:NAD+ oxidoreductase that uses the redox span between ferredoxin (E′ = −500 mV) and NADH (E′ = −320 mV) to establish an ion gradient over the membrane [63]. This is equivalent to one-third to one-half of an ATP. Conversely, the generated Δμ˜Na+/H+, can drive the endergonic electron flow from NADH to ferredoxin. The genes are organised in the order rnfCDGEAB, as found in many Clostridia [63]. The genome of Th. phaeum lacks Rnf genes [32] (Table 3).
The genome of C. ultunese Esp/BST does not harbour genes that are related to Ech (energy-converting hydrogenase) hydrogenase that have been found in S. schinkii and Th. phaeum [28,32]. Likewise, the genomes of T. acetatoxydans [30] and P. lettingae do not encode Ech hydrogenase (Table 3). The Ech hydrogenases belong to the group of [Ni-Fe] hydrogenases, which catalyse the reversible exergonic oxidation of reduced ferredoxin to the reduction of protons that are coupled to vectoral ion transport across the membrane [64].
3.6. Soluble Electron Transfer Proteins
The genome of C. ultunense Esp/BST encodes several ferredoxins, one rubredoxin, and flavodoxins (Table 3). A similar composition of soluble electron transfer proteins has been found for the genomes of P. lettingae and Th. phaeum, including flavodoxins, ferredoxins, and rubredoxin (Table 3). However, the genomes of S. schinkii do not harbour any flavodoxin-related genes [28,30,32] (Table 3). Of all SAOB genomes, only P. lettingae encodes a cytochrome (Table 3). One electron transfer flavoprotein (ETF) complex was found to be encoded in the genome of C. ultunense Esp/BST (EtfA,B CUESP_1366-1368, AZSU01_20759-20761). The genomes of P. lettingae (Tlet_1692-1693; Tlet_1832-1833), and T. acetatoxydans (TepiRe1_2532-2533) were also found to harbour ETF complex genes. No ETF-related genes were identified in the genomes of Th. phaeum and S. schinkii (Table 3).
3.7. Hydrogenases
The genome of C. ultunense Esp/BST encodes one putative [Fe-Fe] hydrogenase (CUESP_2849-2850; AZSU_01_60160-60162) that is adjacent to genes coding for a cytoplasmatic formate dehydrogenase (CUESP_2852-2856; AZSU01_60163-60167). The short intergenic regions indicate a potential operon. A similar gene organisation is found in the genome of Th. phaeum (Tph_c18420-_c18430) [32] (Table 3). Putative [Fe-Fe] hydrogenases have also been identified in the genomes of T. acetatoxydans [30], S. schinkii [28], and P. lettingae (Tlet_0952-0957; Tlet_1518-1522), with S. schinkii harbouring four duplicates, the highest number. In contrast to T. acetatoxydans and P. lettingae, the genome of S. schinkii contains both cytoplasmatic and membrane-bound formate hydrogenases, but these are located separately from any hydrogenase-related genes [28] (Table 3). The genome of C. ultunense Esp/BST, and the genomes of T. acetatoxydans and P. lettingae, do not contain genes that are related to [Ni-Fe] hydrogenases. However, those complexes have been found in the genome of both S. schinkii and Th. phaeum (see [64] for classification of hydrogenases). Genes that were predicted to encode a putative NAD(P)-binding oxidoreductase/heterodisulphide reductase complex and a putative formate lyase complex seem to be unique to the genomes of S. schinkii and Th. phaeum [28,32], respectively, as no such complexes are predicted for the genomes of C. ultunense Esp/BST, T. acetatoxydans, and P. lettingae (Table 3).
3.8. Adenosine Triphosphate (ATP) Synthase and Pyrophosphatase (PPase)
Next to the ATP synthase complex (CUESP_1228-1235; AZSU_20626-20633), the genome of C. ultunense Esp/BST encodes a putative membrane-bound ion translocating pyrophosphatase (CUESP_0504; AZSU01__10499). ATP synthase and putative pyrophosphatase (PPase) genes are also predicted for the genomes of the other SAOB (Table 3), except for T. acetatoxydans, the genome of which does not encode an ATP synthase complex [30].
4. Discussion
4.1. Attributes Linked to Anaerobic Digestion Environments
It has been shown repeatedly that the presence and abundance of SAOB in engineered methanogenic processes are strongly positively correlated with high ammonia concentrations. Factors such as acetate concentration, dilution rate, and methanogenic community structure have also been suggested to promote SAO activity [5,6,7,8,9,10,65,66,67,68,69,70,71,72]. However, certain competitive abilities can be accredited to S. schinkii, as this SAOB has been found at high abundance under both low- and high-ammonia conditions [9,10,68,73]. Potential competiveness for the substrate acetate has been suggested as one explanation, as S. schinkii expresses a putative acetate uptake system [28], which is otherwise only encoded by the genome of the thermophilic Th. phaeum.
Another factor affecting the abundance of SAOB might be the proportion of proteinaceous material that is provided in the feedstock, as indicated by the genomic equipment of SAOB. A large proportion of the transport systems in all SAOB are predicted to shuffle oligopeptides and amino acids over the membrane, rather than carbohydrates. Furthermore, a number of genes that are needed for de novo synthesis of amino acids are lacking, which explains the observed dependency on yeast extract when cultivating SAOB under laboratory conditions. Thus, higher availability of proteins in the feedstock might be beneficial, as it provides both the precursors for biosynthesis and energy sources. Apart from Th. phaeum, none of the SAOB harbours genes for ammonium transporters and the mesophilic SAOB also lack the high ammonium affinity GS, both further emphasising an adaptation to amino acid-rich environments. At this time, there are no indications of habitats other than engineered AD processes when searching for 16S rRNA gene traces in the nucleotide (nr/nt) database. C. ultunense Esp/BST has been identified in a fermenting woad vat, which is another artificial environment [74]. However, all SAOB are spore-forming, which enables (re)-entry and (re)-population of biogas processes depleted in SAOB, and is beneficial when conditions become lethal, e.g., through exposure to oxygen. Moreover, none of the known SAOBs appears to be a strictly obligate anaerobe, as indicated by genes conferring protection against super oxide radicals. This attribute facilitates the transfer between cultivation bottles and bio-augmentation of laboratory-scale reactors, as well as phenotypic characterisation and metabolic engineering.
One more interesting attribute of the SAOB is their apparent robustness against phages. In nutrient-rich and cell-dense environments such as AD processes, viruses can be expected to be high in numbers and diversity [75]. Despite the high virus abundance in AD environments, the genomes of C. ultunense Esp/BST and other SAOB appear to be only weakly infected. This indicates a certain robustness against phage attacks, as bacterial genomes can contain up to 20% prophage-related genes [76,77]. The significantly higher number of CRISPR loci acquired by the SAOB points to strong phage-microbial interactions. CRISPRs have been found in almost half of all the bacterial genomes and almost all of the archaeal genomes sequenced [78]. The cas/CRISPR system is a prokaryotic defence mechanism, enabling the organism to respond to and eliminate invading genetic material, such as bacteriophages [79]. The CRISPR-associated sequence (cas) genes are often directly adjacent to the CRISPR loci. The number of CRISPR loci ranges from one to 21, but up to three loci occur most frequently [32]. The adaptive nature of CRISPR triggered by invading phages confers robustness and the ability to encounter new phages. Thus, the strong impact of bacteriophages on microbial diversity and community composition in AD processes, as recently shown by Zhang et al. [75], might be less pronounced in SAOB populations. There are also indications that phages selectively attack the larger populations within a microbial community [80,81]. Moreover, the CRISPR/cas loci that were identified in SAOB might potentially enable strain-typing applications for controlling naturally and metabolically engineered SAOB strains.
4.2. Acetate Oxidation in Mesophilic and Thermophilic Syntrophic Acetate-Oxidising Bacteria
Cultivation experiments, enzyme activity studies and mRNA-based investigations previously led to the assumption that C. ultunense Esp/BST belongs to the physiological group of acetogens, indicating that the WL pathway plays a central metabolic role, as has also been reported for the SAOB Th. phaeum, S. schinkii, and T. acetatoxydans [13,24,25,26,29,30,32]. Consequently, in AD processes, these SAOB were thought to be generally able to perform two metabolic functions: acetate production from smaller organic molecules employing the reductive direction of the WL pathway and syntrophic acetate oxidation using the oxidative direction. However, our study revealed that at least two pathways for mesophilic acetate oxidation must exist, as the genome of C. ultunense Esp/BST lacks key enzymes of the WL pathway (Figure 4 and Figure 5).
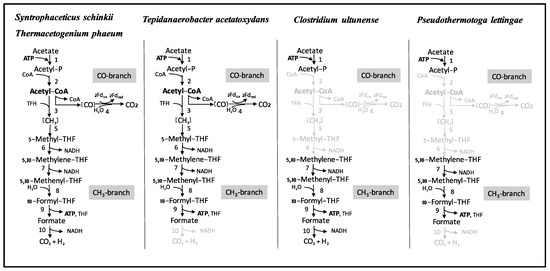
Figure 4.
Wood-Ljungdahl (WL) pathway proteins encoded by the genomes of the SAOB S. schinkii, Th. phaeum, T. acetatoxydans, C. ultunense Esp/BST, and P. lettingae. (1) Acetate kinase, (2) phosphotransacetylase, (3) acetyl CoA synthetase complex (including corrinoid protein), (4) carbon monoxide dehydrogenase, (5) methyl transferase, (6) methylene THF reductase, (7) methylene tetrahydrofolate (THF) dehydrogenase, (8) methenyl THF cyclohydrolase, (9) formyl THF synthetase, (10) formate dehydrogenase. Functions displayed in light grey are not encoded.
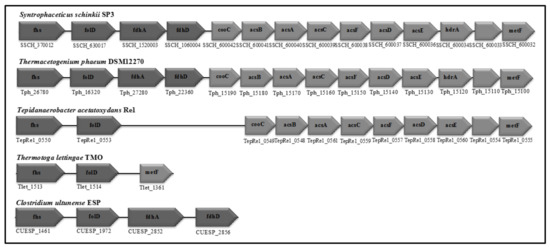
Figure 5.
Organisation of the WL pathway genes in the SAOB S. schinkii, Th. phaeum, T. acetatoxydans, C. ultunense strain Esp (also valid for strain BST) and P. lettingae. acsA,B,C,D,F, Carbon monoxide dehydrogenase/Acetyl-CoA synthetase subunits; acsE, Methyltransferase; hdr, heterodisulphide reductase; metF, methylene tetrahydrofolate reductase; folD, bifunctional methenyl tetrahydrofolate cyclohydrolase/methylene tetrahydrofolate dehydrogenase; fhs, formyl tetrahydrofolate reductase; cooC, accessory protein; fdhA/fdhD, formate dehydrogenase. Light grey: Genes are organized in a putative operon. Dark grey: Genes are scattered in the genome.
In the case of the thermophilic P. lettingae, the lack of WL pathway genes was expected, as the metabolism has been described as non-acetogenic [22] (Figure 4 and Figure 5). Both Th. phaeum and S. schinkii harbour the complete set of WL pathway genes (Figure 4 and Figure 5), and transcriptomic profiling of an acetate-oxidising co-culture has clearly confirmed the involvement of the oxidative WL pathway in acetate oxidation in S. schinkii [28,32]. For Th. phaeum, no such expression data are available, but enzyme activity studies have confirmed expression of key functions of the WL pathway, indicating that this pathway is a potential acetate oxidation route [27]. Moreover, Th. phaeum and S. schinkii are closely related (92.1% 16s RNA gene identity), with 50% of the genomes in synteny. Gene organisation of the WL pathway genes in Th. phaeum is identical to that in S. schinkii (Figure 5). Genome-guided analysis of T. acetatoxydans has revealed a truncated WL pathway [30] (Figure 4). However, a role of the WL pathway in acetate oxidation has been questioned, as net ATP synthesis is strongly impaired due to the lack of F0/F1 ATP synthase [30], and substrate level phosphorylation via the oxidative WL pathway does not gain any net ATP (Figure 4). There has been speculation about acetate oxidation in T. acetatoxydans, including a metabolic route via an oxidative tricarboxylic acid cycle [30]. However, no evidence on gene expression or activity level has been supplied so far. In the case of C. ultunense Esp/BST and P. lettingae, this proposed metabolic route cannot be applied, as both genomes lack key enzymes, such as fumarate reductase and succinyl-CoA:acetate CoA transferase. However, the free energy released (ΔG0′ = −35 kJ/mol) in SAO indicates that there is no such reaction, which can be coupled to ATP synthesis (ΔG0′ = +60 to 70 kJ/mol) [82] directly, no matter which metabolic reactions contribute to SAO.
All SAOB are potentially able to activate acetate to acetyl phosphate via acetate kinase (Figure 4). In the case of S. schinkii and Th. phaeum, phosphotransacetylase activity enables the acetyl moiety to enter the WL pathway (Figure 4), as strongly supported by transcriptomic data obtained for S. schinkii [28]. For C. ultunense Esp/BST and P. lettingae, the metabolic fate of the acetyl phosphate still needs to be explained. However, mRNA expression and enzyme activity studies clearly indicate the involvement of formyltetrahydrofolate synthetase in the acetate oxidation pathway of S. schinkii, T. phaeum, C. ultunense Esp/BST, and T. acetatoxydans [27,28,29]. For P. lettingae, no such expression or activity data have been published so far.
4.3. Energy Conservation Associated with Syntrophic Acetate Oxidation
The SAOB differ markedly in their energy conserving systems, indicating different strategies. No core set could be identified, except for [Fe-Fe] hydrogenases, which are known to be involved in H2 evolution, and which are encoded by all SAOB genomes (Figure 6).
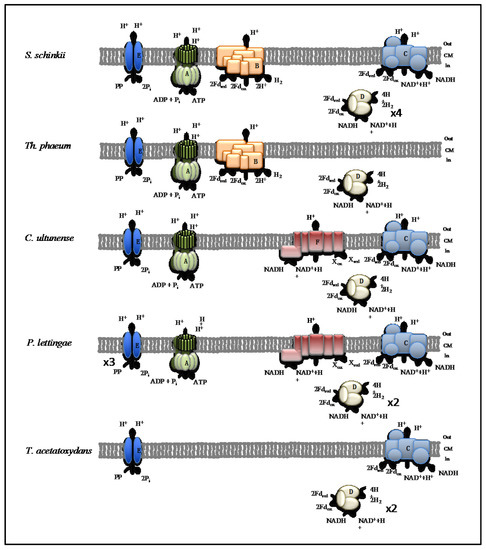
Figure 6.
Energy conservation systems potentially involved in syntrophic acetate oxidation. (A) F0/F1 ATP synthase, (B) Ech (energy-converting hydrogenase) hydrogenase, (C) Proton-translocating ferredoxin:NAD+ oxidoreductase (Rnf) complex, (D) [Fe-Fe] hydrogenases, (E) ion-translocating pyrophosphatase, (F) NADH:quinone hydrogenase (NQR) complex. Numbers indicate predicted gene duplications. Na+ translocation might occur instead of H+ translocation.
As observed for many anaerobic bacteria, ferredoxin and rubredoxin seem to be the main electron carriers in energy metabolism. Ferredoxin contains iron-sulphur clusters as an electron-mediating cofactor. Flavodoxins contain a tightly bound FMN as the electron-receiving component. The redox potential is strongly affected by the apoprotein [83]. Rubredoxin contains redox-active iron, but it lacks inorganic sulphide. In the case of C. ultunense, Th. phaeum and P. lettingae, flavodoxins can potentially substitute for ferredoxin, e.g., under iron shortage. Both ferredoxin and flavodoxin can serve as electron donors in reactions with standard redox potential as low as −500 mV, and can therefore serve as either an electron donor or an electron acceptor in hydrogen production or consumption [84]. The respiratory Ech hydrogenase and Rnf complex can both couple ferredoxin oxidation to proton translocation. Ech hydrogenase transfers electrons from ferredoxin to protons, and Rnf complex mediates electron transfer from ferredoxin to NAD+, covering the redox span from E0′ = −500 to −413 mV and E0′ = −500 to −320 mV, respectively [63,85]. The proton gradient that was established by either complex can be used for ATP production and for providing energy for endergonic reactions, such as oxidation of methyl THF to methylene THF by methylene THF reductase, as needed in the oxidative WL pathway. The Rnf complex is the most widespread electron transport complex in anaerobic microorganisms [63]. However, it appears unlikely that the complex supports energy conservation in the oxidative WL pathway, as the standard potential of the NADH that is produced might not be sufficiently negative to reduce protons. This is in agreement with the observed down regulation of the Rnf genes in S. schinkii when oxidising acetate and the lack of Rnf genes in the genome of Th. phaeum [28,32] (Figure 6).
Instead, energy conservation by the oxidative WL pathway most likely involves Ech hydrogenase, which can couple oxidation of ferredoxin directly to the formation of a proton gradient (Figure 6). However, the oxidative WL pathway produces NADH as redox equivalent from three enzymatic reactions (reaction 6, 7, 10; Figure 5), and only one reaction reduces ferredoxin (reaction 4, Figure 5) [86]. The genomes of S. schinkii and Th. phaeum are predicted to encode potential electron-bifurcating ferredoxin- and NAD+-dependent [Fe-Fe] hydrogenases [28,32], which might couple the endergonic formation of hydrogen from NADH to the exergonic formation of hydrogen from reduced ferredoxin (Figure 6). In that case, a proton motive force would be generated due to the cytosolic consumption of protons. Transcriptomic studies that were performed on S. schinkii support the involvement of Ech hydrogenase and bifurcating [Fe-Fe] hydrogenase in acetate oxidation [28]. For Th. phaeum, a potential link between formate oxidation and bifurcating hydrogen formation has also been suggested [32]. A similar gene organisation has been found in C. ultunense Esp/BST, indicating the potential connection of both activities. With regard to T. acetatoxydans, SAO-related energy conservation could potentially be achieved by a bifurcating [Fe-Fe] hydrogenase [30], rather than by respiratory complexes, as no such complexes have been predicted, except for a putative Rnf complex (Figure 6). This might also apply for C. ultunense Esp/BST and P. lettingae: The proton motive force is very likely formed by chemical or scalar protons rather than vectoral protons, as next to Rnf complex both genomes harbour only a putative bifurcating [Fe-Fe] hydrogenase (Figure 6). However, both of the genomes encode a putative NQR complex, which indicates that another electron acceptor rather than protons can potentially be utilised. This respiratory complex transfers electrons from NADH to quinone covering a redox span between −320 and +90 mV. The net redox reaction is similar to that carried out by H+ pumping NADH-quinone oxidoreductase complex that is found in mitochondria and many respiring bacteria, but the complexes are not homologous [60].
In Vibrio cholera, the NQR complex generates first an electrochemical Na gradient, followed by formation of a proton motive force by a respiratory complex that is connected via quinol. Interestingly, in C. ultunense Esp/BST and in P. lettingae, the genome lacks the quinone-binding subunit NQRA, indicating that the final electron acceptor could potentially have a more negative redox potential than quinone. The last redox carrier is very likely FMN, the redox potential of which strongly depends on the apoprotein [83]. The NQR complex might follow downstream of the Rnf complex, both together forming a potential respiratory chain (Figure 6). However, the involvement of the NQR complex in acetate oxidation and the final inorganic or organic electron acceptor(s) still need to be uncovered. Interestingly, the genome of A. metalliredigens, the current closest sequenced relative of C. ultunense Esp/BST, also encodes a putative NQR complex (Figure 3). A. metalliredigens has been described as a strictly anaerobic metal-reducing bacterium, only distantly related to other commonly studied iron-reducing microorganisms [87]. It can utilise lactate or acetate (in the presence of yeast extract) when any of the electron acceptors Fe(III)-citrate, Fe(III)-EDTA, Co(III)-EDTA, or Cr(VI) is present [87]. For C. ultunense Esp/BST, acetate oxidation using Fe(III) as an electron acceptor has been excluded [24,25]. In P. lettingae, acetate oxidation has been observed in the presence of thiosulphate [22]. In order to provide reduced ferredoxin for anabolic and catabolic pathways, both the Rnf complex and Ech hydrogenases can reverse the electron flow using the electrochemical gradient and drive the thermodynamically unfavourable reduction of ferredoxin with NADH or hydrogen. In T. acetatoxydans, C. ultunense Esp/BST, and P. lettingae, the Rnf complex might connect the NADH pool fed by reactions of acetate oxidation to hydrogen evolution by hydrogenases, which requires ferredoxin. The ETF complex seems not play a role in energy conservation connected to SAO. This complex is mainly involved in oxidation of specific substrate by means of dehydrogenase reactions [88]. In the the case of C. ultunense Esp/BST and P. lettingae, the activity is likely to be associated to the beta-oxidation of fatty acids, as it is encoded together with an acyl-dehydrogenase subunit. The ETF found encoded in the genome of T. acetatoxydans likely receives electrons from oxidation of lactate, as it is part of an operon encoding a lactate uptake permease and a lactate dehydrogenase subunit.
Moreover, additional energy might be conserved by the ion-translocating pyrophosphatase found in all SAOB (Figure 6). These PPases are integral membrane proteins that couple the hydrolysis of pyrophosphate (PPi) to the transport of monovalent cations (Na+/H+) against the electrochemical potential gradient, generating an ion motive force [89]. Thus, PPi that is formed as a by-product of biosynthetic reactions in which nucleoside triphosphates are converted to nucleoside monophosphates could be hydrolysed by the membrane-bound PPase and contribute to the formation of a proton motive force and subsequently to the synthesis of ATP. In S. schinkii, PPase expression was observed when oxidising acetate syntrophically [28].
5. Summary
Very little is known about the metabolic capacity and flexibility of SAOB, mainly being due to strict cultivation requirements and difficulties in reconstituting the thermodynamically unfavourable acetate oxidation process under laboratory conditions. Therefore, genome-based studies and comparisons are important, as they provide insights into questions regarding metabolic and physiological potential and restrictions, and they provide the basis for metabolic engineering, control, and monitoring. The results of the present study clearly indicate unexpected metabolic diversity between the known SAOB with regard to acetate oxidation, energy conservation, and metabolic flexibility, and a certain environmental robustness that is beneficial for AD processes. The metabolic diversity also implies differences in regulation of the SAO pathways with respect to potential inducers, repressors, and regulators, which has potential consequences regarding the abundance and activity of SAOB in anaerobic digestion processes.
Supplementary Materials
The following are available online at http://www.mdpi.com/2073-4425/9/4/225/s1.
Acknowledgments
This work was supported by the University of the Punjab, Lahore, Pakistan. Uppsala Genome Center performed the sequencing, supported by the Science for Life Laboratory (Uppsala), the National Bioinformatics Infrastructure in Sweden (NBIS) supporting the SLU bioinformatics Infrastructure at SLU (SLUBI) and Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX), Uppsala, Sweden. The contribution of S.M. and E.B.-R. was supported by EU FP7 ALLBIO project, grant number 289452. E.B.-R. was also partly supported by Swedish VR project, grant number 2015-03443. A.S. and B.M. were supported by the Swedish Energy Agency (project No. P36651-1). B.M. was further supported by FORMAS, grant number 942-2015-1008. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author Contributions
S.M., B.M., E.B. and A.S. contributed to the conception and design of this project. S.M. was involved in acquisition and bioinformatics analysis of the data. B.M. und S.M. analysed and interpreted the data. S.M. prepared the figures and tables. B.M. and S.M. wrote the manuscript and prepared figures and tables. A.S. and E.B. provided financial support. All authors were involved in critical revision of the manuscript, have given final approval of the version to be published and agree to be accountable for all aspects of the work.
Conflicts of Interest
The authors declare that they have no competing interests.
References
- Chen, Y.; Cheng, J.J.; Creamer, K.S. Inhibition of anaerobic digestion process: A review. Bioresour. Technol. 2008, 99, 4044–4064. [Google Scholar] [CrossRef] [PubMed]
- Rajagopal, R.; Massé, D.I.; Singh, G. A critical review on inhibition of anaerobic digestion process by excess ammonia. Bioresour. Technol. 2013, 143, 632–641. [Google Scholar] [CrossRef] [PubMed]
- Westerholm, M.; Moestedt, J.; Schnürer, A. Biogas production through syntrophic acetate oxidation and deliberate operating strategies for improved digester performance. Appl. Energy 2016, 179, 124–135. [Google Scholar] [CrossRef]
- Sowers, K.R. Methanogenesis. In Encyclopedia of Microbiology, 3rd ed.; Schaechter, M., Ed.; Academic Press: Oxford, UK, 2009; pp. 265–286. ISBN 978-0-12-373944-5. [Google Scholar]
- Angenent, L.T.; Sung, S.; Raskin, L. Methanogenic population dynamics during startup of a full-scale anaerobic sequencing batch reactor treating swine waste. Water Res. 2002, 36, 4648–4654. [Google Scholar] [CrossRef]
- Fotidis, I.A.; Karakashev, D.; Angelidaki, I. The dominant acetate degradation pathway/methanogenic composition in full-scale anaerobic digesters operating under different ammonia levels. Int. J. Environ. Sci. Technol. 2014, 11, 2087–2094. [Google Scholar] [CrossRef]
- Schnürer, A.; Nordberg, A. Ammonia, a selective agent for methane production by syntrophic acetate oxidation at mesophilic temperature. Water Sci. Technol. 2008, 57, 735–740. [Google Scholar] [CrossRef] [PubMed]
- Schnürer, A.; Zellner, G.; Svensson, B.H. Mesophilic syntrophic acetate oxidation during methane formation in biogas reactors. FEMS Microbiol. Ecol. 1999, 29, 249–261. [Google Scholar] [CrossRef]
- Sun, L.; Muller, B.; Westerholm, M.; Schnurer, A. Syntrophic acetate oxidation in industrial CSTR biogas digesters. J. Biotechnol. 2014, 171, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Westerholm, M.; Leven, L.; Schnurer, A. Bioaugmentation of syntrophic acetate-oxidizing culture in biogas reactors exposed to increasing levels of ammonia. Appl. Environ. Microbiol. 2012, 78, 7619–7625. [Google Scholar] [CrossRef] [PubMed]
- Shimada, T.; Morgenroth, E.; Tandukar, M.; Pavlostathis, S.G.; Smith, A.; Raskin, L.; Kilian, R.E. Syntrophic acetate oxidation in two-phase (acid-methane) anaerobic digesters. Water Sci. Technol. 2011, 64, 1812–1820. [Google Scholar] [CrossRef] [PubMed]
- Hattori, S.; Luo, H.; Shoun, H.; Kamagata, Y. Involvement of formate as an interspecies electron carrier in a syntrophic acetate-oxidizing anaerobic microorganism in coculture with methanogens. J. Biosci. Bioeng. 2001, 91, 294–298. [Google Scholar] [CrossRef]
- Schnürer, A.; Svensson, B.H.; Schink, B. Enzyme activities in and energetics of acetate metabolism by the mesophilic syntrophically acetate-oxidizing anaerobe Clostridium ultunense. FEMS Microbiol. Lett. 1997, 154, 331–336. [Google Scholar] [CrossRef]
- Schnürer, A.; Houwen, F.; Svensson, B. Mesophilic syntrophic acetate oxidation during methane formation by a triculture at high ammonium concentration. Arch. Microbiol. 1994, 162, 70–74. [Google Scholar] [CrossRef]
- Chauhan, A.; Ogram, A. Phylogeny of acetate-utilizing microorganisms in soils along a nutrient gradient in the florida everglades. Appl. Environ. Microbiol. 2006, 72, 6837–6840. [Google Scholar] [CrossRef] [PubMed]
- Gray, N.D.; Sherry, A.; Grant, R.J.; Rowan, A.K.; Hubert, C.R.J.; Callbeck, C.M.; Aitken, C.M.; Jones, D.M.; Adams, J.J.; Larter, S.R.; et al. The quantitative significance of Syntrophaceae and syntrophic partnerships in methanogenic degradation of crude oil alkanes. Environ. Microbiol. 2011, 13, 2957–2975. [Google Scholar] [CrossRef] [PubMed]
- Horn, M.A.; Matthies, C.; Küsel, K.; Schramm, A.; Drake, H.L. Hydrogenotrophic methanogenesis by moderately acid-tolerant methanogens of a methane-emitting acidic peat. Appl. Environ. Microbiol. 2003, 69, 74–83. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.M.; Head, I.M.; Gray, N.D.; Adams, J.J.; Rowan, A.K.; Aitken, C.M.; Bennett, B.; Huang, H.; Brown, A.; Bowler, B.F.J.; et al. Crude-oil biodegradation via methanogenesis in subsurface petroleum reservoirs. Nature 2008, 451, 176–180. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Conrad, R. Thermoanaerobacteriaceae oxidize acetate in methanogenic rice field soil at 50 °C. Environ. Microbiol. 2010, 12, 2341–2354. [Google Scholar] [CrossRef] [PubMed]
- Mayumi, D.; Dolfing, J.; Sakata, S.; Maeda, H.; Miyagawa, Y.; Ikarashi, M.; Tamaki, H.; Takeuchi, M.; Nakatsu, C.H.; Kamagata, Y. Carbon dioxide concentration dictates alternative methanogenic pathways in oil reservoirs. Nat. Commun. 2013, 4, 1998. [Google Scholar] [CrossRef] [PubMed]
- Rui, J.; Qiu, Q.; Lu, Y. Syntrophic acetate oxidation under thermophilic methanogenic condition in Chinese paddy field soil. FEMS Microbiol. Ecol. 2011, 77, 264–273. [Google Scholar] [CrossRef] [PubMed]
- Balk, M.; Weijma, J.; Stams, A.J.M. Thermotoga lettingae sp. nov., a novel thermophilic, methanol-degrading bacterium isolated from a thermophilic anaerobic reactor. Int. J. Syst. Evol. Microbiol. 2002, 52, 1361–1368. [Google Scholar] [PubMed]
- Hattori, S.; Kamagata, Y.; Hanada, S.; Shoun, H. Thermacetogenium phaeum gen. nov., sp. nov., a strictly anaerobic, thermophilic, syntrophic acetate-oxidizing bacterium. Int. J. Syst. Evol. Microbiol. 2000, 50, 1601–1609. [Google Scholar] [CrossRef] [PubMed]
- Schnurer, A.; Schink, B.; Svensson, B.H. Clostridium ultunense sp. nov., a mesophilic bacterium oxidizing acetate in syntrophic association with a hydrogenotrophic methanogenic bacterium. Int. J. Syst. Bacteriol. 1996, 46, 1145–1152. [Google Scholar] [CrossRef] [PubMed]
- Westerholm, M.; Roos, S.; Schnürer, A. Syntrophaceticus schinkii gen. nov., sp. nov., an anaerobic, syntrophic acetate-oxidizing bacterium isolated from a mesophilic anaerobic filter. FEMS Microbiol. Lett. 2010, 309, 100–104. [Google Scholar] [CrossRef] [PubMed]
- Westerholm, M.; Roos, S.; Schnürer, A. Tepidanaerobacter acetatoxydans sp. nov., an anaerobic, syntrophic acetate-oxidizing bacterium isolated from two ammonium-enriched mesophilic methanogenic processes. Syst. Appl. Microbiol. 2011, 34, 260–266. [Google Scholar] [CrossRef] [PubMed]
- Hattori, S.; Galushko, A.S.; Kamagata, Y.; Schink, B. Operation of the co dehydrogenase/acetyl coenzyme a pathway in both acetate oxidation and acetate formation by the syntrophically acetate-oxidizing bacterium Thermacetogenium Phaeum. J. Bacteriol. 2005, 187, 3471–3476. [Google Scholar] [CrossRef] [PubMed]
- Manzoor, S.; Bongcam-Rudloff, E.; Schnurer, A.; Muller, B. Genome-guided analysis and whole transcriptome profiling of the mesophilic syntrophic acetate oxidising bacterium Syntrophaceticus schinkii. PLoS ONE 2016, 11, e0166520. [Google Scholar] [CrossRef] [PubMed]
- Müller, B.; Sun, L.; Schnürer, A. First insights into the syntrophic acetate-oxidizing bacteria—A genetic study. MicrobiologyOpen 2013, 2, 35–53. [Google Scholar] [CrossRef] [PubMed]
- Müller, B.; Manzoor, S.; Niazi, A.; Bongcam-Rudloff, E.; Schnürer, A. Genome-guided analysis of physiological capacities of Tepidanaerobacter acetatoxydans provides insights into environmental adaptations and syntrophic acetate oxidation. PLoS ONE 2015, 10, e0121237. [Google Scholar] [CrossRef] [PubMed]
- Manzoor, S.; Müller, B.; Niazi, A.; Schnürer, A.; Bongcam-Rudloff, E. Working draft genome sequence of the mesophilic acetate oxidizing bacterium Syntrophaceticus schinkii strain Sp3. Stand. Genom. Sci. 2015, 10, 99. [Google Scholar] [CrossRef] [PubMed]
- Oehler, D.; Poehlein, A.; Leimbach, A.; Müller, N.; Daniel, R.; Gottschalk, G.; Schink, B. Genome-guided analysis of physiological and morphological traits of the fermentative acetate oxidizer Thermacetogenium phaeum. BMC Genom. 2012, 13, 723. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Zhou, H.; Zhang, L.; Zhang, J.; Wang, Y.; Wang, S.; Zhou, Z.; Yan, X. Draft genome sequence of Clostridium ultunense strain BS (DSMZ 10521), recovered from a mixed culture. Genome Announc. 2014, 2. [Google Scholar] [CrossRef] [PubMed]
- Manzoor, S.; Müller, B.; Niazi, A.; Bongcam-Rudloff, E.; Schnürer, A. Draft genome sequence of Clostridium ultunense strain Esp, a syntrophic acetate-oxidizing bacterium. Genome Announc. 2013, 1. [Google Scholar] [CrossRef] [PubMed]
- Zehnder, A.J.; Huser, B.A.; Brock, T.D.; Wuhrmann, K. Characterization of an acetate-decarboxylating, non-hydrogen-oxidizing methane bacterium. Arch. Microbiol. 1980, 124, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Scilifelab. Available online: https://www.scilifelab.se/platforms/ngi/ (accessed on 26 October 2016).
- Andrews, S. FastQC a Quality Control Tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 16 March 2016).
- Chevreux, B.; Wetter, T.; Suhai, S. Genome sequence assembly using trace signals and additional sequence information. Comput. Sci. Biol. Proc. German Conf. Bioinform. 1999, 99, 45–56. [Google Scholar]
- Darling, A.E.; Mau, B.; Perna, N.T. ProgressiveMauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 2010, 5, e11147. [Google Scholar] [CrossRef] [PubMed]
- Boetzer, M.; Henkel, C.V.; Jansen, H.J.; Butler, D.; Pirovano, W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 2011, 27, 578–579. [Google Scholar] [CrossRef] [PubMed]
- Boetzer, M.; Pirovano, W. Toward almost closed genomes with GapFiller. Genome Biol. 2012, 13, R56. [Google Scholar] [CrossRef] [PubMed]
- Gordon, D. Viewing and editing assembled sequences using Consed. Curr. Protoc. Bioinform. 2003, 2. [Google Scholar] [CrossRef]
- Milne, I.; Stephen, G.; Bayer, M.; Cock, P.J.A.; Pritchard, L.; Cardle, L.; Shaw, P.D.; Marshall, D. Using tablet for visual exploration of second-generation sequencing data. Brief. Bioinform. 2013, 14, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Liolios, K.; Mavromatis, K.; Tavernarakis, N.; Kyrpides, N.C. The Genomes on Line Database (GOLD) in 2007: Status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 2008, 36, D475–D479. [Google Scholar] [CrossRef] [PubMed]
- Vallenet, D.; Labarre, L.; Rouy, Z.; Barbe, V.; Bocs, S.; Cruveiller, S.; Lajus, A.; Pascal, G.; Scarpelli, C.; Médigue, C. MaGe: A microbial genome annotation system supported by synteny results. Nucleic Acids Res. 2006, 34, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Hyatt, D.; Chen, G.-L.; Locascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef] [PubMed]
- Bocs, S.; Cruveiller, S.; Vallenet, D.; Nuel, G.; Medigue, C. AMIGene: Annotation of microbial genes. Nucleic Acids Res. 2003, 31, 3723–3726. [Google Scholar] [CrossRef] [PubMed]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the phast phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 1981, 17, 368–376. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Alauzet, C.; Marchandin, H.; Courtin, P.; Mory, F.; Lemee, L.; Pons, J.L.; Chapot-Chartier, M.P.; Lozniewski, A.; Jumas-Bilak, E. Multilocus analysis reveals diversity in the genus Tissierella: Description of Tissierella carlieri sp. nov. in the new class Tissierellia classis nov. Syst. Appl. Microbiol. 2014, 37, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Sun, Y.; Ma, S.; Chen, L.; Zhang, H.; Deng, Y. Isolation and characterization of Keratinibaculum paraultunense gen. nov., sp. nov., a novel thermophilic, anaerobic bacterium with keratinolytic activity. FEMS Microbiol. Lett. 2013, 345, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Paredes-Sabja, D.; Shen, A.; Sorg, J.A. Clostridium difficile spore biology: Sporulation, germination, and spore structural proteins. Trends Microbiol. 2014, 22, 406–416. [Google Scholar] [CrossRef] [PubMed]
- Warner, J.B.; Lolkema, J.S. CcpA-dependent carbon catabolite repression in bacteria. Microbiol. Mol. Biol. Rev. 2003, 67, 475–490. [Google Scholar] [CrossRef] [PubMed]
- Bernson, V.S.M. Acetyl-CoA hydrolase; activity, regulation and physiological, significance of the enzyme in brown adipose tissue from hamster. Eur. J. Biochem. 1976, 67, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Wohlfarth, G.; Buckel, W. A sodium ion gradient as energy source for Peptostreptococcus asaccharolyticus. Arch. Microbiol. 1985, 142, 128–135. [Google Scholar] [CrossRef] [PubMed]
- Juarez, O.; Barquera, B. Insights into the mechanism of electron transfer and sodium translocation of the Na+-pumping NADH:quinone oxidoreductase. Biochim. Biophys. Acta 2012, 1817, 1823–1832. [Google Scholar] [CrossRef] [PubMed]
- Casutt, M.S.; Nedielkov, R.; Wendelspiess, S.; Vossler, S.; Gerken, U.; Murai, M.; Miyoshi, H.; Moller, H.M.; Steuber, J. Localization of ubiquinone-8 in the Na+-pumping NADH:quinone oxidoreductase from Vibrio cholerae. J. Biol. Chem. 2011, 286, 40075–40082. [Google Scholar] [CrossRef] [PubMed]
- Goker, M.; Spring, S.; Scheuner, C.; Anderson, I.; Zeytun, A.; Nolan, M.; Lucas, S.; Tice, H.; Del Rio, T.G.; Cheng, J.F.; et al. Genome sequence of the Thermotoga thermarum type strain (LA3T) from an African solfataric spring. Stand. Genom. Sci. 2014, 9, 1105–1117. [Google Scholar] [CrossRef] [PubMed]
- Biegel, E.; Schmidt, S.; González, J.M.; Müller, V. Biochemistry, evolution and physiological function of the Rnf complex, a novel ion-motive electron transport complex in prokaryotes. Cell. Mol. Life Sci. 2011, 68, 613–634. [Google Scholar] [CrossRef] [PubMed]
- Vignais, P.M.; Billoud, B. Occurrence, classification, and biological function of hydrogenases: An overview. Chem. Rev. 2007, 107, 4206–4272. [Google Scholar] [CrossRef] [PubMed]
- Ahring, B.K.; Schmidt, J.E.; Winther-Nielsen, M.; Macario, A.J.; de Macario, E.C. Effect of medium composition and sludge removal on the production, composition, and architecture of thermophilic (55 °C) acetate-utilizing granules from an upflow anaerobic sludge blanket reactor. Appl. Environ. Microbiol. 1993, 59, 2538–2545. [Google Scholar] [PubMed]
- Hao, L.-P.; Lü, F.; He, P.-J.; Li, L.; Shao, L.-M. Predominant contribution of syntrophic acetate oxidation to thermophilic methane formation at high acetate concentrations. Environ. Sci. Technol. 2011, 45, 508–513. [Google Scholar] [CrossRef] [PubMed]
- Karakashev, D.; Batstone, D.J.; Trably, E.; Angelidaki, I. Acetate oxidation is the dominant methanogenic pathway from acetate in the absence of Methanosaetaceae. Appl. Environ. Microbiol. 2006, 72, 5138–5141. [Google Scholar] [CrossRef] [PubMed]
- Moestedt, J.; Müller, B.; Westerholm, M.; Schnürer, A. Ammonia threshold for inhibition of anaerobic digestion of thin stillage and the importance of organic loading rate. Microb. Biotechnol. 2016, 9, 180–194. [Google Scholar] [CrossRef] [PubMed]
- Muller, B.; Sun, L.; Westerholm, M.; Schnurer, A. Bacterial community composition and fhs profiles of low- and high-ammonia biogas digesters reveal novel syntrophic acetate-oxidising bacteria. Biotechnol. Biofuels 2016, 9, 48. [Google Scholar] [CrossRef] [PubMed]
- Petersen, S.P.; Ahring, B.K. Acetate oxidation in a thermophilic anaerobic sludge-digestor: The importance of non-acetoclastic methanogenesis from acetate. FEMS Microbiol. Ecol. 1991, 9, 149–157. [Google Scholar] [CrossRef][Green Version]
- Shigematsu, T.; Tang, Y.; Kobayashi, T.; Kawaguchi, H.; Morimura, S.; Kida, K. Effect of dilution rate on metabolic pathway shift between aceticlastic and nonaceticlastic methanogenesis in chemostat cultivation. Appl. Environ. Microbiol. 2004, 70, 4048–4052. [Google Scholar] [CrossRef] [PubMed]
- Sprott, G.D.; Patel, G.B. Ammonia toxicity in pure cultures of methanogenic bacteria. Syst. Appl. Microbiol. 1986, 7, 358–363. [Google Scholar] [CrossRef]
- Westerholm, M.; Dolfing, J.; Sherry, A.; Gray, N.D.; Head, I.M.; Schnürer, A. Quantification of syntrophic acetate-oxidizing microbial communities in biogas processes. Environ. Microbiol. Rep. 2011, 3, 500–505. [Google Scholar] [CrossRef] [PubMed]
- Milanovic, V.; Osimani, A.; Taccari, M.; Garofalo, C.; Butta, A.; Clementi, F.; Aquilanti, L. Insight into the bacterial diversity of fermentation woad dye vats as revealed by PCR-DGGE and pyrosequencing. J. Ind. Microbiol. Biotechnol. 2017, 44, 997–1004. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Gao, Q.; Zhang, Q.; Wang, T.; Yue, H.; Wu, L.; Shi, J.; Qin, Z.; Zhou, J.; Zuo, J.; et al. Bacteriophage-prokaryote dynamics and interaction within anaerobic digestion processes across time and space. Microbiome 2017, 5, 57. [Google Scholar] [CrossRef] [PubMed]
- Brüssow, H.; Canchaya, C.; Hardt, W.D. Phages and the evolution of bacterial pathogens: From genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 2004, 68, 560–602. [Google Scholar] [CrossRef] [PubMed]
- Casjens, S.; Palmer, N.; van Vugt, R.; Huang, W.M.; Stevenson, B.; Rosa, P.; Lathigra, R.; Sutton, G.; Peterson, J.; Dodson, R.J.; et al. A bacterial genome in flux: The twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 2000, 35, 490–516. [Google Scholar] [CrossRef] [PubMed]
- Rath, D.; Amlinger, L.; Rath, A.; Lundgren, M. The CRISPR-Cas immune system: Biology, mechanisms and applications. Biochimie 2015, 117, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Horvath, P.; Barrangou, R. CRISPR/Cas, the immune system of bacteria and archaea. Science 2010, 327, 167–170. [Google Scholar] [CrossRef] [PubMed]
- Weinbauer, M.G.; Rassoulzadegan, F. Are viruses driving microbial diversification and diversity? Environ. Microbiol. 2004, 6, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Winter, C.; Smit, A.; Herndl, G.J.; Weinbauer, M.G. Impact of virioplankton on archaeal and bacterial community richness as assessed in seawater batch cultures. Appl. Environ. Microbiol. 2004, 70, 804–813. [Google Scholar] [CrossRef] [PubMed]
- Schink, B. Energetics of syntrophic cooperation in methanogenic degradation. Microbiol. Mol. Biol. Rev. 1997, 61, 262–280. [Google Scholar] [PubMed]
- Sancho, J. Flavodoxins: Sequence, folding, binding, function and beyond. Cell. Mol. Life Sci. 2006, 63, 855–864. [Google Scholar] [CrossRef] [PubMed]
- Buckel, W.; Thauer, R.K. Energy conservation via electron bifurcating ferredoxin reduction and proton/Na+ translocating ferredoxin oxidation. Biochim. Biophys. Acta (BBA)-Bioenerg. 2013, 1827, 94–113. [Google Scholar] [CrossRef] [PubMed]
- Thauer, R.K.; Kaster, A.-K.; Goenrich, M.; Schick, M.; Hiromoto, T.; Shima, S. Hydrogenases from methanogenic archaea, nickel, a novel cofactor, and H2 storage. Annu. Rev. Biochem. 2010, 79, 507–536. [Google Scholar] [CrossRef] [PubMed]
- Ragsdale, S.W.; Pierce, E. Acetogenesis and the Wood–Ljungdahl pathway of CO2 fixation. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2008, 1784, 1873–1898. [Google Scholar] [CrossRef] [PubMed]
- Ye, Q.; Roh, Y.; Carroll, S.L.; Blair, B.; Zhou, J.; Zhang, C.L.; Fields, M.W. Alkaline anaerobic respiration: Isolation and characterization of a novel alkaliphilic and metal-reducing bacterium. Appl. Environ. Microbiol. 2004, 70, 5595–5602. [Google Scholar] [CrossRef] [PubMed]
- Tsai, M.H.; Saier, M.H., Jr. Phylogenetic characterization of the ubiquitous electron transfer flavoprotein families ETF-α and ETF-β. Res. Microbiol. 1995, 146, 397–404. [Google Scholar] [CrossRef]
- Baykov, A.A.; Malinen, A.M.; Luoto, H.H.; Lahti, R. Pyrophosphate-fueled Na+ and H+ transport in prokaryotes. Microbiol. Mol. Biol. Rev. 2013, 77, 267–276. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).