Abstract
Polyploidy has played a crucial role in the evolution of many plant taxa, namely in higher latitudinal zones. Surprisingly, after several decades of an intensive research on polyploids, there are still common polyploid species whose evolutionary history is virtually unknown. Here, we addressed the origin of sweet vernal grass (Anthoxanthum odoratum) using flow cytometry, DNA sequencing, and in situ hybridization-based cytogenetic techniques. An allotetraploid and polytopic origin of the species has been verified. The chromosome study reveals an extensive variation between the European populations. In contrast, an autopolyploid origin of the rarer tetraploid vernal grass species, A. alpinum, has been corroborated. Diploid A. alpinum played an essential role in the polyploidization of both European tetraploids studied.
1. Introduction
Anthoxanthum L., the vernal grass, is a genus of the family Poaceae, comprising annual and perennial taxa that are often self-incompatible [1] and show large diversity in ploidy and morphology (e.g., [2,3]). Despite being a relatively small genus standing out of main research attention within the Poaceae, it offers a great opportunity to study different modes of polyploidization and their consequences for the further evolution of the entire genus.
The genus Anthoxanthum is distributed mainly in temperate and arctic-alpine regions of northern Eurasia, the Mediterranean, Macaronesia, and East Africa. The delimitation of several species within the genus is controversial (e.g., [2,4,5,6,7]), but the most traditional taxonomic concepts [5,6,8] recognize seven Anthoxanthum species in Europe, three annuals (A. aristatum Bois., A. gracile Biv., and A. ovatum Lag.) and four perennials (comprising A. alpinum A. Löve et D. Löve, A. maderense Teppner, A. odoratum L., and A. amarum Brot.). Following Pimentel et al. [9], where two sections of Anthoxanthum are recognized (sect. Ataxia and sect. Anthoxanthum), two additional species (besides all European) are included in the sect. Anthoxanthum in Africa: the East African-endemic A. nivale K.Schum. [10,11] and an endemic species of Ethiopian mountains A. aethiopicum Hedberg [12]. In addition to species appearing in the abovementioned taxonomic literature, one additional taxon (called “Mediterranean diploid” hereafter) was recognized [3,13,14]. Most species are diploids, but polyploidy has been found in some populations of A. alpinum (tetraploids), all populations of A. odoratum (tetraploids), A. amarum (high polyploids), as well as A. aethiopicum (tetraploid) and A. nivale (tetraploids, octoploids, and dodecaploids) [3,10,11,12,13,15,16,17,18].
Sweet vernal grass (A. odoratum) represents the most widespread Anthoxanthum species that grows throughout Europe and has been introduced in North and South America, Australia, Tasmania, and New Zealand. It is also known from KwaZulu-Natal and Eastern Cape in South Africa [19] and is introduced as a fodder grass to some countries.
There has been a long-standing debate as to whether 4x A. odoratum originated via auto- or allo-polyploidization. An autotetraploid origin of A. odoratum has been suggested much earlier than the second type based on the meiotic investigation and high frequency of quadrivalents [20,21,22]. Additionally, Hedberg [23] supported this hypothesis based on the occurrence of triploid individuals within diploid populations. However, after the discovery of another diploid karyotype (“Mediterranean diploid”, [14]) and based on the crossing experiments [24,25], allo-polyploidization turned out to be a more likely hypothesis. Assuming the additivity of genome sizes, Chumová et al. have also found the autopolyploid hypothesis unlikely in the previous study [3].
Mainly on the basis of genetic and phenotypic evidence Borrill [1] has concluded that A. odoratum could have arisen from the hybridization of A. alpinum and A. ovatum or their closely related ancestors. To verify Borrills’ conclusions, Jones [14] did a chromosome study on A. odoratum. His results were that “neither karyotype nor meiotic pairing contains any support for the supposition that A. odoratum is an autotetraploid and unless it be assumed that the species has undergone an extreme degree of chromosome differentiation, it must be concluded that it has a hybrid origin”. As a possible scenario based on the karyotypes of the diploids A. aristatum, A. ovatum, and an unnamed species from Crete (“Mediterranean diploid”), he suggested that A. odoratum could have evolved from such crosses as A. alpinum × A. ovatum, A. alpinum × “Mediterranean diploid” or A. ovatum × “Mediterranean diploid”. Additionally, Teppner [26] and Hedberg [25,27] proposed a hypothesis of doubling after hybridization between two different diploids (A. alpinum and the “Mediterranean diploid”). Our previous data based on the genome sizes [3] have also shown the diploid A. alpinum (5.52 pg) to be one parent of A. odoratum and the “Mediterranean diploid” (7.42 pg) the second one, if we accept the additive model of genome size values. Nevertheless, our data cannot exclude the participation of A. aristatum/ovatum or A. maderense in the genesis of the tetraploid as the second parent of A. odoratum.
To confirm the allopolyploid hypothesis and to suggest possible scenarios of the origin of A. odoratum, we have built on our previous studies [3,13] and involved analyses of nuclear and plastid DNA, and fluorescence and genomic in situ hybridization (FISH and GISH, respectively) that have been used in many studies to resolve the origins of polyploids before (e.g., [28,29,30,31]).
Furthermore, we involved tetraploid A. alpinum in the analyses. This cytotype is distributed in the Southern Jura, on the western part of the Alps and in the Massif Central, where it replaces the diploid form [26,32]. Its autopolyploid origin has been suggested several times [3,24,26,32,33,34] and, therefore, seemed to provide a good comparative material in this case.
In fact, the tetraploid accessions of Anthoxanthum have been analyzed in the frame given by comprehensive study on diploids [13] supplemented with nuclear low-copy gene phylogeny and GISH analyses. Such an approach, combining data on diploids with new data for tetraploids, allows us to reveal the relationships between tetraploids and diploids, determine the most likely origin of tetraploids and elucidate the consequences of polyploid evolution in Anthoxanthum.
2. Materials and Methods
2.1. Plant Material
The Anthoxanthum odoratum and tetraploid A. alpinum material was collected in 2006–2019 throughout the species’ range in Europe (Figure 1, Supplementary Table S1). Ploidy level of all plants was determined by flow cytometry to separate them from the diploid morphologically similar species, and both cytotypes were validated by chromosome counts. In addition to the new collections, all populations of these two species from our previous study [3] were included here and 39 diploid populations (each population represented by one specimen) from Chumová et al. [13] were used in the molecular and genomic analyses.
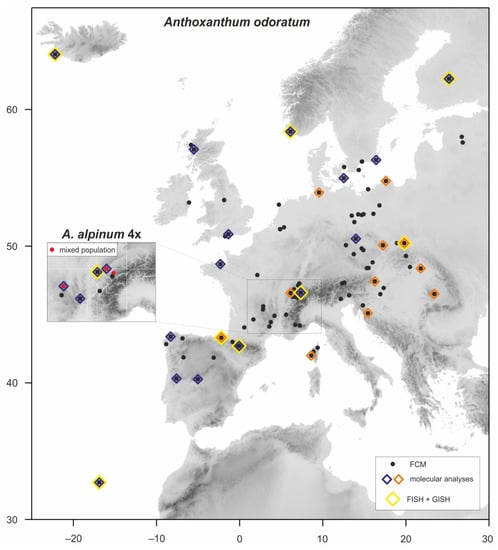
Figure 1.
Map showing the locations of 103 populations of Anthoxanthum odoratum and 8 populations of tetraploid A. alpinum (in the inset cutout) included in the study. A supplementary color legend highlights the use of selected individuals in various analyses.
2.2. Flow Cytometry
Holoploid and monoploid genome sizes [35] were estimated by means of propidium iodide FCM. For each plant, one young, intact leaf, approximately 1 cm in length, was chopped along with an appropriate amount of an internal reference standard using a new razor blade in a Petri dish containing 0.5 mL of ice-cold Otto I buffer (0.1 M citric acid, 0.5% Tween 20) [36,37]. The resulting suspension was filtered through a 42-µm nylon mesh and incubated at room temperature for at least 5 min. After incubation, the suspension was stained by using 1 mL of Otto II buffer (0.4 M Na2HPO4·12 H2O) supplemented with the intercalating fluorescent dye propidium iodide, RNAase IIA (both at the final concentrations of 50 µg·mL−1) and β-mercaptoethanol (2 µL·mL−1). The samples were stained for 5 min at room temperature and analyzed using a Partec CyFlow cytometer (Partec GmbH, Münster, Germany) equipped with a 532 nm diode-pumped solid-state laser Cobolt Samba (Cobolt AB, Solna, Sweden) as the source of excitation light. Fluorescence intensity of 5000 particles was recorded, and the data were analyzed using Partec FloMax Software version 2.4d. Pisum sativum ‘Ctirad’ (2C = 8.76 pg; [38]) served as the reference standard. In total, 275 Anthoxanthum plants (242 individuals of A. odoratum and additional 33 individuals of tetraploid A. alpinum) were subjected to FCM analysis, and their DNA tetraploid level (sensu Suda et al. [39]) was inferred from their estimated DNA C-values, using karyologically verified plants as reference points.
To test the association between genome sizes and geographic locations (latitude, longitude and altitude) of sampled populations the Pearson’s correlation in R 3.6.3 [40] was used.
2.3. Molecular Data Collection
All molecular analyses were performed on 39 selected diploid individuals used in previous study [13], 28 (cpDNA)/23 (GBSSI) selected individuals of A. odoratum and 4 individuals of A. alpinum 4x (Figure 1, Supplementary Table S1). We analyzed two plastid regions (trnL-trnF and rpl32-trnL) and the Granule-bound starch synthase (GBSSI or waxy) gene of nuclear DNA (nrDNA). Total genomic DNA was extracted from 0.5 g of dried leaf tissue using the DNeasy Plant Mini Kit (Qiagen, Valencia, CA, USA). The plastid region trnL-trnF was amplified and sequenced using primers c and f (trnL intron plus trnL-trnF intergenic spacer, hereafter trnL-F; [41]). The PCR conditions for the trnL-F region were as follows: 1 min of denaturation at 94 °C, followed by 34 cycles at 94 °C for 50 s, 50 s at 52.5 °C, 90 s at 72 °C, and a final extension of 10 min at 72 °C. The plastid region rpl32-trnL was amplified and sequenced using primers trnL(UAG) and rpL32-F under the PCR conditions following [42]. Finally, the GBSSI gene was amplified and sequenced using primers F-for and M-bac under the PCR conditions following [43]: 1 min of denaturation at 94 °C, followed by 35 cycles at 94 °C for 50 s, 50 s at 65 °C, 2 min at 72 °C and a final extension of 20 min at 72 °C. Due to the occurrence of within-individual polymorphisms in some of the directly sequenced PCR products of the GBSSI gene, PCR products for GBSSI were cloned using the pGEM-T Easy Vector System (Promega, Madison, WI, USA) following the manufacturer’s instructions but downscaled to half-volume reactions. Details of the procedure were as described by Záveská et al. [44]. A total of 16 colonies from each individual were used as templates for PCR and sequencing. PCRs of all loci were done with MyTaq polymerase (Bioline, London, UK) or AmpliTaq Gold DNA Polymerase (Applied Biosystems, Foster City, CA, USA), following the manufacturer’s instructions, but with the annealing temperatures mentioned above. PCR products were purified with the Jetquick PCR Purification Spin Kit (Genomed, Warsaw, Poland) and directly sequenced at Macrogen Inc. (Seoul, South Korea) or at the DNA sequencing laboratory of the Biological Section of Charles University, with the original PCR primer sets in both directions.
2.4. Molecular Data Analyses (cpDNA and nrDNA)
The two datasets of sequences, i.e., the concatenated chloroplast DNA (cpDNA) dataset and the GBSSI dataset, were aligned independently using MAFFT 7 [45], and then the alignments were improved manually in BioEdit v.7.0.0. [46]. Polymorphisms found in a particular alignment in only one sequence were considered as polymerase errors and corrected [47]. GBSSI sequences from the cloned PCR products were inspected for presence of PCR or in vivo recombinants according to Záveská et al. [44], and a maximum of four GBSSI alleles per tetraploid were kept for further analyses.
Phylogenetic trees were reconstructed using Bayesian analyses (BA) accomplished with MrBayes v.3.1.2. [48]. The datasets were tested for the best substitution model using jModelTest v.0.1.1 [49] with default settings. We chose GTR + G + I model (i.e., MrBayes setting to nst = 6 rates = invgamma) according to the Bayesian information criterion (BIC; [50,51]) for both of them. Two parallel runs with four chains each were used, sampling every 100th tree for 8 million generations for each dataset. The first 10% of samples (8000 trees) were discarded as burn-in, and the remaining 72,000 trees per run were summarized. Nodes with posterior probability (PP) values of 0.95 and above were regarded as significant and those with PP values below 0.95 regarded as non-significant.
2.5. Chromosome Preparations
In total, eight populations of A. odoratum and one population of tetraploid A. alpinum were chosen for FISH and GISH analysis (see Table S1). Mitotic chromosome spreads were prepared from root tips as described by [3]. Briefly, root tips were harvested from germinating seeds or cultivated plants, pre-treated with ice-cold water for 16 h, fixed in ethanol/acetic acid (3:1) fixative for 24 h at 4 °C and stored at −20 °C until further use. Selected root tips were rinsed in distilled water (twice for 5 min) and citrate buffer (10 mM sodium citrate, pH 4.8; twice for 5 min), and digested in 0.3% cellulase, cytohelicase and pectolyase (all Sigma–Aldrich) in citrate buffer at 37 °C for 3 h. After digestion, individual root tips were dissected on a microscope slide in 20 μL acetic acid and spread on the slide placed on a metal hot plate (50 °C) for c. 30 s. Then, the preparation was fixed in freshly prepared ethanol/acetic acid (3:1) fixative by dropping the fixative around the drop of acetic acid and into it. The preparation was dried using a hair dryer and staged using a phase contrast microscope. Chromosome preparations were treated with 100 μg·mL−1. RNase in 2 × sodium saline citrate (SSC; 20 × SSC: 3 M sodium chloride, 300 mM trisodium citrate, pH 7.0) for 60 min and with 0.1 mg·mL−1 pepsin in 0.01 M HCl at 37 °C for 5 min; then postfixed in 4% formaldehyde in 2 × SSC for 10 min, washed in 2 × SSC twice for 5 min, and dehydrated in an ethanol series (70%, 90%, and 100%, 2 min each).
2.6. DNA Probes
The BAC clone T15P10 (AF167571) of Arabidopsis thaliana bearing 45S rRNA gene repeats was used for in situ localization of nucleolar organizer regions (NOR), and the A. thaliana clone pCT 4.2 (M65137), corresponding to a 500 bp 5S rRNA repeat, was used for localization of 5S rDNA loci. For GISH, total genomic DNA (gDNA) was extracted from healthy young leaves according to [52] followed by RNase treatment (50 µg·mL−1). Extracted gDNA was checked for protein, starch or RNA contamination using a Beckmann photospectrometer and ran on a 1% (w/v) agarose gel in 1 × Tris-acetate-EDTA (TAE) buffer. All DNA probes were labeled with biotin-dUTP or digoxigenin-dUTP by nick translation as described in Mandáková and Lysak [53].
2.7. In Situ Hybridization
Selected labelled DNA probes were pooled together, ethanol precipitated, dissolved in a 20 µL mixture containing 50% formamide, 10% dextran sulfate and 2 × SSC, and pipetted onto each of the microscopic slides. The slides were heated at 80 °C for 2 min and incubated at 37 °C overnight. Hybridized probes were visualized through fluorescently-labeled antibodies against biotin-dUTP (red) and digoxigenin-dUTP (green) as in Mandáková and Lysak [53]. Chromosomes were counterstained with 4’,6-diamidino-2-phenylindole (DAPI, 2 µg·mL−1) in Vectashield antifade. Fluorescence signals were analyzed and photographed using a Zeiss Axioimager epifluorescence microscope and a CoolCube camera (MetaSystems, Altlussheim, Germany). Individual images were merged and processed using the Photoshop CS software (Adobe Systems). To test the widest possible combination of putative parental species, we performed three series of GISH (A. alpinum as one parent + one member of diploids of the A. aristatum/ovatum complex, the “Mediterranean diploid” and A. maderense as the second parent) in several replicates using plants of different provenance.
3. Results
3.1. Intraspecific Variation in Nuclear Genome Size
The genome sizes of both tetraploids formed two non-overlapping groups (Figure 2, Supplementary Table S1), which allowed all individuals to be clearly distinguished, although three mixed populations were found. For Anthoxanthum odoratum, 242 plants from 103 populations were analyzed. Mean 2C-values varied from 12.05 to 13.83 pg (mean 12.89 ± 0.34 pg), intraspecific variation in genome size is thus 14.8%. In total, 33 plants from 8 populations were analyzed in Anthoxanthum alpinum, mean 2C-values are lower, ranging between 10.73 and 11.22 pg (mean 11.00 ± 0.14 pg DNA) and observed intraspecific variation is 4.6%.
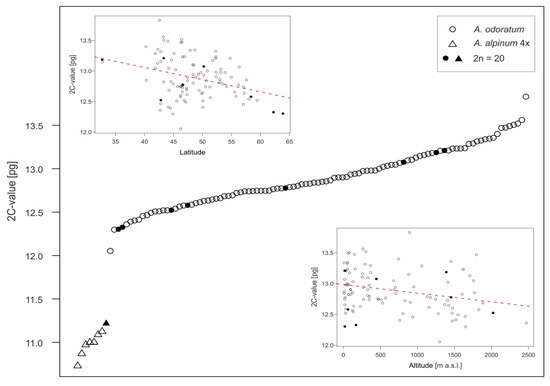
Figure 2.
Absolute genome size variation of two tetraploid species studied, A. odoratum (open circles) and A. alpinum (open triangles). Closed symbols indicate individuals with a counted number of chromosomes. Two insets show the correlation of A. odoratum genome size with latitude (top left) and altitude (bottom right).
In A. odoratum, which was collected from sufficiently large geographic area, the intraspecific variation was non-randomly distributed and showed highly significant negative correlation with latitude and a less pronounced but still significant negative association with altitude (Table 1, Figure 2). Narrow geographic distribution precluded performing the same analyses for 4x A. alpinum.

Table 1.
Pearson’s correlation with corresponding p-values (significant values in bold) for population distribution data (latitude, longitude, and altitude) and mean population genome sizes (2C-values) for Anthoxanthum odoratum.
3.2. Molecular Analyses–Plastid DNA
The lengths of the trnL-trnF and the rpl32-trnL intergenic spacers in the Anthoxanthum full dataset (including 71 specimens plus 3 Hierochloë specimens used as outgroups) were 982 and 823 bp, respectively. The combined alignment was 1805 bp long and comprised 129 variable characters including 103 parsimony-informative sites. GenBank accession numbers are provided in Supplementary Table S1.
Hierarchical genetic relationships resolved by Bayesian inference based on the cpDNA dataset shows three main groups of haplotypes within the ingroup. A strongly supported basal group (PP = 1.00) corresponds to the Mediterranean annual diploid species Anthoxanthum gracile. The first well-supported bigger group (PP = 0.99, “A. alpinum clade”, Figure 3) includes all diploids of A. alpinum as well as all tetraploids of A. alpinum (labels in bold violet in Figure 3).
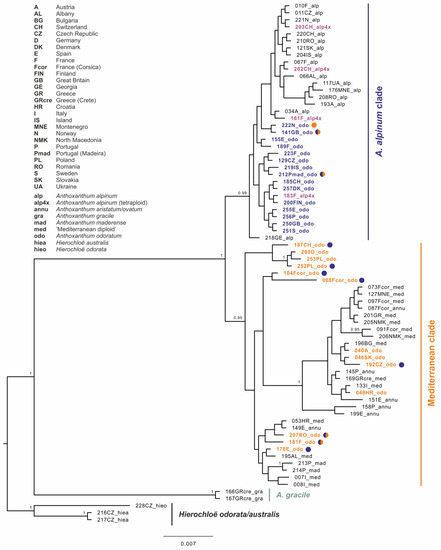
Figure 3.
Maximum clade credibility tree obtained from the Bayesian analysis of cpDNA markers (trnL-trnF and rpl32-trnL intergenic spacers). The numbers by nodes represent Bayesian posterior probabilities if over 0.95. Colored circles indicate individuals carrying a different GBSSI haplotype than the corresponding cpDNA-GBBSI haplotype corresponding to “A. alpinum clade” (blue), “Mediterranean clade” (orange) and both clades (half-colored circles).
It also includes a large proportion of tetraploid A. odoratum (labels in bold blue in Figure 3). In sister position to the A. alpinum clade, haplotypes of all populations of the “Mediterranean diploid”, annual taxa and A. maderense were resolved with the rest of A. odoratum (“Mediterranean clade”, PP = 0.95, A. odoratum labels in bold orange in Figure 3). Several smaller strongly supported groups can be found in this group compared to the poorly structured A. alpinum clade, but do not usually correspond to the different species.
3.3. Molecular Analyses–nrDNA–GBSSI
GBSSI alignment in the Anthoxanthum full dataset (68 specimens/90 clones + 2 Hierochloë as outgroups) was 1166 bp long and comprised 383 variable characters including 268 parsimony-informative sites. In 11 individuals, two different sequence-types were detected and used after cloning analyses. GenBank accession numbers are provided in Supplementary Table S1.
The topology of the Bayesian gene tree based on GBSSI data (Figure 4) revealed three main ingroup haplotype groups with similar pattern to those based on plastid data, but with additional “Iberian annual clade” as a sister group to A. gracile. Therefore, we kept similar naming and coloring patterns, and we highlight the differences in GBSSI patterns in A. odoratum from those of cpDNA in the Figure 3, where colored circles indicate different positions of the individual within the GBSSI tree. As in the chloroplast tree, A. gracile exhibits on the base, all A. alpinum plants belong to the “A. alpinum clade” and all diploid Mediterranean taxa to the “Mediterranean clade” except for a few annual individuals forming separate lineages. Anthoxanthum odoratum splits evenly in both bigger clades and each of the two clones in a different one in some cases (individuals from Great Britain, France, Madeira–Portugal, and Romania; indicated by arrows in Figure 4).
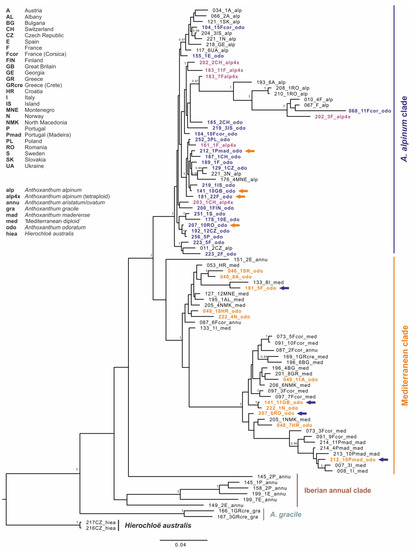
Figure 4.
Maximum clade credibility tree obtained from the Bayesian analysis of nuclear DNA marker GBSSI. The numbers by nodes represent Bayesian posterior probabilities if over 0.95. Colored arrows indicate individuals that exhibit two alleles of GBSSI in different clades.
3.4. Chromosome Localization of rDNA Loci
All of the analyzed Anthoxanthum populations have a tetraploid chromosome number (2n = 4x = 20; Figure 5 and Figure 6, Figure S1). We uncovered an extensive inter-population variation in the number of 45S (5–9 loci) and 5S rDNA (4–7 loci) and their position on chromosomes (Figure 5, Supplementary Table S1). Geographically more closely related populations show a more similar rDNA pattern, e.g., populations from the Pyrenees and from Madeira (Figure 5B–D), or populations from Norway and Finland (Figure 5G,H) which supports their closer relationship.
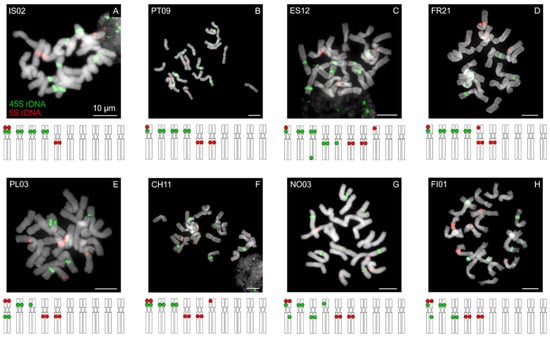
Figure 5.
Chromosome localization of rDNA in Anthoxanthum odoratum. Mitotic chromosome complements of A. odoratum (all 2n = 4x = 20) hybridized with 45S (green fluorescence) and 5S (red) rDNA probes. Chromosomes were counterstained with DAPI. Scale bars, 10 µm. Populations from: Island (A), Madeira–Portugal (B), Spain (C), France (D), Poland (E), Switzerland (F), Norway (G), Finland (H) locality details in Supplementary Table S1.
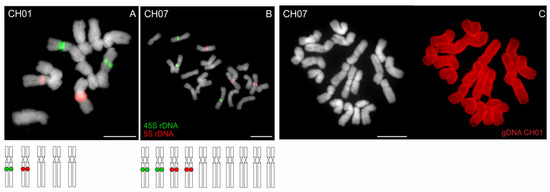
Figure 6.
Chromosome localization of rDNA and GISH in Anthoxanthum alpinum. (A) Diploid population of A. alpinum (2n = 10; CH01 from Switzerland) exhibits one 45S- (green fluorescence) and one 5S rDNA-bearing (red fluorescence) chromosome pair. (B) Tetraploid population of A. alpinum (2n = 20; CH07, Supplemetary Table S1) shows exactly twice the number of rDNA loci compared to the diploid cytotype supporting an autopolyploid origin of the tetraploid. (C) GISH in tetraploid A. alpinum (CH07) using a probe corresponding to gDNA of diploid A. alpinum (CH01, red fluorescence) further suggests an autopolyploid origin of the tetraploid. Chromosomes were counterstained with DAPI. Scale bars, 10 µm.
3.5. GISH in Polyploid Anthoxanthum Species’
To uncover the origin of A. odoratum, labelled gDNA of all putative diploid parental species was hybridized on mitotic chromosomes of eight tetraploid populations. Following progenitors were tested: diploid A. alpinum (RO03, IS01 or UA01), and one of the three Mediterranean diploid species (or its ancestor/s)–“Mediterranean diploid” (BG03, GR04), A. aristatum/ovatum (ES07, ES09), or A. maderense (PT01). The hybridization pattern was highly variable between individual populations of A. odoratum, including numerous intergenomic translocations (Figure 7). GISH probe corresponding to A. alpinum (red fluorescence in Figure 7 and Figure S1) covered from ¼ (PT09 and Norwegian NO01) up to ½ (in the Icelandic IS02) of the allopolyploid genome, i.e., 5–10 chromosomes were detected using gDNA of A. alpinum. The Mediterranean-like genome fraction (green fluorescence in Figure 7 and Figure S1) virtually forms from ½ to ¾ of the hybrid genome (i.e., 10–15 chromosomes) regardless of which of the Mediterranean gDNA probe was used.
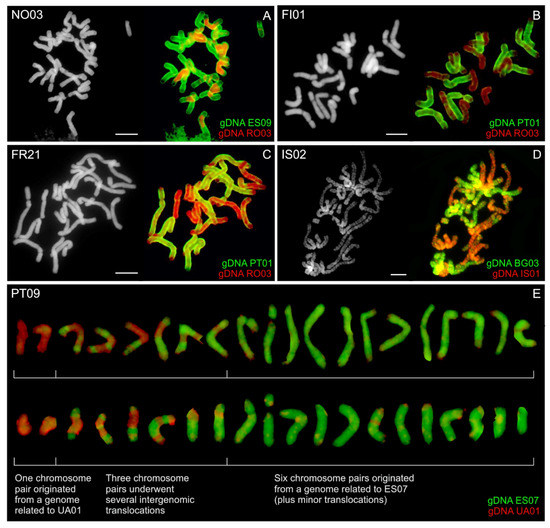
Figure 7.
GISH in Anthoxanthum odoratum. Mitotic chromosomes of five selected population of A. odoratum ((A)—NO03, Norway; (B)—FI01, Finland; (C)—FR21, France; (D)—IS02, Iceland; (E)—PT09, Madeira–Portugal) were hybridized using gDNA of A. alpinum (RO03, IS01 or UA01, red fluorescence in (A–E) and A. aristatum/ovatum (ES09 or ES07; green fluorescence in (A,E)), A. maderense (PT01; green fluorescence in (B,C)) and “Mediterranean diploid” (BG03, green fluorescence in (D)). Chromosomes were counterstained with DAPI. Scale bars, 10 µm.
4. Discussion
4.1. Intraspecific Variation in Nuclear Genome Size
The issue of intraspecific variation in genome size has been of interest for a long time and still remains somewhat controversial (e.g., [54,55]). In Anthoxanthum, intraspecific variation in genome size was observed in all European taxa [3], A. odoratum is no exception and with the intraspecific variation 14.8% (Figure 2), it belongs to those species with greater variability.
Several mechanisms are likely responsible for the observed variation in genome size, at least part of the variation can be ascribed to chromosomal heterogeneity, because of presence of aneuploidy and the supernumerary chromosomes was revealed in Anthoxanthum in previous studies [3,56]. On the other hand, there is almost no correlation between the number of B-chromosomes and genome size variability in Anthoxanthum annuals [56], indicating that the changes mainly in A-chromosomes are responsible for a huge fraction of genome size variability. It is consistent with the fact that we have not found any B-chromosomes during chromosome counting in this study.
The relatively continuous variation in genome size suggested that differences in the size of individual chromosomes have to be involved. Interestingly, the intraspecific variation in genome size in A. odoratum is non-randomly distributed and showed highly significant negative correlation with latitude and a less pronounced but still significant negative association with altitude (Figure 2). This finding is highly congruent with the pattern revealed in other diploid Anthoxanthum species [3].
Because nuclear genome size can influence several phenotypic and developmental characteristics irrespective of the information coded in the DNA (i.e., the nucleotypic effect; [57]), it can be speculated that the variation found in nuclear genome size represents adaptation to different environmental conditions. Such adaptation may underlie correlations between genome size and abiotic conditions found in several grass genera as Dactylis [58,59], Festuca [60], Koeleria [61], or Zea [62]. Obviously, an important source of genome size variability in grasses, i.e., the differential proliferation of the repetitive DNA elements (e.g., [63,64,65]), must also be taken into account. However, data for Anthoxanthum are lacking.
4.2. Intraspecific Variation in Molecular and Cytogenomic Analyses
Striking intraspecific variation was found in A. odoratum in molecular, as well as cytogenomic analyses, which is not surprising due to its polyploid origin. The sequences of A. odoratum are highly polymorphic and are present in the clades with A. alpinum, as well as with the diploid Mediterranean taxa in the molecular analyses (Figure 3 and Figure 4). In two thirds of cases, individuals exhibit the same pattern within the chloroplast and the nuclear maximum clade credibility tree. However, in the remaining cases, individuals are placed into “A. alpinum clade” in the chloroplast tree and into the “Mediterranean clade” in the nuclear tree and vice versa, or their GBSSI clones fall into both clades (Figure 3 and Figure 4). In addition, the “Mediterranean clade” is further subdivided into more or less well-supported groups which have been omitted here for simplicity, but the topology depicts relationships among geographically isolated diploid populations [13], and A. odoratum can be found in each of them.
In contrast, tetraploid A. alpinum shares the pattern in chloroplast and nuclear trees, and in all cases can be found within the “A. alpinum clade”.
In terms of cytogenomics, apart from the fact that 20 chromosomes were counted in all samples analyzed, it is difficult to find two sweet vernal grasses with identical rDNA- and GISH-pattern which demonstrates an extensive variation in chromosome structure in A. odoratum. Our cytogenetic data suggests that the allotetraploid experienced post-polyploid population-specific intergenomic translocations, probably accompanied by a number of other chromosomal rearrangements that shaped today’s karyotypes. Our findings are in agreement with Jones who pointed out an extreme degree of chromosome differentiation in A. odoratum [14]. This study is only a stepping stone for subsequent detailed cytogenetic examination using chromosome-specific markers (e.g., oligopainting).
Although the variability in rDNA location and number among populations remains unresolved, a similar pattern has been observed in some species of the genus Paspalum [66] and Deschampsia [67,68]. Complexity may be further increased by differences in rDNA variation across the species’ range, which has also been documented in the latter genus ([69] as opposed to [68]).
4.3. On the Origin of Anthoxanthum odoratum
Although initially it was thought that A. odoratum could be autopolyploid, the opinion slowly began to change and everything pointed to the allopolyploid origin of this taxon, as outlined in the introduction. To confirm the hypothesis of allopolyploid origin, we tried to involve analyses of plastid and nuclear DNA (Figure 3 and Figure 4) that have shown allopolyploid and polytopic origin of A. odoratum, as the species exhibits in both main clades (and all subclades in the Mediterranean one) within the trees. Moreover, some of the samples carried at least two alleles of the GBSSI waxy-gene occurring in both (A. alpinum and Mediterranean) clades. The observation has similar conclusions to those of Pimentel et al. (2013), with no clear evidence of the parental species. Then we have involved the fluorescence and genomic in situ hybridization (Figure 5 and Figure 7, Figure S1) that have been used in many studies to resolve the origins of polyploids before (e.g., [31]) and in Anthoxanthum species by Drapikowska et al. [70]. rDNA pattern (FISH) is highly variable within individual populations of A. odoratum (Figure 5) as mentioned above, and only suggests close relationships of some populations. Even if we combine the rDNA pattern of the potential diploid ancestors (diploid A. alpinum and the Mediterranean diploid taxa; [13]), we are not able to get the pattern of any A. odoratum, ruling out any suggestion of a likely origin.
GISH data (Figure 7) indicate allopolyploid origin and have a highly variable pattern within individual populations, too. On its basis, with a number of visible chromosomal translocations, we can only assume that one of the parental species is A. alpinum or its ancestor and the other one is one of the Mediterranean diploids or their common ancestor. The pattern is virtually unchanged on each individual of A. odoratum using genomic probes from the three species mentioned, so we can just take into consideration the annual life cycle of A. aristatum/ovatum and geographical distance of A. maderense to suggest that the most likely parent is the “Mediterranean diploid”. The pattern points to various scenarios of the tetraploid origin ranging from equal to noticeably unbalanced contribution of both parents. In two thirds of accessions analyzed it theoretically seems to possible fusion of a reduced gamete of A. alpinum-like genome (bearing five chromosomes) with an unreduced gamete of “Mediterranean diploid”-like genome (bearing 15 chromosomes; Figure 7E). On the other hand, the possible long existence of tetraploids may have contributed to the fact that the current GISH pattern does not correspond to the state at their origin, but rather to their later evolution.
Current distributional ranges of putative parental taxa points to hybridization events in the past that could begin by divergence of A. alpinum (2.02 Mya; [13]). Interestingly, similar timing of polyploidization events was shown by Tusime et al. [11] in the African polyploid Anthoxanthum nivale.
4.4. Autopolyploid Origin of Anthoxanthum alpinum
The autopolyploid origin of tetraploid A. alpinum was first suggested on the basis of the analyses of its karyotype by Teppner [26] and by Hedberg [24]. In addition, investigations on the flowering phenology [32] and on the sensitivity to a specific rust showed greater similarity in the two cytotypes of A. alpinum, compared to related species “Mediterranean diploid” and A. odoratum [33], thus corroborating the autopolyploid origin of A. alpinum. The close genetic similarity testing in 1999 by Zeroual-Humbert-Droz and Felber [34] using isozymes between the diploid and tetraploid cytotype confirmed the autopolyploid origin of the tetraploid, too. The study of Chumová et al. [3] provides additional support for this hypothesis by revealing almost identical mean monoploid genome sizes of diploid (1Cx = 2.76 pg) and tetraploid (1Cx = 2.75 pg) cytotypes of A. alpinum. The position within the “A. alpinum clade” on chloroplast and nuclear tree (Figure 3 and Figure 4), as well as the pattern shown in Figure 6 as the result of fluorescence and genomic in situ hybridization on diploid and tetraploid A. alpinum from Switzerland, also supports this hypothesis.
4.5. Consequences of Allopolyploidy in A. odoratum
Despite long-lasting debate on the polyploid origin of A. odoratum, our study represents the first rigorous evidence on allotetraploidy in its evolutionary history. We have shown the contribution of A. alpinum (or its most recent common ancestor) as one parental species and the participation of a second, as yet ambiguous parental diploid species from the Mediterranean. The gathered evidence points very likely to so-called “Mediterranean diploid” (this study and [3,13]) that was repeatedly recorded in the past as diploid A. odoratum [16,24,32,33]. This name can no longer be used, because we have definitively rejected the hypothesis of an autotetraploid origin of A. odoratum. The species name “odoratum” is restricted to the allotetraploid taxon only. Unlike A. alpinum, where the species name “alpinum” can be used for both (diploid and tetraploid) cytotypes due to its autopolyploid origin. In line with our previous studies [3,13], we call for a thorough and detailed description of the “Mediterranean diploid” and its proper recognition for science as a new species. Its taxonomic value is well supported by vicariant species range compared to tetraploid A. odoratum and other diploid species [13], by perennial life-cycle [14] and the genome size [3].
5. Conclusions
By combining flow cytometry, molecular analyses and fluorescence and genomic in situ hybridization, we pointed out the large intraspecific variability in Anthoxanthum odoratum and were able to confirm its allopolyploid origin. We also provide further evidence for the autopolyploid origin of tetraploid. A. alpinum. We have suggested the possible parental species of both tetraploids, noting that in some cases these are rather the ancestors of existing species, so their exact identification is probably impossible.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/genes12070966/s1, Table S1: Overview of Anthoxanthum odoratum populations used in the study. In addition to the georeferenced data, the results of the genome size estimation and the codes used in each analysis are added for each population. Moreover, the table includes information on diploid accessions from previous studies used for phylogenetic analyses; Figure S1: The GISH pattern of particular individuals of A. odoratum and one tetraploid A. alpinum (bottom right pane) obtained by hybridization of gDNA from different combinations of putative parental species. Individuals from each population of A. odoratum are in rows, while the three different parental combinations are arranged in columns.
Author Contributions
Z.C. and P.T. conceived and designed the study and collected plant material. Z.C., T.M. and P.T. performed the laboratory work and subsequent analyses. Z.C. drafted the manuscript with contributions from all the co-authors. All authors have read and agreed to the published version of the manuscript.
Funding
The research was financially supported by Charles University (GAUK projects 9661/2009 and 5414/2013), The Czech Science Foundation (grant no. 19-03442S) and CEITEC 2020 project (grant no. LQ1601). Additional support was supplied by long-term research development project no. RVO 67985939 provided by the Academy of Sciences of the Czech Republic.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study (DNA sequences) are provided in GenBank under accession numbers MZ399726-MZ399797 and MZ408552-MZ408642 (details in Supplementary Table S1).
Acknowledgments
We are grateful to F. Kolář, P. Kúr, M. Pimentel, S. Píšová, J. Prančl, E. Sahuquillo, R. Urfusová, T. Urfus, and E. Záveská for providing some Anthoxanthum samples, to V. Kučabová, L. Flašková, E. Záveská, and K. Krak for the technical support in the laboratory or during analyses and to Štěpánka Hrdá and Blanka Hamplová from OMICS Biocev for their help with sequencing.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Borrill, M. Experimental Studies of Evolution in Anthoxanthum (Gramineae). Genetica 1963, 34, 183–210. [Google Scholar] [CrossRef]
- Pimentel, M.; Sahuquillo, E. Relationships among Some Populations of Anthoxanthum alpinum and A. odoratum (Poaceae, Pooideae): A Morphological/Anatomical Approach. Aliso 2007, 23, 472–484. [Google Scholar] [CrossRef][Green Version]
- Chumová, Z.; Krejčíková, J.; Mandáková, T.; Suda, J.; Trávníček, P. Evolutionary and Taxonomic Implications of Variation in Nuclear Genome Size: Lesson from the Grass Genus Anthoxanthum (Poaceae). PLoS ONE 2015, 10, e0133748. [Google Scholar] [CrossRef] [PubMed]
- Paunero, E. Las Especies Españoles Del Género Anthoxanthum L. An. Inst. Botánico AJ Cavanilles 1953, 12, 401–442. [Google Scholar]
- Tutin, T. Anthoxanthum L. In Flora Europaea; Tutin, T., Heywood, V., Burges, N., Moore, D., Valentine, D., Walters, S., Webb, D., Eds.; Cambridge University Press: Cambridge, UK, 1980; Volume 5. [Google Scholar]
- Teppner, H. Anthoxanthum maderense Spec. Nova and A. odoratum (Poaceae-Aveneae) from Madeira and Their Chromosome Morphology. Phyton Ann. Rei Bot. 1998, 38, 307–321. [Google Scholar]
- Pimentel, M.; Catalan, P.; Sahuquillo, E. Morphological and Molecular Taxonomy of the Annual Diploids Anthoxanthum aristatum and A. ovatum (Poaceae) in the Iberian Peninsula. Evidence of Introgression in Natural Populations. Bot. J. Linn. Soc. 2010, 164, 53–71. [Google Scholar] [CrossRef][Green Version]
- Valdés Valdés, B. Revisión de Las Especies Anuales Del Género Anthoxanthum (Graminae). Lagascalia 1973, 3, 99–141. [Google Scholar]
- Pimentel, M.; Sahuquillo, E.; Torrecilla, Z.; Popp, M.; Catalan, P.; Brochmann, C. Hybridization and Long-Distance Colonization at Different Time Scales: Towards Resolution of Long-Term Controversies in the Sweet Vernal Grasses (Anthoxanthum). Ann. Bot. 2013, 112, 1015–1030. [Google Scholar] [CrossRef]
- Clayton, W.D. Gramineae (Part I). In Flora of Tropical East Africa; Redhead, E.M., Polhill, E.M., Eds.; Ministry for Overseas Development: London, UK, 1970. [Google Scholar]
- Tusiime, F.M.; Gizaw, A.; Wondimu, T.; Masao, C.A.; Abdi, A.A.; Muwanika, V.; Trávníček, P.; Nemomissa, S.; Popp, M.; Eilu, G.; et al. Sweet Vernal Grasses (Anthoxanthum) Colonized African Mountains along Two Fronts in the Late Pliocene, Followed by Secondary Contact, Polyploidization and Local Extinction in the Pleistocene. Mol. Ecol. 2017, 26, 3513–3532. [Google Scholar] [CrossRef]
- Hedberg, I. Cytotaxonomic Reconnaissance of Tropical African Anthoxanthum L. (Gramineae). Bot. Not. 1976, 129, 85–90. [Google Scholar]
- Chumová, Z.; Záveská, E.; Mandáková, T.; Krak, K.; Trávníček, P. The Mediterranean: The Cradle of Anthoxanthum (Poaceae) Diploid Diversity. Ann. Bot. 2017, 120, 285–302. [Google Scholar] [CrossRef] [PubMed]
- Jones, K. Chromosomes and the Origin of Anthoxanthum odoratum L. Chromosoma 1964, 15, 248–274. [Google Scholar] [CrossRef]
- Hedberg, O. Afro-Alpine Vascular Plants. Symb. Bot. Ups. 1957, 15, 54. [Google Scholar]
- Felber, F. Distribution des cytodèmes d’Anthoxanthum odoratum L. s lat. en Suisse. Les relations Alpes-Jura. Bot. Helvetica 1986, 96, 145–158. [Google Scholar]
- Phillips, S. Poaceae (Graminae). In Flora of Ethiopia and Eritreia; Hedberg, I., Edwards, S., Eds.; Swedish Science Press: Upsalla, Sweden, 1995; pp. 40–42. [Google Scholar]
- Teppner, H. Poaceae in the Greenhouses of the Botanic Garden Institute of Botany in Graz (Austria, Europe). Fritschiana 2002, 31, 1–42. [Google Scholar]
- Mashau, A.C. A Synopsis of Anthoxanthum (Poaceae: Pooideae: Poeae) in Southern Africa and Description of a New Subspecies. Kew Bull. 2016, 71, 18. [Google Scholar] [CrossRef][Green Version]
- Katterman, G. Über Die Bildung Polyvalenter Chromosomenverbände Bei Einigen Gramineen. Planta 1931, 12, 734–774. [Google Scholar] [CrossRef]
- Parthasarathy, N. Cytogenetical Studies in Oryzeae and Phalaridineae. Ann. Bot. Lond. NS 1939, 3, 43–76. [Google Scholar] [CrossRef]
- Östergren, G. Chromosome Numbers in Anthoxanthum. Hereditas 1942, 33, 242–243. [Google Scholar]
- Hedberg, I. Cytotaxonomic Studies on Anthoxanthum odoratum L. s. Lat. 2. Investigations of Some Swedish and of a Few Swiss Population Samples. Symb. Bot. Ups. 1967, 18, 5–97. [Google Scholar]
- Hedberg, I. Cytotaxonomic Studies on Anthoxanthum odoratum L. s. Lat. IV. Karyotypes, Meiosis and Origin of Tetraploid A. odoratum. Hereditas 1970, 48, 471–502. [Google Scholar]
- Hedberg, I. The Genesis of Tetraploid Anthoxanthum odoratum. Symb. Bot. Ups. 1986, 27, 147–154. [Google Scholar]
- Teppner, H. Caryotypes of European, Perennial Species of Gramineae Genus Anthoxanthum. Osterreichische Bot. Z. 1970, 118, 280–292. [Google Scholar] [CrossRef]
- Hedberg, I. Morphological, Cytotaxonomic and Evolutionary Studies in Anthoxanthum odoratum L. s. Lat.—A Critical Review. Sommerfeltia 1990, 11, 97–107. [Google Scholar]
- Hodkinson, T.R.; Chase, M.W.; Takahashi, C.; Leitch, I.J.; Bennett, M.D.; Renvoize, S.A. The Use of DNA Sequencing (ITS and TrnL-F), AFLP, and Fluorescent in Situ Hybridization to Study Allopolyploid Miscanthus (Poaceae). Am. J. Bot. 2002, 89, 279–286. [Google Scholar] [CrossRef]
- Ellneskog-Staam, P.; Salomon, B.; von Bothmer, R.; Anamthawat-Jonsson, K. The Genome Composition of Hexaploid Psammopyrum athericum and Octoploid Psammopyrum pungens (Poaceae: Triticeae). Genome 2003, 46, 164–169. [Google Scholar] [CrossRef] [PubMed]
- Loureiro, J.; Kopecky, D.; Castro, S.; Santos, C.; Silveira, P. Flow Cytometric and Cytogenetic Analyses of Iberian Peninsula Festuca Spp. Plant Syst. Evol. 2007, 269, 89–105. [Google Scholar] [CrossRef]
- Mahelka, V.; Kopecký, D. Gene Capture from across the Grass Family in the Allohexaploid Elymus repens (L.) Gould (Poaceae, Triticeae) as Evidenced by ITS, GBSSI, and Molecular Cytogenetics. Mol. Biol. Evol. 2010, 27, 1370–1390. [Google Scholar] [CrossRef]
- Felber, F. Phenology of Flowering in Diploid and Tetraploid Populations of Anthoxanthum alpinum and Anthoxanthum odoratum. Can. J. Bot. Rev. Can. Bot. 1988, 66, 2258–2264. [Google Scholar] [CrossRef]
- Felber, F. Sensitivity of the Four Cytodemes of Anthoxanthum odoratum L. s. Lat. (Poaceae) to Puccinia sardonensis Gaumann (Uredinales). Taxon 1987, 36, 573–577. [Google Scholar] [CrossRef]
- Zeroual-Humbert-Droz, C.; Felber, F. Evidence from Isozyme Analysis of Autopolyploidy in Anthoxanthum alpinum A. & D. Löve. Bot. Helvetica 1999, 109, 217–227. [Google Scholar]
- Greilhuber, J.; Dolezel, J.; Lysak, M.A.; Bennett, M.D. The Origin, Evolution and Proposed Stabilization of the Terms “genome Size” and “C-Value” to Describe Nuclear DNA Contents. Ann. Bot. 2005, 95, 255–260. [Google Scholar] [CrossRef]
- Otto, S.P. The Evolutionary Consequences of Polyploidy. Cell 2007, 131, 452–462. [Google Scholar] [CrossRef]
- Doležel, J.; Greilhuber, J.; Suda, J. Estimation of Nuclear DNA Content in Plants Using Flow Cytometry. Nat. Protoc. 2007, 2, 2233–2244. [Google Scholar] [CrossRef] [PubMed]
- Trávníček, P.; Ponert, J.; Urfus, T.; Jersáková, J.; Vrána, J.; Hřibová, E.; Doležel, J.; Suda, J. Challenges of Flow-Cytometric Estimation of Nuclear Genome Size in Orchids, a Plant Group with Both Whole-Genome and Progressively Partial Endoreplication. Cytom. A 2015, 87A, 958–966. [Google Scholar] [CrossRef]
- Suda, J.; Krahulcová, A.; Trávníček, P.; Krahulec, F. Ploidy Level versus DNA Ploidy Level: An Appeal for Consistent Terminology. Taxon 2006, 55, 447–450. [Google Scholar] [CrossRef]
- R Core Team. A Language and Environment for Statistical Computing R Foundation for Statistical Computing; R Core Team: Vienna, Austria, 2020. [Google Scholar]
- Taberlet, P.; Gielly, L.; Pautou, G.; Bouvet, J. Universal Primers for Amplification of 3 Noncoding Regions of Chloroplast DNA. Plant Mol. Biol. 1991, 17, 1105–1109. [Google Scholar] [CrossRef] [PubMed]
- Shaw, J.; Lickey, E.B.; Schilling, E.E.; Small, R.L. Comparison of Whole Chloroplast Genome Sequences to Choose Noncoding Regions for Phylogenetic Studies in Angiosperms: The Tortoise and the Hare III. Am. J. Bot. 2007, 94, 275–288. [Google Scholar] [CrossRef] [PubMed]
- Mason-Gamer, R.J.; Weil, C.F.; Kellogg, E.A. Granule-Bound Starch Synthase: Structure, Function, and Phylogenetic Utility. Mol. Biol. Evol. 1998, 15, 1658–1673. [Google Scholar] [CrossRef] [PubMed]
- Záveská, E.; Fér, T.; Šída, O.; Krak, K.; Marhold, K.; Leong-Škorničková, J. Phylogeny of Curcuma (Zingiberaceae) Based on Plastid and Nuclear Sequences: Proposal of the New Subgenus Ecomata. Taxon 2012, 61, 747–763. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Hall, T. BioEdit, Biological Sequence Alignment Editor for Win95/98/NT/2K/XP, Version 7.0.4.1. 2004. Available online: http://www.Mbio.Ncsu.Edu/BioEdit/Bioedit.Html (accessed on 12 December 2020).
- Popp, M.; Erixon, P.; Eggens, F.; Oxelman, B. Origin and Evolution of a Circumpolar Polyploid Species Complex in Silene (Caryophyllaceae) Inferred from Low Copy Nuclear RNA Polymerase Introns, RDNA, and Chloroplast DNA. Syst. Bot. 2005, 30, 302–313. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian Inference of Phylogenetic Trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [PubMed]
- Posada, D.J. ModelTest: Phylogenetic Model Averaging. Mol. Biol. Evol. 2008, 25, 1253–1256. [Google Scholar] [CrossRef]
- Schwarz, G. Estimating the Dimension of a Model. Ann. Stat. 1978, 6, 461–464. [Google Scholar] [CrossRef]
- Posada, D.; Buckley, T.R. Model Selection and Model Averaging in Phylogenetics: Advantages of Akaike Information Criterion and Bayesian Approaches over Likelihood Ratio Tests. Syst. Biol. 2004, 53, 793–808. [Google Scholar] [CrossRef] [PubMed]
- Dellaporta, S.L.; Wood, J.; Hicks, J.B. A Plant DNA Minipreparation: Version II. Plant Mol. Biol. Report. 1983, 1, 19–21. [Google Scholar] [CrossRef]
- Mandáková, T.; Lysak, M.A. Painting of Arabidopsis chromosomes with chromosome-specific BAC clones. In Current Protocols in Plant Biology; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2016; ISBN 978-1-119-07736-7. [Google Scholar]
- Greilhuber, J. Intraspecific Variation in Genome Size in Angiosperms: Identifying Its Existence. Ann. Bot. 2005, 95, 91–98. [Google Scholar] [CrossRef]
- Šmarda, P.; Bureš, P. Understanding Intraspecific Variation in Genome Size in Plants. Preslia 2010, 82, 41–61. [Google Scholar]
- Chumová, Z.; Mandáková, T.; Trávníček, P. Are B-Chromosomes Responsible for the Extraordinary Genome Size Variation in Selected Anthoxanthum Annuals? Plant Syst. Evol. 2016, 302, 731–738. [Google Scholar] [CrossRef]
- Bennett Michael David Nuclear DNA Content and Minimum Generation Time in Herbaceous Plants. Proc. R. Soc. Lond. B Biol. Sci. 1972, 181, 109–135. [CrossRef]
- Creber, H.M.C.; Davies, M.S.; Francis, D.; Walker, H.D. Variation in DNA C Value in Natural Populations of Dactylis glomerata L. New Phytol. 1994, 128, 555–561. [Google Scholar] [CrossRef]
- Reeves, G.; Francis, D.; Davies, M.S.; Rogers, H.J.; Hodkinson, T.R. Genome Size Is Negatively Correlated with Altitude in Natural Populations of Dactylis glomerata. Ann. Bot. 1998, 82, 99–105. [Google Scholar] [CrossRef]
- Šmarda, P.; Bureš, P. Intraspecific DNA Content Variability in Festuca pallens on Different Geographical Scales and Ploidy Levels. Ann. Bot. 2006, 98, 665–678. [Google Scholar] [CrossRef]
- Pečinka, A.; Suchánková, P.; Lysak, M.A.; Trávníček, B.; Doležel, J. Nuclear DNA Content Variation among Central European Koeleria Taxa. Ann. Bot. 2006, 98, 117–122. [Google Scholar] [CrossRef]
- Laurie, D.A.; Bennett, M.D. Nuclear DNA Content in the Genera Zea and Sorghum. Intergeneric, Interspecific and Intraspecific Variation. Heredity 1985, 55, 307–313. [Google Scholar] [CrossRef]
- Stritt, C.; Wyler, M.; Gimmi, E.L.; Pippel, M.; Roulin, A.C. Diversity, Dynamics and Effects of Long Terminal Repeat Retrotransposons in the Model Grass Brachypodium distachyon. New Phytol. 2020, 227, 1736–1748. [Google Scholar] [CrossRef]
- Zwyrtková, J.; Němečková, A.; Čížková, J.; Holušová, K.; Kapustová, V.; Svačina, R.; Kopecký, D.; Till, B.J.; Doležel, J.; Hřibová, E. Comparative Analyses of DNA Repeats and Identification of a Novel Fesreba Centromeric Element in Fescues and Ryegrasses. BMC Plant Biol. 2020, 20, 280. [Google Scholar] [CrossRef]
- Giraud, D.; Lima, O.; Huteau, V.; Coriton, O.; Boutte, J.; Kovarik, A.; Leitch, A.R.; Leitch, I.J.; Aïnouche, M.; Salmon, A. Evolutionary Dynamics of Transposable Elements and Satellite DNAs in Polyploid Spartina Species. Plant Sci. 2021, 302, 110671. [Google Scholar] [CrossRef]
- Vaio, M.; Speranza, P.; Valls, J.F.; Guerra, M.; Mazzella, C. Localization of the 5S and 45S RDNA Sites and CpDNA Sequence Analysis in Species of the Quadrifaria Group of Paspalum (Poaceae, Paniceae). Ann. Bot. 2005, 96, 191–200. [Google Scholar] [CrossRef]
- González, M.L.; Urdampilleta, J.D.; Fasanella, M.; Premoli, A.C.; Chiapella, J.O. Distribution of RDNA and Polyploidy in Deschampsia antarctica E. Desv. in Antarctic and Patagonic Populations. Polar Biol. 2016, 39, 1663–1677. [Google Scholar] [CrossRef]
- González, M.L.; Chiapella, J.O.; Urdampilleta, J.D. The Antarctic and South American Species of Deschampsia: Phylogenetic Relationships and Cytogenetic Differentiation. Syst. Biodivers. 2021, 1–40. [Google Scholar] [CrossRef]
- Amosova, A.V.; Bolsheva, N.L.; Zoshchuk, S.A.; Twardovska, M.O.; Yurkevich, O.Y.; Andreev, I.O.; Samatadze, T.E.; Badaeva, E.D.; Kunakh, V.A.; Muravenko, O.V. Comparative Molecular Cytogenetic Characterization of Seven Deschampsia (Poaceae) Species. PLoS ONE 2017, 12, e0175760. [Google Scholar] [CrossRef]
- Drapikowska, M.; Susek, K.; Hasterok, R.; Szkudlarz, P.; Celka, Z.; Jackowiak, B. Variability of Stomata and 45s and 5s Rdnas Loci Characteristics in Two Species of Anthoxanthum Genus: A. aristatum and A. odoratum (Poaceae). Acta Biol. Hung. 2013, 64, 352–363. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).