3.2. Evaluation Metrics
In general, the signal-to-noise ratio is the way that the noise signal is measured in either a signal or an image. In other words, a better way to assess the amount of noise in an image is to measure the ratio of pure pixels (called mean of the signal or mean of the pixel values) to noisy pixels, which is the standard deviation of the signal (called the noise standard deviation), as shown in Equation (7).
where
n is the total number of pixels in the micrograph.
In order to reduce radiation damage to the biomolecules of interest during the imaging process of the microscopy, a limited electron dose was used, as high-energy electrons can greatly damage specimens during imaging, resulting in extremely noisy micrographs. Moreover, the micrographs contained two-dimensional projections of a particle in different orientations. Generally, cryo-EM images have low contrast, due to the similarity of the electron density of the protein to that of the surrounding solution, as well as the limited electron dose used in data collection. In addition, the micrographs might have contained sections of ice, deformed particles, protein aggregates, etc., which would complicate particle picking. The cryo-EM images (micrographs) of the protein particles were taken by electron microscope, which contained randomly arranged particles along with non-particles—bits of frost, deformed particles, protein aggregates, and so on. These images suffered from heavy background noise and low contrast, due to a limited electron dose used in imaging. For these reasons, we observed that the micrographs had an extremely low signal-to-noise ratio.
Typically, for the first pre-processing stage, we used the EMAN2 software [
29] to adjust the global intensity of the cryo-EM and convert them from MRC file format to the PNG image format in order to apply standard image-processing tools to them. In terms of selecting the best results, we used different scaling factors with EMAN2 [
29] to adjust the intensity of the cryo-EM images. Then, we computed different evaluation metrics, such as the peak signal-to-noise ratio (PSNR), signal-to-noise ratio (SNR), and mean squared error (MSE), to evaluate the improvement of the quality of cryo-EM images in the whole dataset in each different scaling factor and compare the results with the original micrographs (i.e., without the using the intensity adjustment scaling factor).
Based on the SNR evaluation metric in Equation (7), where noise signal was measured, the PSNR often measured ratios between the maximum signal (pure pixels) and noise (corrupted pixels). PNSR uses a logarithmic decibel scale to measure the ratio between the maximum signal and noise that has a very wide dynamic range, as shown in Equation (8).
where
is the maximum possible pixel value of the micrograph and
is mean squared error given in Equation (9):
where
is the micrograph image size,
is the original micrograph, and
is the pre-processed micrograph.
For the pre-processing stage, we used common image pre-processing criteria, such as peak signal-to-noise ratio (PSNR), signal-to-noise ratio (SNR), and mean squared error (MSE), to evaluate the improvement of the quality of cryo-EM images [
33]. For the particle clustering and detection stages, we used the accuracy, precision, recall, and F1-score (i.e., the geometry means of precision and recall) to evaluate the particle clustering/detection results.
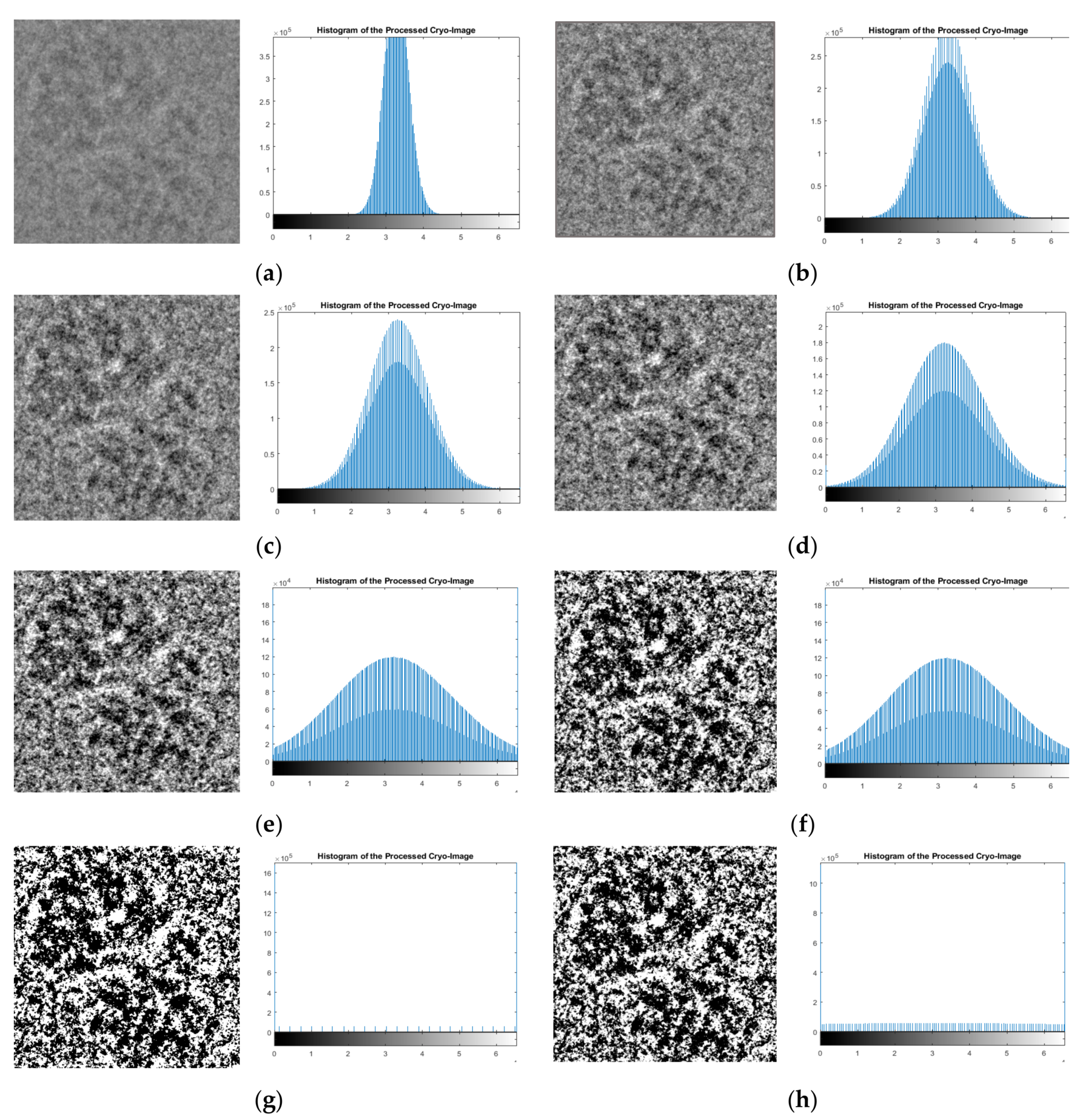
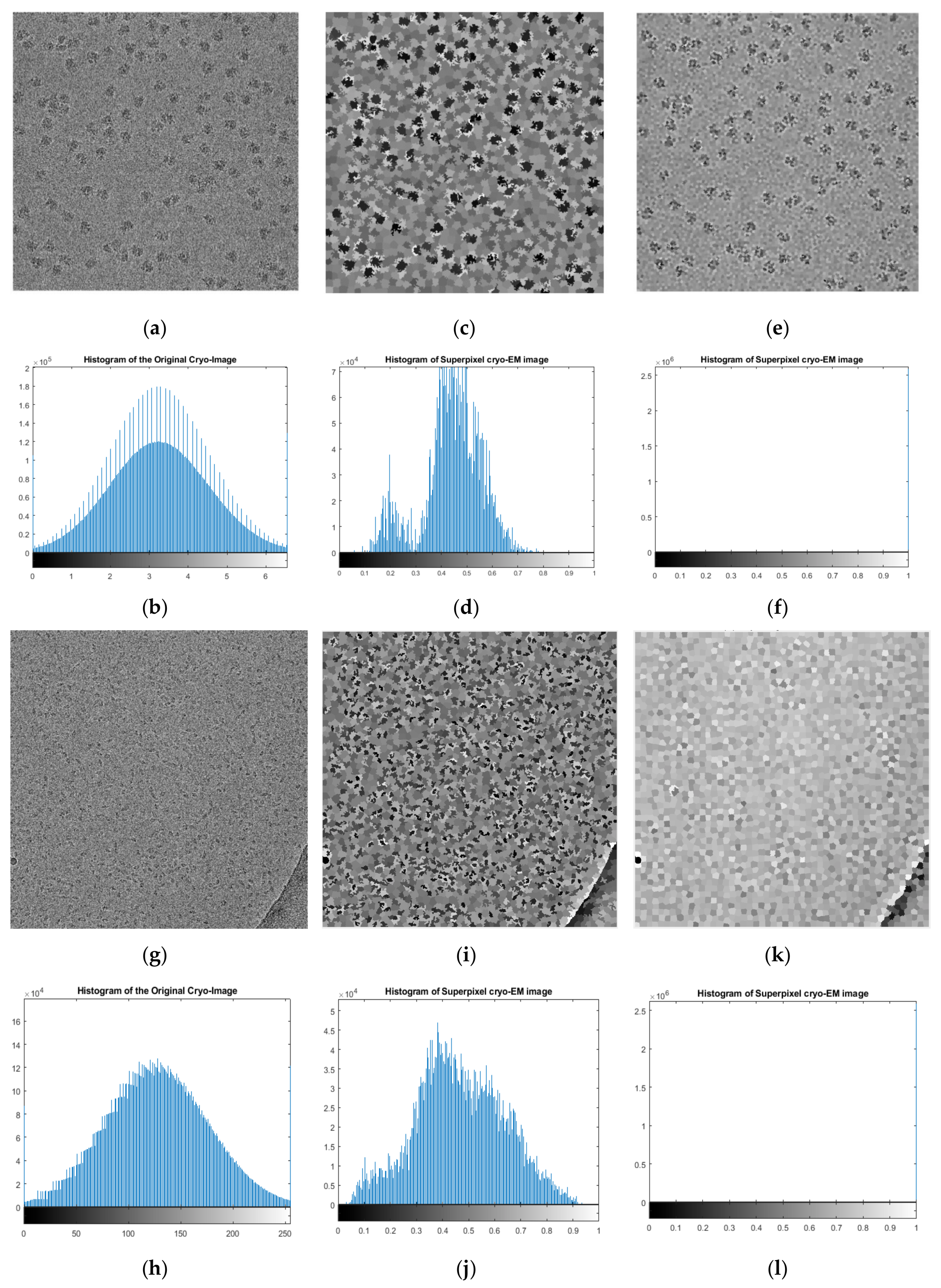
Table 1 reports the average quality measurements of the cryo-EM images with/without EMAN2 [
29] intensity adjustment. The average quality measurements (PSNR, SNR, and MSE) of the original cryo-EM images were 28.06, 6.99 dB, and 26218.13, respectively. The intensity adjustment with many scaling factors improved the quality. The best scaling factor, which increased the PSNR and SNR while simultaneously decreasing the MSE, was the “sane” option, which picked a good range of scaling factors automatically. The three scores of using the “sane” scaling factor were improved by 29.27, 8.10 dB, and 0.198643.
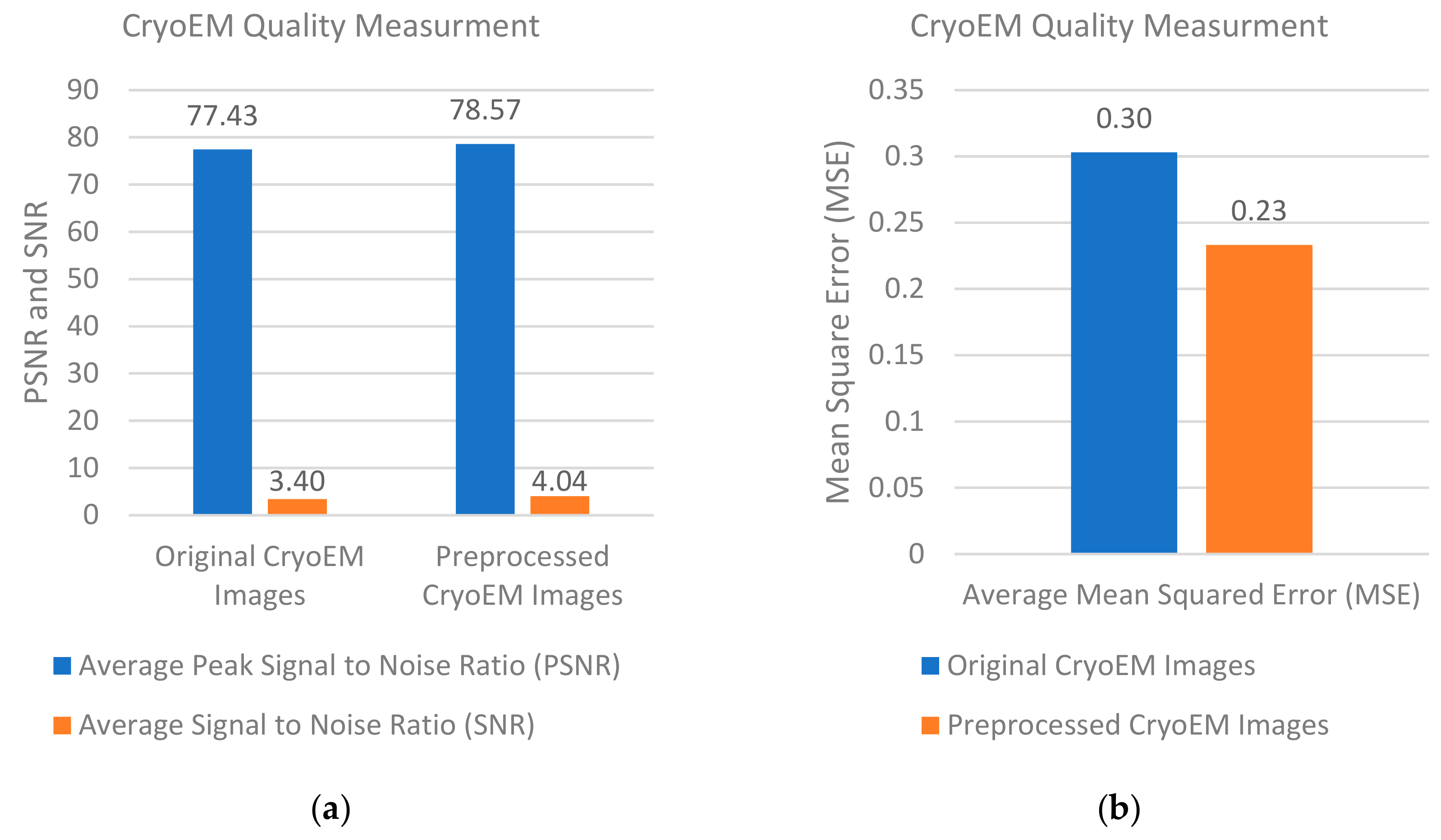
Figure 12a compares the average PSNR and SNR scores of the cryo-EM images before and after all the pre-processing steps in the pre-processing stage.
Figure 12b shows the MSE scores of the cryo-EM images before and after all the pre-processing steps.
The average PSNR score increased from 77.43 to 78.57 and the average SNR score increased from 3.40 to 4.05. The average MSE was reduced from 0.302 to 0.233. The range of PSNR scores increased from [77.429–77.43] for the original cryo-EM images to [78.52–78.64] for the pre-processed ones. The range of the SNR scores increased from [3.36–3.44] for the original images to [4.04–4.052] for the pre-processed ones. The range of MSE scores decreased from [0.3026–0.3033] to [0.23–0.237] after the pre-processing steps. According to Student’s t test, the p-values of the changes of PSNR, SNR, and MSE scores caused by the pre-processing were (-15.70) -28.31, and -19.53, respectively, indicating that the pre-processing steps significantly improved the quality of cryo-EM images.
3.3. Particle Clustering, Detection, and Picking Results
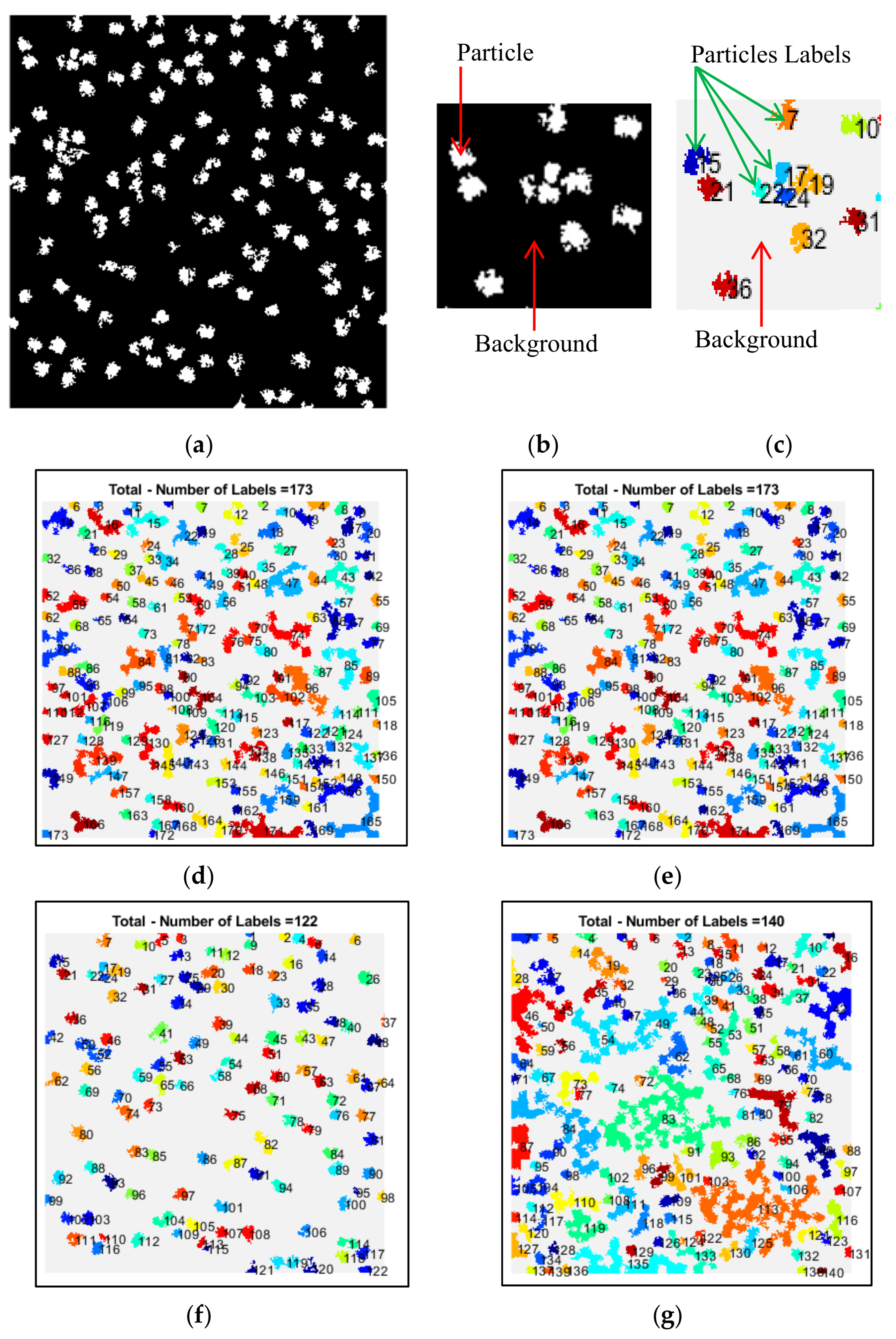
In order to evaluate the performance of automated particle clustering and picking, we generated a true reference by manually picking the particles on the images.
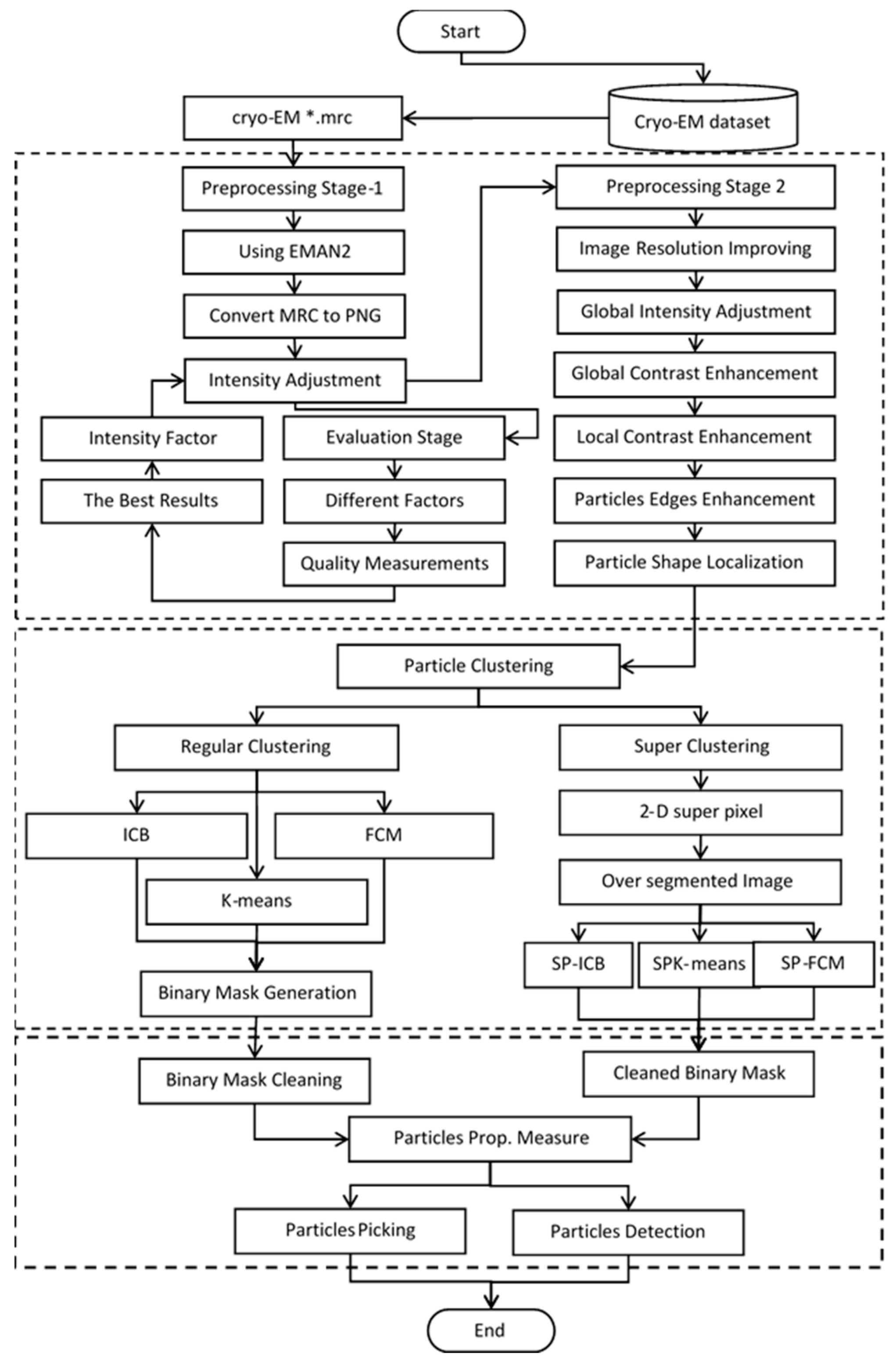
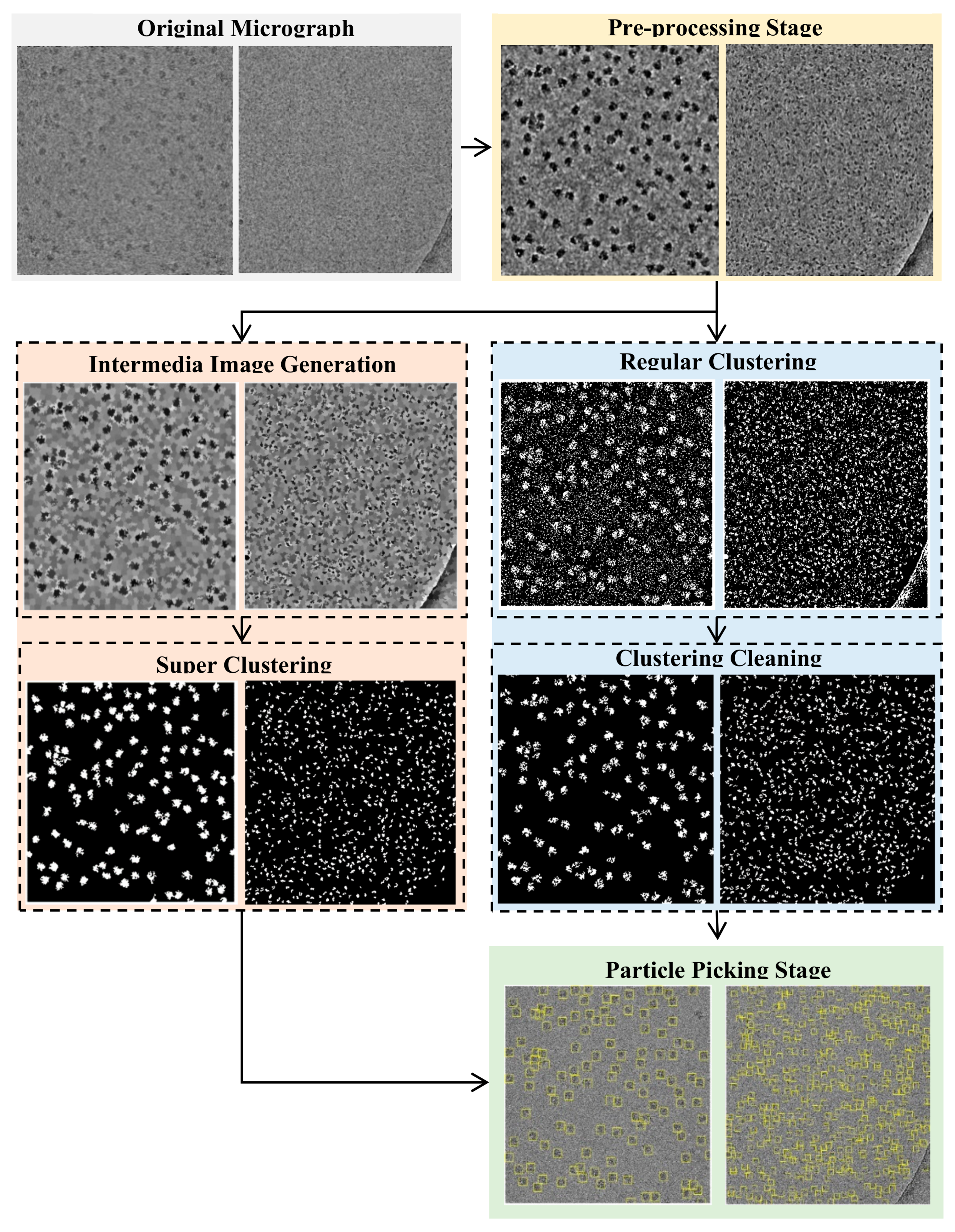
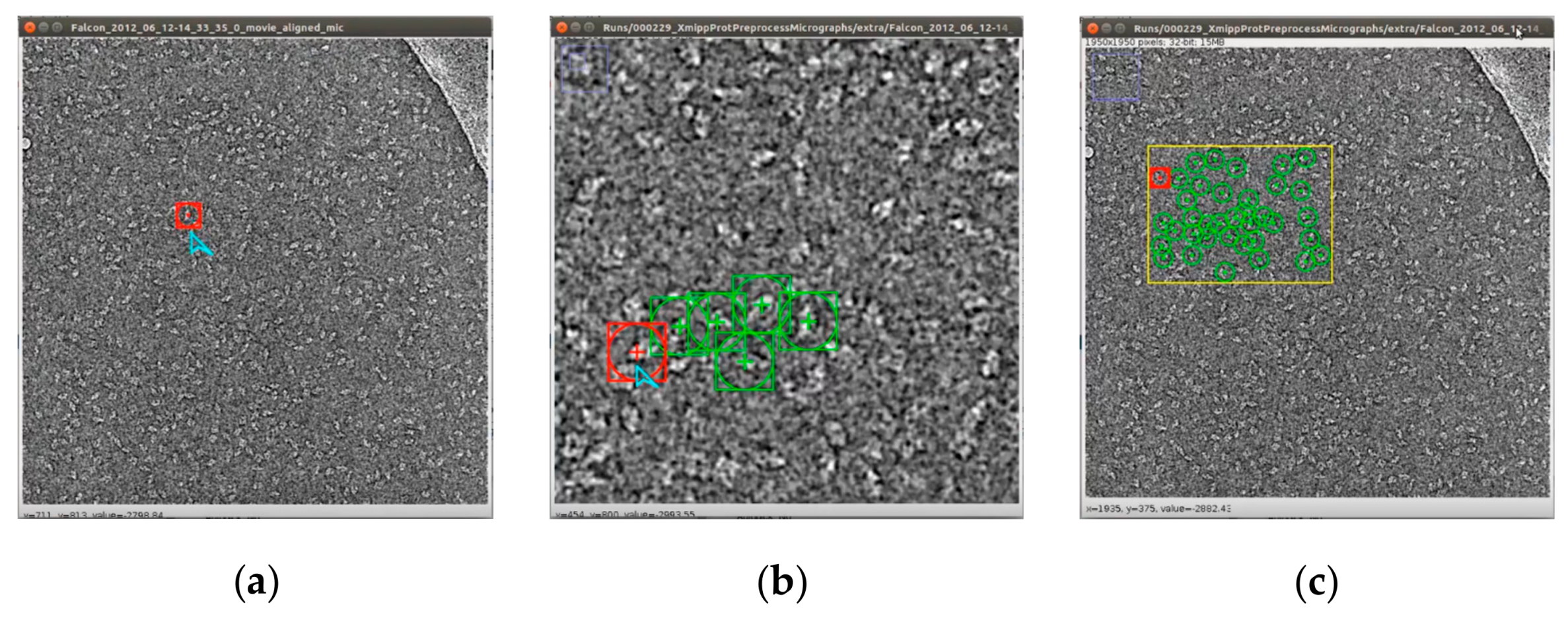
Figure 13 illustrates the entire workflow of the super-clustering approach for fully automated complex and irregular single particle picking in cryo-EM.
The super-clustering approach was designed for fully automated single particle picking in cryo-EM. Our framework contained three stages. The first stage was micrograph pre-processing (shown on the yellow box of
Figure 13), the second stage was the clustering stage, which had two different approaches, i.e., the regular clustering approach (shown in the blue box of
Figure 13) and the super-clustering approach (shown in the orange box of
Figure 13), and the third stage was the single particle picking (shown in the green box of
Figure 13).
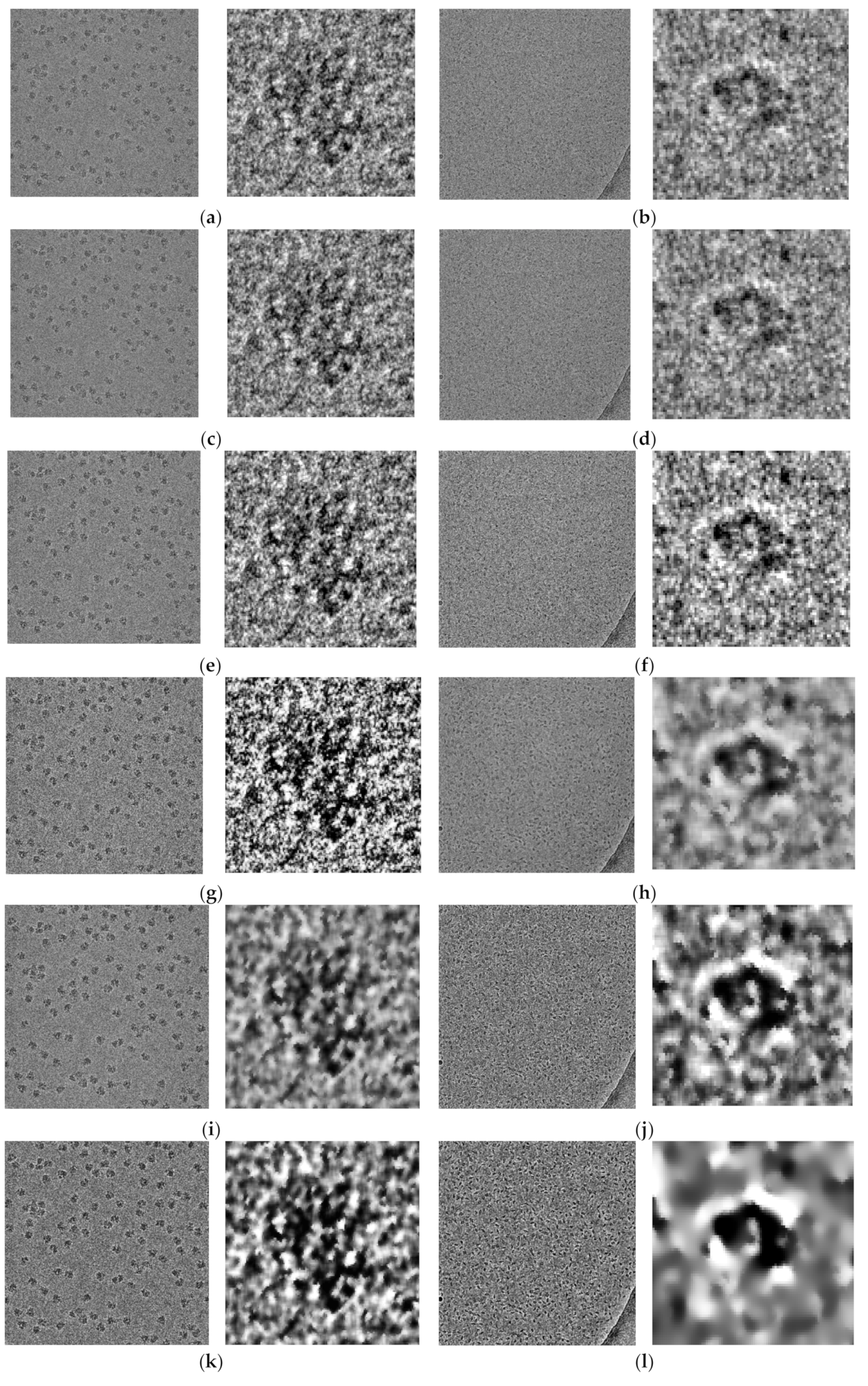
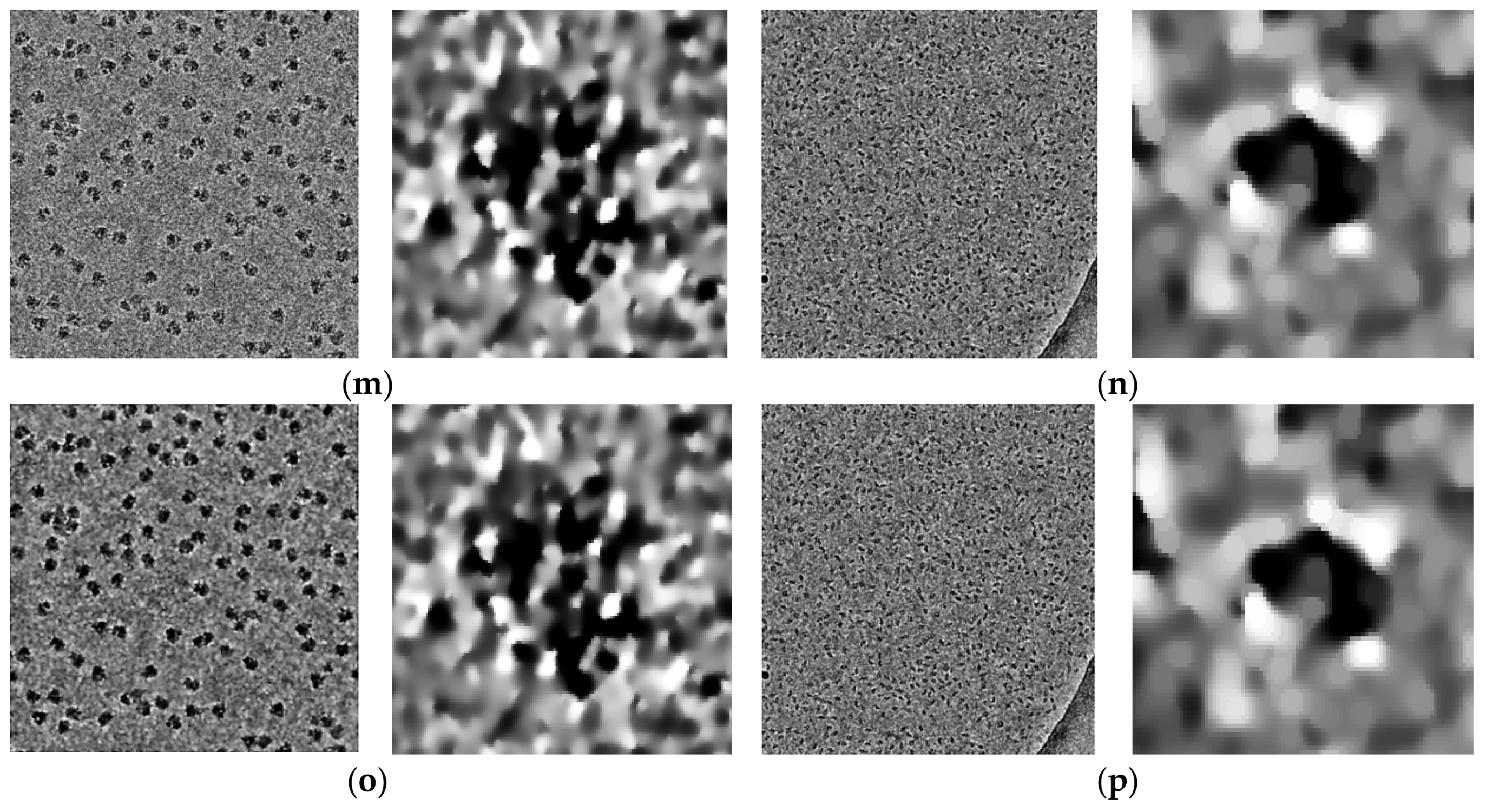
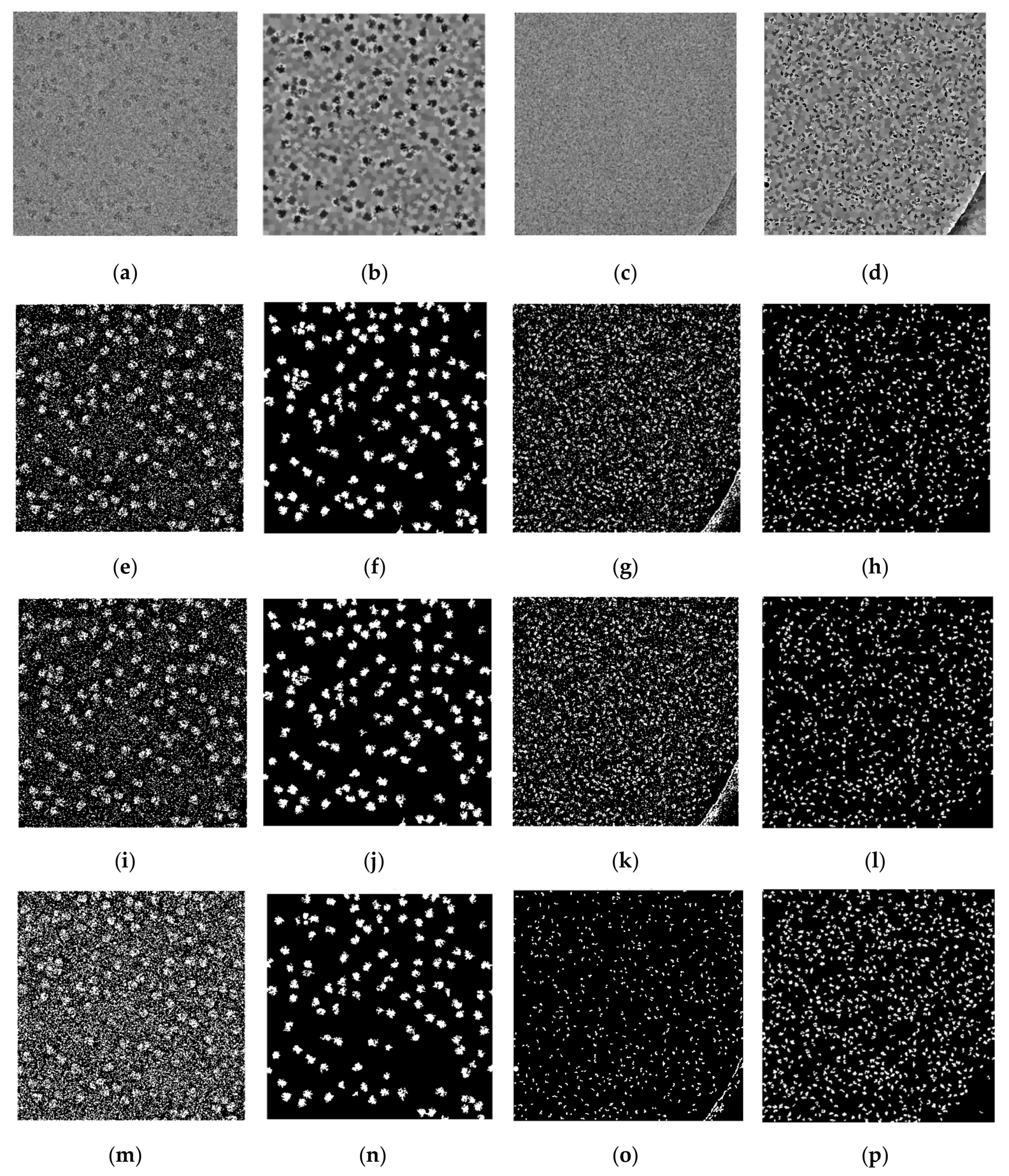
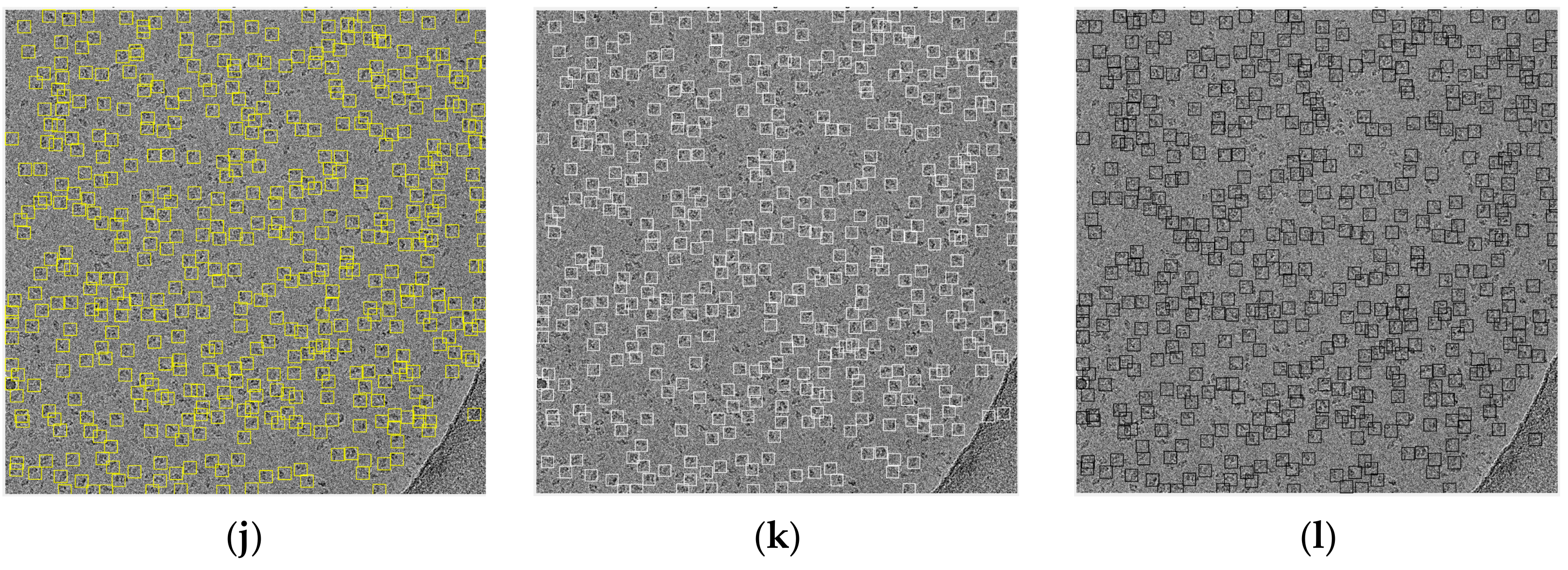
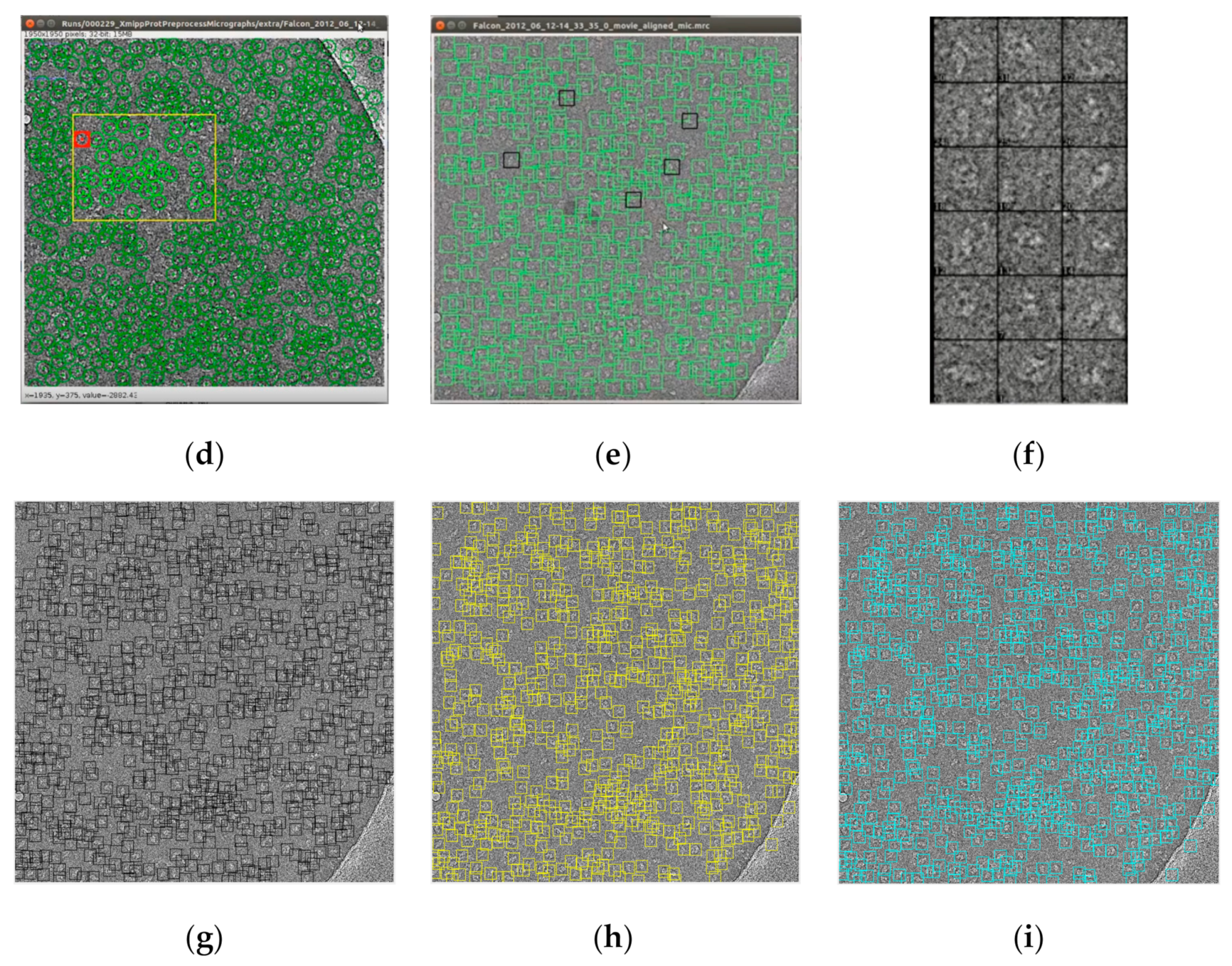
Figure 14 shows some examples of the fully automated complex single particle shape detection and picking using the super-clustering methods and the base cluster methods. The true particles that failed to be detected (false negatives) are denoted by red dots. Yellow dots represent the non-particle background (e.g., icy) objects that were falsely detected as particles (false positives).
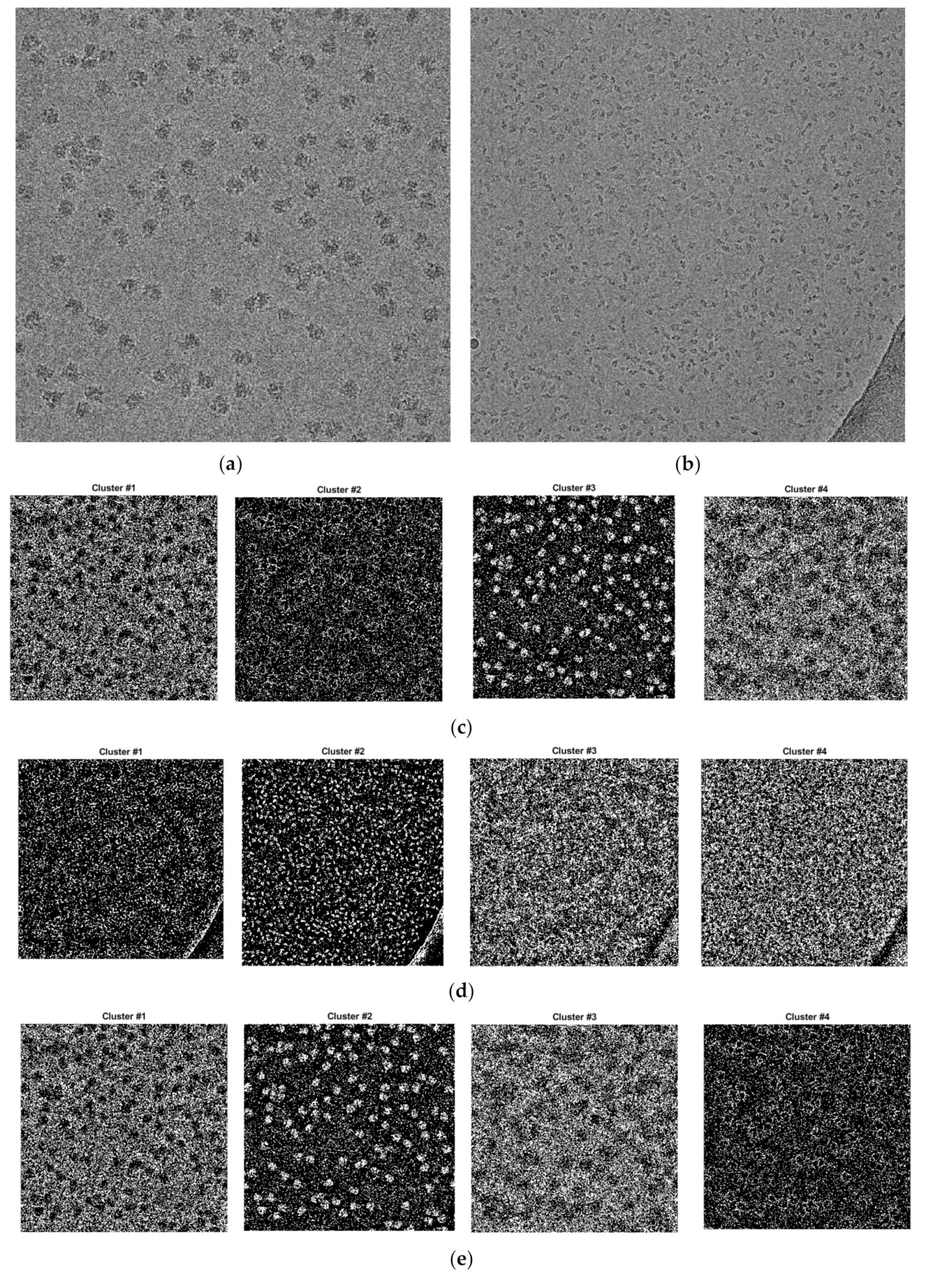
Compared with the results of the base clustering methods (k-means, FCM, and IBC), the performances of the super-clustering methods (SP-IBC, SP-K-means, and SP-FCM) were significantly improved. The number of false negatives and false positives was significantly reduced.
Table 2 reports the recall, precision, accuracy, F1 score, and running time of the base single particle picking methods (IBC, k-means, and FCM).
Table 3 shows the results of the super-clustering methods (SP-IBC, SP-k-means, and SP-FCM).
Table 3 shows the results of the super-clustering methods (SP-IBC, SP-k-means, and SP-FCM). The three super-clustering methods were fully automated. The mean average of particle picking accuracy increased by 12.03%, 3.5%, and 2.1% for the IBC, k-means, and FCM, respectively. Also, the average time taken (pre-processing, running time of clustering, and particle picking) over the whole dataset decreased by 5.68 s for the SP-IBC, 86.24 s for the SP-k-means, and 59.58 s for the SP-FCM.
Generally, the super-clustering methods achieved better performance than their corresponding base methods according to almost all the metrics. SP-k-means clustering achieved a higher accuracy (95.48%) than SP-FCM (94.08%) and SP-IBC (88.98%). SP-IBC ran substantially faster than the other methods and all three super-clustering methods were fully automated.
3.4. Comparison with Other Particle Picking Methods
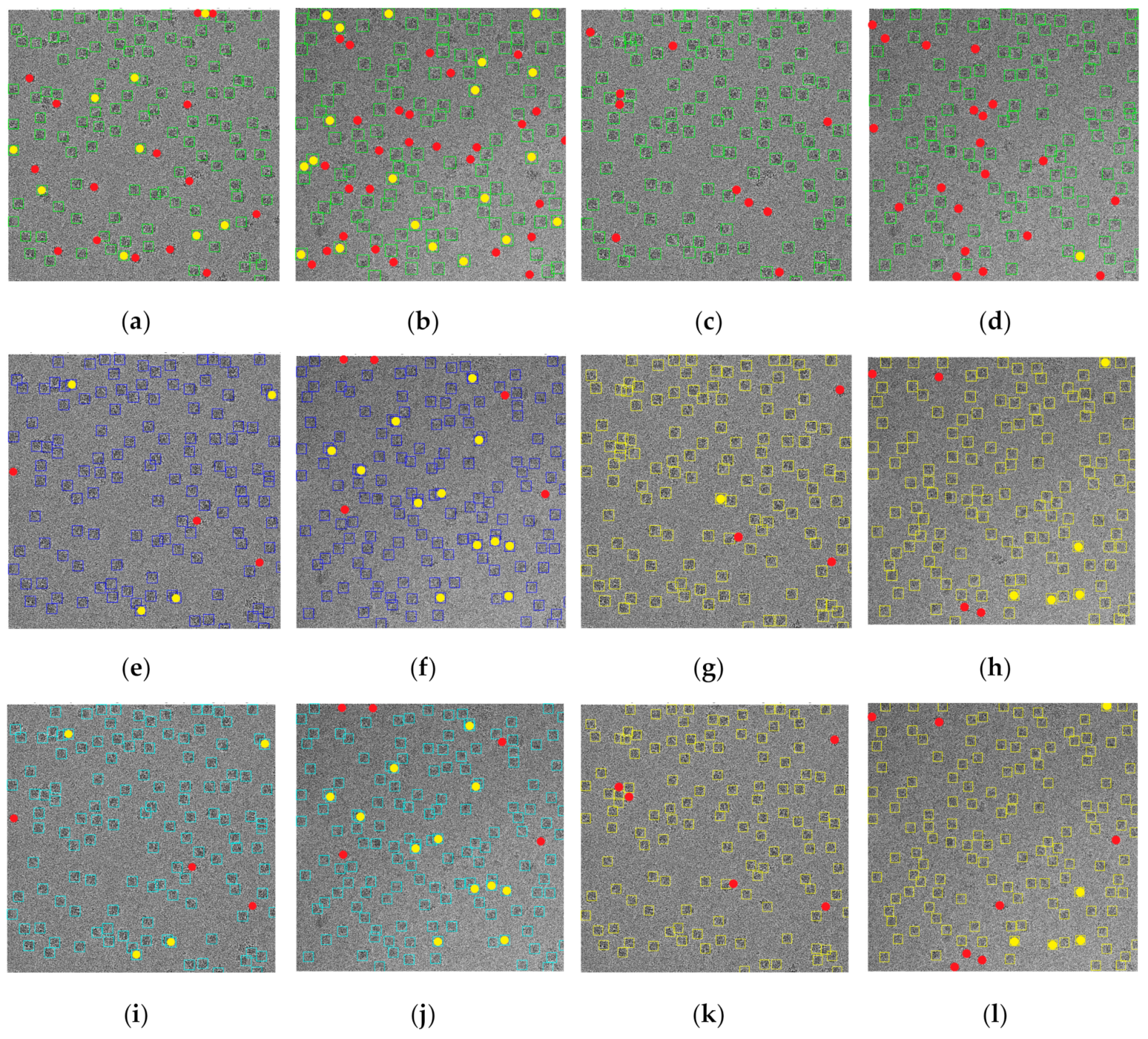
We compared SuperCryoEMPicker with two other methods, Scipion [
34] and EMAN2 [
29], in terms of computational efficiency, detection quality, and automation. Both Scipion [
34] and EMAN2 [
29] needed a reference set of particles to be selected manually (
Figure 15a for Scipion;
Figure 15e for EMAN2 [
29]), which were used to train the methods to pick more particles (
Figure 15d using Scipion and
Figure 15e using EMAN2 [
29]). Use of the arbitrarily manually selected particles resulted in most of the true particles being selected (
Figure 15c using Scipion [
34] and
Figure 15e,f using EMAN2). However, some false positives, likely corresponding to thick ice, were also incorrectly selected (
Figure 15d using Scipion [
34] and
Figure 15e using EMAN2 [
29]). Increasing the number of the manually selected particles could reduce the number of false positives, but at the expense of increasing the number of false negatives (
Figure 15e for EMAN2 [
29]). In comparison, SuperCryoEMPicker successfully captured all true particles on the images without using any manually selected samples for training (
Figure 15g–i).
Quantitative assessment of the comparison is shown in
Figure 16 and
Table 4.
Figure 16a,b shows a micrograph from the beta-galactosidase dataset [
30] after particle picking using EMAN2 [
29], where only five particle references were selected (more references selected means more time taken to complete the task but more accurate results), and our super-clustering approach.
Figure 16a shows the particle picking performance results using EMAN2 [
29]. In terms of evaluating each particle picking tool, in addition to our fully automated particle picking approach, three criteria were selected to label and evaluate the particle picking performance results: True Positive (TP) picking, where the correct particles were marked by the yellow circles; False Negative (FN) picking, where the missed particles were marked by red circles; False Positive (FP) picking, where the incorrectly picked particles were marked by blue circles.
Figure 16b shows the same criteria of the particle picking results using the super-clustering approach.
Table 4 illustrates the statistical evaluation of the performance results based on the TP, FN, and FP for each single particle picking algorithm, as well as the particle shape class and total number of particles (ground truth) in each image. Note that the super-clustering approach for fully automated single particle picking performed better in regard to detecting the shapes; it achieved 99.13% sensitivity, 98.45% precision, and 97.61% accuracy.