Integration of Bioinformatic Predictions and Experimental Data to Identify circRNA-miRNA Associations
Abstract
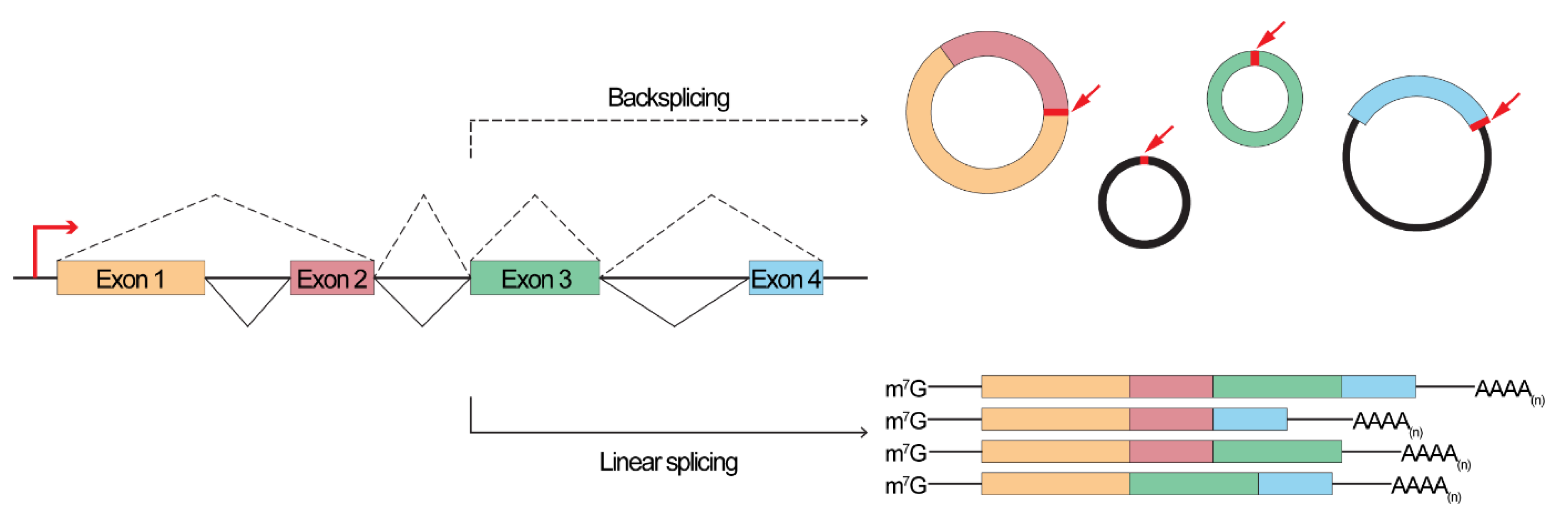
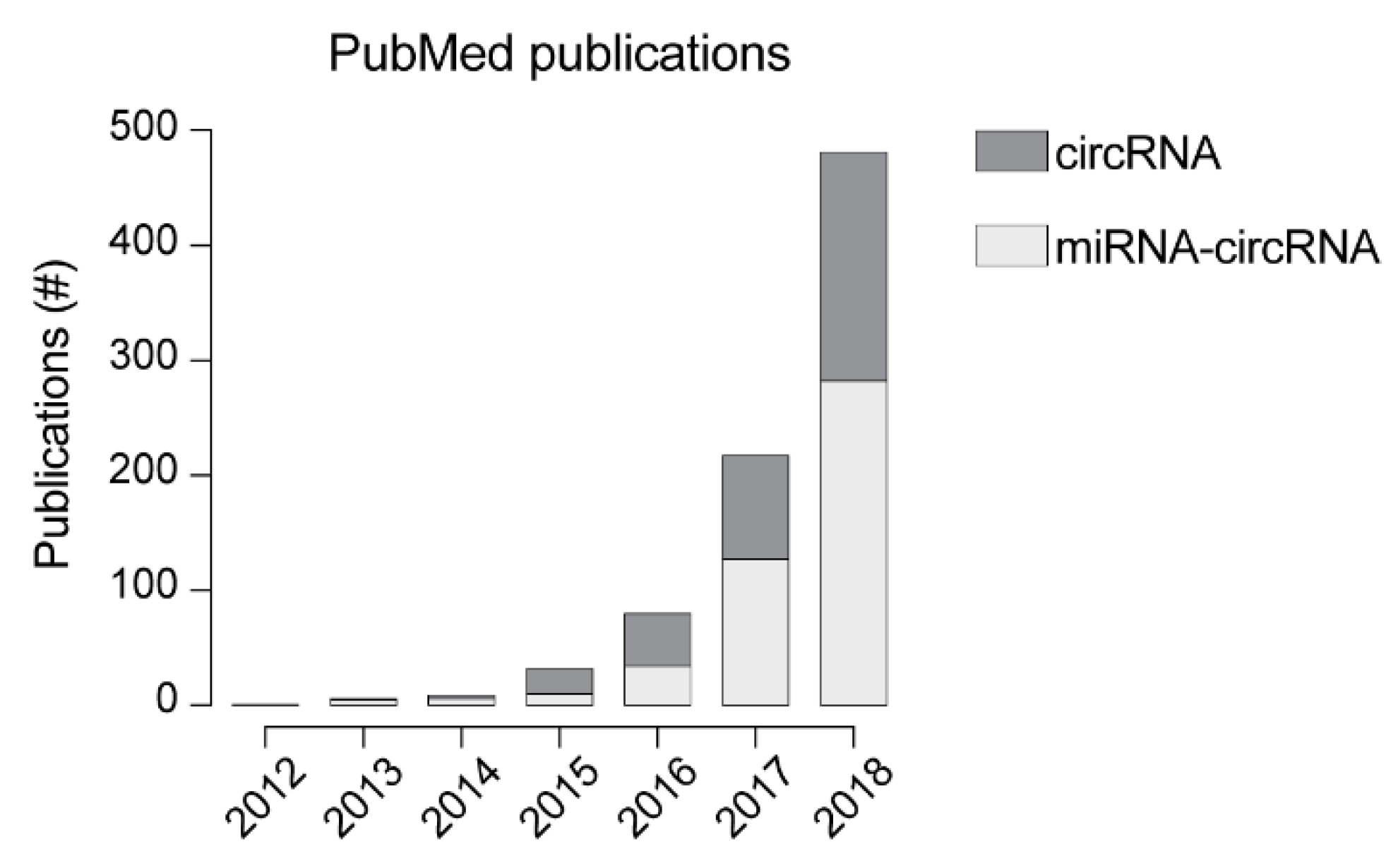
1. Introduction
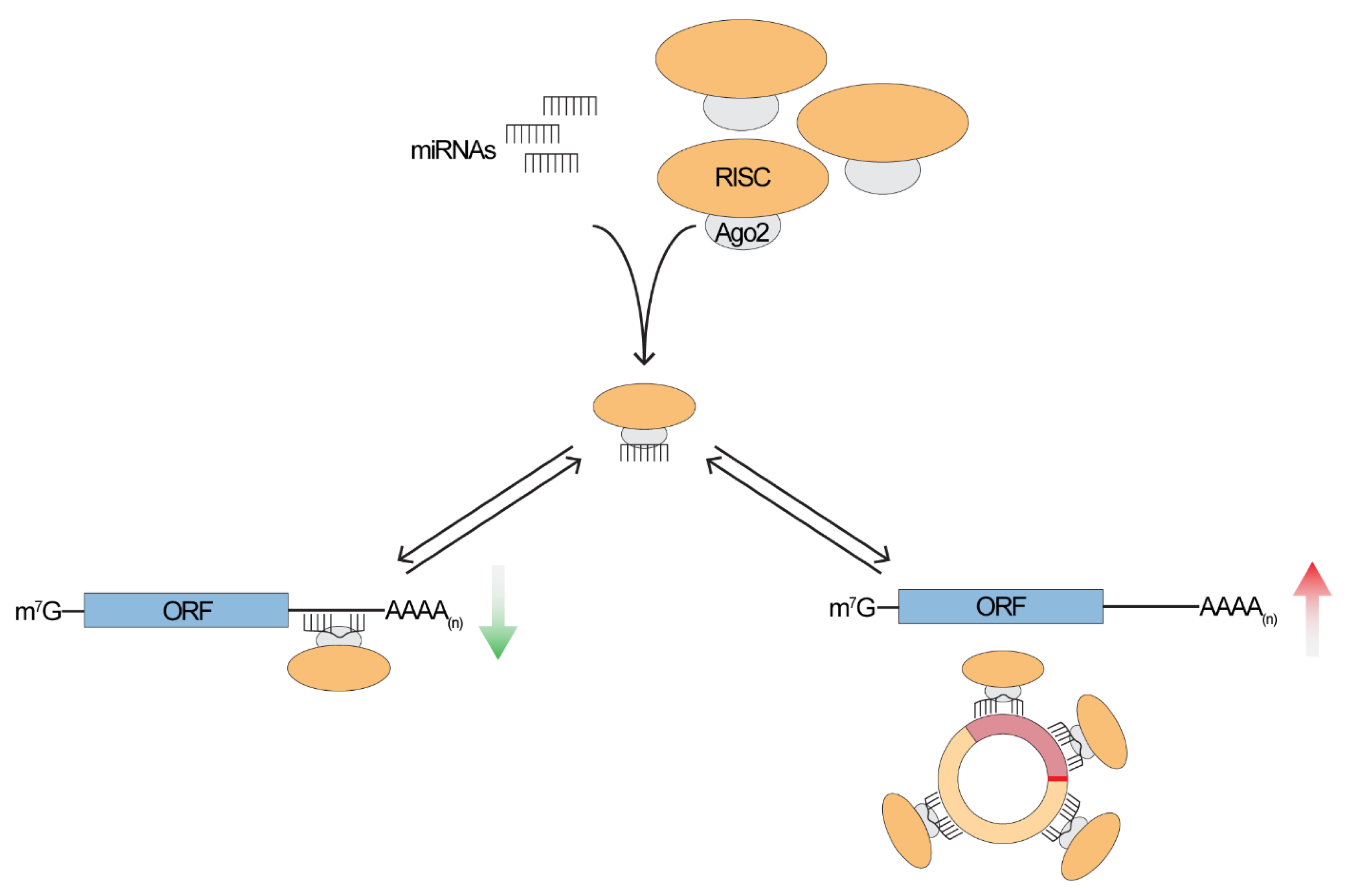
2. To Sponge or Not to Sponge, That is the Question
3. Predicting circRNA-miRNA Binding Sites
3.1. Investigating Known circRNAs
3.2. Characterizing Novel circRNAs
4. Integrating Seed Prediction on Custom Sequences with Experimental Data
Ranking MREs
5. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Carthew, R.W.; Sontheimer, E.J. Origins and mechanisms of miRNAs and siRNAs. Cell 2009, 136, 642–655. [Google Scholar] [CrossRef] [PubMed]
- Kosik, K.S. Circles reshape the RNA world. Nature 2013, 495, 322–324. [Google Scholar] [CrossRef] [PubMed]
- Kos, A.; Dijkema, R.; Arnberg, A.C.; van der Meide, P.H.; Schellekens, H. The hepatitis delta (δ) virus possesses a circular RNA. Nature 1986, 323, 558. [Google Scholar] [CrossRef] [PubMed]
- Capel, B.; Swain, A.; Nicolis, S.; Hacker, A.; Walter, M.; Koopman, P.; Goodfellow, P.; Lovell-Badge, R. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 1993, 73, 1019–1030. [Google Scholar] [CrossRef]
- Hacker, A.; Capel, B.; Goodfellow, P.; Lovell-Badge, R. Expression of Sry, the mouse sex determining gene. Development 1995, 121, 1603–1614. [Google Scholar] [PubMed]
- Jeske, Y.W.; Bowles, J.; Greenfield, A.; Koopman, P. Expression of a linear Sry transcript in the mouse genital ridge. Nat. Genet. 1995, 10, 480–482. [Google Scholar] [CrossRef] [PubMed]
- Nigro, J.M.; Cho, K.R.; Fearon, E.R.; Kern, S.E.; Ruppert, J.M.; Oliner, J.D.; Kinzler, K.W.; Vogelstein, B. Scrambled exons. Cell 1991, 64, 607–613. [Google Scholar] [CrossRef]
- Zaphiropoulos, P.G. Circular RNAs from transcripts of the rat cytochrome P450 2C24 gene: Correlation with exon skipping. Proc. Natl. Acad. Sci. USA 1996, 93, 6536–6541. [Google Scholar] [CrossRef]
- Salzman, J.; Gawad, C.; Wang, P.L.; Lacayo, N.; Brown, P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 2012, 7, e30733. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef] [PubMed]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Lasda, E.; Parker, R. Circular RNAs: Diversity of form and function. RNA 2014, 20, 1829–1842. [Google Scholar] [CrossRef] [PubMed]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 2014, 56, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, A.; Memczak, S.; Wyler, E.; Torti, F.; Porath, H.T.; Orejuela, M.R.; Piechotta, M.; Levanon, E.Y.; Landthaler, M.; Dieterich, C.; et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015, 10, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Starke, S.; Jost, I.; Rossbach, O.; Schneider, T.; Schreiner, S.; Hung, L.H.; Bindereif, A. Exon circularization requires canonical splice signals. Cell Rep. 2015, 10, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, Z. Efficient backsplicing produces translatable circular mRNAs. RNA 2015, 21, 172–179. [Google Scholar] [CrossRef]
- Conn, S.J.; Pillman, K.A.; Toubia, J.; Conn, V.M.; Salmanidis, M.; Phillips, C.A.; Roslan, S.; Schreiber, A.W.; Gregory, P.A.; Goodall, G.J. The RNA binding protein quaking regulates formation of circRNAs. Cell 2015, 160, 1125–1134. [Google Scholar] [CrossRef]
- Zhang, X.O.; Dong, R.; Zhang, Y.; Zhang, J.L.; Luo, Z.; Zhang, J.; Chen, L.L.; Yang, L. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 2016, 26, 1277–1287. [Google Scholar] [CrossRef]
- Salzman, J.; Chen, R.E.; Olsen, M.N.; Wang, P.L.; Brown, P.O. Cell-type specific features of circular RNA expression. PLoS Genet. 2013, 9, e1003777. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, X.O.; Chen, T.; Xiang, J.F.; Yin, Q.F.; Xing, Y.H.; Zhu, S.; Yang, L.; Chen, L.L. Circular intronic long noncoding RNAs. Mol. Cell 2013, 51, 792–806. [Google Scholar] [CrossRef]
- Li, F.; Zhang, L.; Li, W.; Deng, J.; Zheng, J.; An, M.; Lu, J.; Zhou, Y. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β-catenin pathway. Oncotarget 2015, 6, 6001–6013. [Google Scholar] [CrossRef]
- Rybak-Wolf, A.; Stottmeister, C.; Glazar, P.; Jens, M.; Pino, N.; Giusti, S.; Hanan, M.; Behm, M.; Bartok, O.; Ashwal-Fluss, R.; et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell 2015, 58, 870–885. [Google Scholar] [CrossRef]
- Suzuki, H.; Zuo, Y.; Wang, J.; Zhang, M.Q.; Malhotra, A.; Mayeda, A. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 2006, 34, e63. [Google Scholar] [CrossRef]
- Vincent, H.A.; Deutscher, M.P. Substrate recognition and catalysis by the exoribonuclease RNase R. J. Biol. Chem. 2006, 281, 29769–29775. [Google Scholar] [CrossRef]
- Westholm, J.O.; Miura, P.; Olson, S.; Shenker, S.; Joseph, B.; Sanfilippo, P.; Celniker, S.E.; Graveley, B.R.; Lai, E.C. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 2014, 9, 1966–1980. [Google Scholar] [CrossRef]
- Conn, V.M.; Hugouvieux, V.; Nayak, A.; Conos, S.A.; Capovilla, G.; Cildir, G.; Jourdain, A.; Tergaonkar, V.; Schmid, M.; Zubieta, C.; et al. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat. Plants 2017, 3, 17053. [Google Scholar] [CrossRef]
- Tang, B.; Hao, Z.; Zhu, Y.; Zhang, H.; Li, G. Genome-wide identification and functional analysis of circRNAs in Zea mays. PLoS ONE 2018, 13, e0202375. [Google Scholar] [CrossRef]
- Wang, Y.; Gao, Y.; Zhang, H.; Wang, H.; Liu, X.; Xu, X.; Zhang, Z.; Kohnen, M.V.; Hu, K.; Wang, H.; et al. Genome-wide profiling of circular RNAs in the rapidly growing shoots of moso bamboo (Phyllostachys edulis). Plant Cell Physiol. 2019, 60, 1354–1373. [Google Scholar] [CrossRef]
- Dori, M.; Haj Abdullah Alieh, L.; Cavalli, D.; Massalini, S.; Lesche, M.; Dahl, A.; Calegari, F. Sequence and expression levels of circular RNAs in progenitor cell types during mouse corticogenesis. Life Sci. Alliance 2019, 2, e201900354. [Google Scholar] [CrossRef]
- Ragan, C.; Goodall, G.J.; Shirokikh, N.E.; Preiss, T. Insights into the biogenesis and potential functions of exonic circular RNA. Sci. Rep. 2019, 9, 2048. [Google Scholar] [CrossRef]
- Xu, K.; Chen, D.; Wang, Z.; Ma, J.; Zhou, J.; Chen, N.; Lv, L.; Zheng, Y.; Hu, X.; Zhang, Y.; et al. Annotation and functional clustering of circRNA expression in rhesus macaque brain during aging. Cell Discov. 2018, 4, 48. [Google Scholar] [CrossRef]
- Li, P.; Chen, S.; Chen, H.; Mo, X.; Li, T.; Shao, Y.; Xiao, B.; Guo, J. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin. Chim. Acta 2015, 444, 132–136. [Google Scholar] [CrossRef]
- Bahn, J.H.; Zhang, Q.; Li, F.; Chan, T.M.; Lin, X.; Kim, Y.; Wong, D.T.; Xiao, X. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin. Chem. 2015, 61, 221–230. [Google Scholar] [CrossRef]
- Memczak, S.; Papavasileiou, P.; Peters, O.; Rajewsky, N. Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS ONE 2015, 10, e0141214. [Google Scholar] [CrossRef]
- Lyu, D.; Huang, S. The emerging role and clinical implication of human exonic circular RNA. RNA Biol. 2017, 14, 1000–1006. [Google Scholar] [CrossRef]
- Holdt, L.M.; Stahringer, A.; Sass, K.; Pichler, G.; Kulak, N.A.; Wilfert, W.; Kohlmaier, A.; Herbst, A.; Northoff, B.H.; Nicolaou, A.; et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 2016, 7, 12429. [Google Scholar] [CrossRef]
- Hansen, T.B.; Kjems, J.; Damgaard, C.K. Circular RNA and miR-7 in cancer. Cancer Res. 2013, 73, 5609–5612. [Google Scholar] [CrossRef]
- Haque, S.; Harries, L.W. Circular RNAs (circRNAs) in health and disease. Genes 2017, 8, 353. [Google Scholar] [CrossRef]
- Abu, N.; Jamal, R. Circular RNAs as promising biomarkers: A mini-review. Front. Physiol. 2016, 7, 355. [Google Scholar] [CrossRef]
- Bonizzato, A.; Gaffo, E.; Te Kronnie, G.; Bortoluzzi, S. CircRNAs in hematopoiesis and hematological malignancies. Blood Cancer J. 2016, 6, e483. [Google Scholar] [CrossRef]
- Kulcheski, F.R.; Christoff, A.P.; Margis, R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J. Biotechnol. 2016, 238, 42–51. [Google Scholar] [CrossRef]
- Li, Z.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L.; et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015, 22, 256–264. [Google Scholar] [CrossRef]
- Du, W.W.; Yang, W.; Liu, E.; Yang, Z.; Dhaliwal, P.; Yang, B.B. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016, 44, 2846–2858. [Google Scholar] [CrossRef]
- Legnini, I.; Di Timoteo, G.; Rossi, F.; Morlando, M.; Briganti, F.; Sthandier, O.; Fatica, A.; Santini, T.; Andronache, A.; Wade, M.; et al. Circ-ZNF609 Is a Circular rna that can be translated and functions in myogenesis. Mol. Cell 2017, 66, 22–37.e29. [Google Scholar] [CrossRef]
- Pamudurti, N.R.; Bartok, O.; Jens, M.; Ashwal-Fluss, R.; Stottmeister, C.; Ruhe, L.; Hanan, M.; Wyler, E.; Perez-Hernandez, D.; Ramberger, E.; et al. Translation of CircRNAs. Mol. Cell 2017, 66, 9–21.e27. [Google Scholar] [CrossRef]
- Huang, S.; Yang, B.; Chen, B.J.; Bliim, N.; Ueberham, U.; Arendt, T.; Janitz, M. The emerging role of circular RNAs in transcriptome regulation. Genomics 2017, 109, 401–407. [Google Scholar] [CrossRef]
- Verduci, L.; Strano, S.; Yarden, Y.; Blandino, G. The circRNA-microRNA code: Emerging implications for cancer diagnosis and treatment. Mol. Oncol. 2019, 13, 669–680. [Google Scholar] [CrossRef]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef]
- Thomson, D.W.; Dinger, M.E. Endogenous microRNA sponges: Evidence and controversy. Nat. Rev. Genet. 2016, 17, 272–283. [Google Scholar] [CrossRef]
- Denzler, R.; Agarwal, V.; Stefano, J.; Bartel, D.P.; Stoffel, M. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol. Cell 2014, 54, 766–776. [Google Scholar] [CrossRef]
- Bosson, A.D.; Zamudio, J.R.; Sharp, P.A. Endogenous miRNA and target concentrations determine susceptibility to potential ceRNA competition. Mol. Cell 2014, 56, 347–359. [Google Scholar] [CrossRef]
- Guo, J.U.; Agarwal, V.; Guo, H.; Bartel, D.P. Expanded identification and charachterization of mammalian circular RNAs. Genome Biol. 2014, 15, 409. [Google Scholar] [CrossRef]
- Alhasan, A.A.; Izuogu, O.G.; Al-Balool, H.H.; Steyn, J.S.; Evans, A.; Colzani, M.; Ghevaert, C.; Mountford, J.C.; Mareneah, L.; Elliott, D.J.; et al. Circular RNA enrichment in platelets is a signature of transcriptome degradation. Blood 2016, 127, e1–e11. [Google Scholar] [CrossRef]
- Piwecka, M.; Glazar, P.; Hernandez-Miranda, L.R.; Memczak, S.; Wolf, S.A.; Rybak-Wolf, A.; Filipchyk, A.; Klironomos, F.; Cerda Jara, C.A.; Fenske, P.; et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 2017, 357, eaam8526. [Google Scholar] [CrossRef]
- Kaur, S.; Mirza, A.H.; Pociot, F. Cell type-selective expression of circular RNAs in human pancreatic islets. Noncoding RNA 2018, 4, 38. [Google Scholar] [CrossRef]
- Holdt, L.M.; Kohlmaier, A.; Teupser, D. Molecular functions and specific roles of circRNAs in the cardiovascular system. Noncoding RNA Res. 2018, 3, 75–98. [Google Scholar] [CrossRef]
- Wang, G.; Liu, W.; Zou, Y.; Wang, G.; Deng, Y.; Luo, J.; Zhang, Y.; Li, H.; Zhang, Q.; Yang, Y.; et al. Three isoforms of exosomal circPTGR1 promote hepatocellular carcinoma metastasis via the miR449a-MET pathway. EBioMedicine 2019, 40, 432–445. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, H.; Li, W.; Yu, J.; Shen, Z.; Ye, G.; Qi, X.; Li, G. CircRNA_100269 is downregulated in gastric cancer and suppresses tumor cell growth by targeting miR-630. Aging 2017, 9, 1585–1594. [Google Scholar] [CrossRef]
- Wang, W.L.; Yang, Z.; Zhang, Y.J.; Lu, P.; Ni, Y.K.; Sun, C.F.; Liu, F.Y. Competing endogenous RNA analysis reveals the regulatory potency of circRNA_036186 in HNSCC. Int. J. Oncol. 2018, 53, 1529–1543. [Google Scholar] [CrossRef]
- Jost, I.; Shalamova, L.A.; Gerresheim, G.K.; Niepmann, M.; Bindereif, A.; Rossbach, O. Functional sequestration of microRNA-122 from Hepatitis C Virus by circular RNA sponges. RNA Biol. 2018, 15, 1032–1039. [Google Scholar] [CrossRef]
- Wang, K.; Gan, T.Y.; Li, N.; Liu, C.Y.; Zhou, L.Y.; Gao, J.N.; Chen, C.; Yan, K.W.; Ponnusamy, M.; Zhang, Y.H.; et al. Circular RNA mediates cardiomyocyte death via miRNA-dependent upregulation of MTP18 expression. Cell Death Differ. 2017, 24, 1111–1120. [Google Scholar] [CrossRef]
- Wang, R.; Zhang, S.; Chen, X.; Li, N.; Li, J.; Jia, R.; Pan, Y.; Liang, H. CircNT5E acts as a sponge of miR-422a to promote glioblastoma tumorigenesis. Cancer Res. 2018, 78, 4812–4825. [Google Scholar] [CrossRef]
- Zhang, F.; Zhang, R.; Zhang, X.; Wu, Y.; Li, X.; Zhang, S.; Hou, W.; Ding, Y.; Tian, J.; Sun, L.; et al. Comprehensive analysis of circRNA expression pattern and circRNA-miRNA-mRNA network in the pathogenesis of atherosclerosis in rabbit. Aging 2018, 10, 2266–2283. [Google Scholar] [CrossRef]
- Cai, X.; Zhao, Z.; Dong, J.; Lv, Q.; Yun, B.; Liu, J.; Shen, Y.; Kang, J.; Li, J. Circular RNA circBACH2 plays a role in papillary thyroid carcinoma by sponging miR-139-5p and regulating LMO4 expression. Cell Death Dis. 2019, 10, 184. [Google Scholar] [CrossRef]
- Go Beyond RNA. Available online: https://www.arraystar.com/circular-rna-research/ (accessed on 15 May 2019).
- Li, S.; Teng, S.; Xu, J.; Su, G.; Zhang, Y.; Zhao, J.; Zhang, S.; Wang, H.; Qin, W.; Lu, Z.J.; et al. Microarray is an efficient tool for circRNA profiling. Brief. Bioinform. 2018. [Google Scholar] [CrossRef]
- Pandey, P.R.; Rout, P.K.; Das, A.; Gorospe, M.; Panda, A.C. RPAD (RNase R treatment, polyadenylation, and poly(A)+ RNA depletion) method to isolate highly pure circular RNA. Methods 2019, 155, 41–48. [Google Scholar] [CrossRef]
- Hansen, T.B.; Veno, M.T.; Damgaard, C.K.; Kjems, J. Comparison of circular RNA prediction tools. Nucleic Acids Res. 2016, 44, e58. [Google Scholar] [CrossRef]
- Glazar, P.; Papavasileiou, P.; Rajewsky, N. circBase: A database for circular RNAs. RNA 2014, 20, 1666–1670. [Google Scholar] [CrossRef]
- Liu, Y.C.; Li, J.R.; Sun, C.H.; Andrews, E.; Chao, R.F.; Lin, F.M.; Weng, S.L.; Hsu, S.D.; Huang, C.C.; Cheng, C.; et al. CircNet: A database of circular RNAs derived from transcriptome sequencing data. Nucleic Acids Res. 2016, 44, D209–D215. [Google Scholar] [CrossRef]
- Dudekula, D.B.; Panda, A.C.; Grammatikakis, I.; De, S.; Abdelmohsen, K.; Gorospe, M. CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016, 13, 34–42. [Google Scholar] [CrossRef]
- Ghosal, S.; Das, S.; Sen, R.; Basak, P.; Chakrabarti, J. Circ2Traits: A comprehensive database for circular RNA potentially associated with disease and traits. Front. Genet. 2013, 4, 283. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015, 4, e05005. [Google Scholar] [CrossRef]
- Miranda, K.C.; Huynh, T.; Tay, Y.; Ang, Y.-S.; Tam, W.-L.; Thomson, A.M.; Lim, B.; Rigoutsos, I. A Pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell 2006, 126, 1203–1217. [Google Scholar] [CrossRef]
- Kertesz, M.; Iovino, N.; Unnerstall, U.; Gaul, U.; Segal, E. The role of site accessibility in microRNA target recognition. Nat. Genet. 2007, 39, 1278–1284. [Google Scholar] [CrossRef]
- Enright, A.J.; John, B.; Gaul, U.; Tuschl, T.; Sander, C.; Marks, D.S. MicroRNA targets in Drosophila. Genome Biol. 2003, 5, R1. [Google Scholar] [CrossRef]
- Betel, D.; Koppal, A.; Agius, P.; Sander, C.; Leslie, C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010, 11, R90. [Google Scholar] [CrossRef]
- Yang, J.H.; Li, J.H.; Shao, P.; Zhou, H.; Chen, Y.Q.; Qu, L.H. starBase: A database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res. 2011, 39, D202–D209. [Google Scholar] [CrossRef]
- Li, J.H.; Liu, S.; Zhou, H.; Qu, L.H.; Yang, J.H. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014, 42, D92–D97. [Google Scholar] [CrossRef]
- Karagkouni, D.; Paraskevopoulou, M.D.; Chatzopoulos, S.; Vlachos, I.S.; Tastsoglou, S.; Kanellos, I.; Papadimitriou, D.; Kavakiotis, I.; Maniou, S.; Skoufos, G.; et al. DIANA-TarBase v8: A decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res. 2018, 46, D239–D245. [Google Scholar] [CrossRef]
- Chou, C.H.; Shrestha, S.; Yang, C.D.; Chang, N.W.; Lin, Y.L.; Liao, K.W.; Huang, W.C.; Sun, T.H.; Tu, S.J.; Lee, W.H.; et al. miRTarBase update 2018: A resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018, 46, D296–D302. [Google Scholar] [CrossRef]
- Peng, S.; Song, C.; Li, H.; Cao, X.; Ma, Y.; Wang, X.; Huang, Y.; Lan, X.; Lei, C.; Chaogetu, B.; et al. Circular RNA SNX29 sponges miR-744 to regulate proliferation and differentiation of myoblasts by activating the Wnt5a/Ca(2+) signaling pathway. Mol. Ther. Nucleic Acids 2019, 16, 481–493. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, X.; Che, S.; Cui, J.; Ma, X.; An, X.; Cao, B.; Song, Y. Endometrial epithelial cell apoptosis is inhibited by a ciR8073-miR181a-neurotensis pathway during embryo implantation. Mol. Ther. Nucleic Acids 2019, 14, 262–273. [Google Scholar] [CrossRef]
- Shang, J.; Chen, W.M.; Wang, Z.H.; Wei, T.N.; Chen, Z.Z.; Wu, W.B. CircPAN3 mediates drug resistance in acute myeloid leukemia through the miR-153-5p/miR-183-5p-XIAP axis. Exp. Hematol. 2019, 70, 42–54.e43. [Google Scholar] [CrossRef]
- Liu, J.; Kong, F.; Lou, S.; Yang, D.; Gu, L. Global identification of circular RNAs in chronic myeloid leukemia reveals hsa_circ_0080145 regulates cell proliferation by sponging miR-29b. Biochem. Biophys. Res. Commun. 2018, 504, 660–665. [Google Scholar] [CrossRef]
- Huang, L.; Chen, M.; Pan, J.; Yu, W. Circular RNA circNASP modulates the malignant behaviors in osteosarcoma via miR-1253/FOXF1 pathway. Biochem. Biophys. Res. Commun. 2018, 500, 511–517. [Google Scholar] [CrossRef]
- Feng, C.; Li, Y.; Lin, Y.; Cao, X.; Li, D.; Zhang, H.; He, X. CircRNA-associated ceRNA network reveals ErbB and Hippo signaling pathways in hypopharingeal cancer. Int. J. Mol. Med. 2019, 43, 127–142. [Google Scholar]
- Jin, X.; Feng, C.Y.; Xiang, Z.; Chen, Y.P.; Li, Y.M. CircRNA expression pattern and circRNA-miRNA-mRNA network in the pathogenesis of nonalcoholic steatohepatitis. Oncotarget 2016, 7, 66455–66467. [Google Scholar] [CrossRef]
- Jiang, C.; Xu, D.; You, Z.; Xu, K.; Tian, W. Dysregulated circRNAs and ceRNA network in esophageal squamous cell carcinoma. Front. Biosci. 2019, 1, 277–290. [Google Scholar]
- Campos-Melo, D.; Droppelmann, C.A.; Volkening, K.; Strong, M.J. Comprehensive luciferase-based reporter gene assay reveals previously masked up-regulatory effects of miRNAs. Int. J. Mol. Sci. 2014, 15, 15592–15602. [Google Scholar] [CrossRef]
- Tan, S.M.; Lieberman, J. Capture and identification of miRNA targets by biotin pulldown and RNA-seq. Methods Mol. Biol. 2016, 1358, 211–228. [Google Scholar] [CrossRef]
- Xie, B.; Zhao, Z.; Liu, Q.; Wang, X.; Ma, Z.; Li, H. CircRNA has_circ_0078710 acts as the sponge of microRNA-31 involved in hepatocellular carcinoma progression. Gene 2019, 683, 253–261. [Google Scholar] [CrossRef]
- Ni, H.; Li, W.; Zhuge, Y.; Xu, S.; Wang, Y.; Chen, Y.; Shen, G.; Wang, F. Inhibition of circHIPK3 prevents angiotensin II-induced cardiac fibrosis by sponging miR-29b-3p. Int. J. Cardiol. 2019, 292, 188–196. [Google Scholar] [CrossRef]
- Guo, J.; Duan, H.; Li, Y.; Yang, L.; Yuan, L. A novel circular RNA circ-ZNF652 promotes hepatocellular carcinoma metastasis through inducing snail-mediated epithelial-mesenchymal transition by sponging miR-203/miR-502-5p. Biochem. Biophys. Res. Commun. 2019, 513, 812–819. [Google Scholar] [CrossRef]
- Li, Y.; Wan, B.; Liu, L.; Zhou, L.; Zeng, Q. Circular RNA circMTO1 suppresses bladder cancer metastasis by sponging miR-221 and inhibiting epithelial-to-mesenchymal transition. Biochem. Biophys. Res. Commun. 2019, 508, 991–996. [Google Scholar] [CrossRef]
- Wang, S.; Li, Q.; Wang, Y.; Li, X.; Wang, R.; Kang, Y.; Xue, X.; Meng, R.; Wei, Q.; Feng, X. Upregulation of circ-UBAP2 predicts poor prognosis and promotes triple-negative breast cancer progression through the miR-661/MTA1 pathway. Biochem. Biophys. Res. Commun. 2018, 505, 996–1002. [Google Scholar] [CrossRef]
- Han, D.; Li, J.; Wang, H.; Su, X.; Hou, J.; Gu, Y.; Qian, C.; Lin, Y.; Liu, X.; Huang, M.; et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology 2017, 66, 1151–1164. [Google Scholar] [CrossRef]
- Cherubini, A.; Barilani, M.; Rossi, R.L.; Jalal, M.M.K.; Rusconi, F.; Buono, G.; Ragni, E.; Cantarella, G.; Simpson, H.; Peault, B.; et al. FOXP1 circular RNA sustains mesenchymal stem cell identity via microRNA inhibition. Nucleic Acids Res. 2019, 47, 5325–5340. [Google Scholar] [CrossRef]
- Wu, Z.; Huang, W.; Wang, X.; Wang, T.; Chen, Y.; Chen, B.; Liu, R.; Bai, P.; Xing, J. Circular RNA CEP128 acts as a sponge of miR-145-5p in promoting the bladder cancer progression via regulating SOX11. Mol. Med. 2018, 24, 40. [Google Scholar] [CrossRef]
- Betel, D.; Wilson, M.; Gabow, A.; Marks, D.S.; Sander, C. The microRNA.org resource: Targets and expression. Nucleic Acids Res. 2008, 36, D149–D153. [Google Scholar] [CrossRef]
- Rennie, W.; Kanoria, S.; Liu, C.; Mallick, B.; Long, D.; Wolenc, A.; Carmack, C.S.; Lu, J.; Ding, Y. STarMirDB: A database of microRNA binding sites. RNA Biol. 2016, 13, 554–560. [Google Scholar] [CrossRef]
- Rennie, W.; Liu, C.; Carmack, C.S.; Wolenc, A.; Kanoria, S.; Lu, J.; Long, D.; Ding, Y. STarMir: A web server for prediction of microRNA binding sites. Nucleic Acids Res. 2014, 42, W114–W118. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Grimson, A.; Farh, K.K.; Johnston, W.K.; Garrett-Engele, P.; Lim, L.P.; Bartel, D.P. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol. Cell 2007, 27, 91–105. [Google Scholar] [CrossRef]
- Loeb, G.B.; Khan, A.A.; Canner, D.; Hiatt, J.B.; Shendure, J.; Darnell, R.B.; Leslie, C.S.; Rudensky, A.Y. Transcriptome-wide miR-155 binding map reveals widespread noncanonical microRNA targeting. Mol. Cell 2012, 48, 760–770. [Google Scholar] [CrossRef]
- Sticht, C.; De La Torre, C.; Parveen, A.; Gretz, N. miRWalk: An online resource for prediction of microRNA binding sites. PLoS ONE 2018, 13, e0206239. [Google Scholar] [CrossRef]
- Rehmsmeier, M.; Steffen, P.; Hochsmann, M.; Giegerich, R. Fast and effective prediction of microRNA/target duplexes. RNA 2004, 10, 1507–1517. [Google Scholar] [CrossRef]
- Hammond, S.M.; Bernstein, E.; Beach, D.; Hannon, G.J. An RNA-directed nuclease mediates post-transcritpional gene silencing in Drosophila cells. Nature 2000, 404, 293–296. [Google Scholar] [CrossRef]
- Hutvágner, G.; Zamore, P.D. A microRNA in a multiple-turnover RNAi enzyme complex. Science 2002, 297, 2056–2060. [Google Scholar] [CrossRef]
- Hafner, M.; Landthaler, M.; Burger, L.; Khorshid, M.; Hausser, J.; Berninger, P.; Rothballer, A.; Ascano, M., Jr.; Jungkamp, A.C.; Munschauer, M.; et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 2010, 141, 129–141. [Google Scholar] [CrossRef]
- Helwak, A.; Kudla, G.; Dudnakova, T.; Tollervey, D. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell 2013, 153, 654–665. [Google Scholar] [CrossRef]
- Jungkamp, A.-C.; Stoeckius, M.; Mecenas, D.; Grün, D.; Mastrobuoni, G.; Kempa, S.; Rajewsky, N. In vivo and transcriptome-wide identification of RNA binding protein target sites. Mol. Cell 2011, 44, 828–840. [Google Scholar] [CrossRef]
- Grosswendt, S.; Filipchyk, A.; Manzano, M.; Klironomos, F.; Schilling, M.; Herzog, M.; Gottwein, E.; Rajewsky, N. Unambiguous identification of miRNA:target site interactions by different types of ligation reactions. Mol. Cell 2014, 54, 1042–1054. [Google Scholar] [CrossRef]
- Chi, S.W.; Zang, J.B.; Mele, A.; Darnell, R.B. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 2009, 460, 479–486. [Google Scholar] [CrossRef]
- Leung, A.K.; Young, A.G.; Bhutkar, A.; Zheng, G.X.; Bosson, A.D.; Nielsen, C.B.; Sharp, P.A. Genome-wide identification of Ago2 binding sites from mouse embryonic stem cells with and without mature microRNAs. Nat. Struct. Mol. Biol. 2011, 18, 237–244. [Google Scholar] [CrossRef]
- Clark, P.M.; Loher, P.; Quann, K.; Brody, J.; Londin, E.R.; Rigoutsos, I. Argonaute CLIP-Seq reveals miRNA targetome diversity across tissue types. Sci. Rep. 2014, 4, 5947. [Google Scholar] [CrossRef]
- Boeckel, J.N.; Jae, N.; Heumuller, A.W.; Chen, W.; Boon, R.A.; Stellos, K.; Zeiher, A.M.; John, D.; Uchida, S.; Dimmeler, S. Identification and characterization of hypoxia-regulated endothelial circular RNA. Circ. Res. 2015, 117, 884–890. [Google Scholar] [CrossRef]
- Moore, M.; Scheel, T.; Luna, J.; Park, C.; Fak, J.; Nishiuchi, E.; Rice, C.; Darnell, R. miRNA-target chimeras reveal miRNA 3′-end pairing as a major determinant of Argonaute target specificity. Nat. Commun. 2015, 6, 8864. [Google Scholar] [CrossRef]
- Ceci, M.; Pio, G.; Kuzmanovski, V.; Dzeroski, S. Semi-supervised multi-view learning for gene network reconstruction. PLoS ONE 2015, 10, e0144031. [Google Scholar] [CrossRef]

| Database | Website | Organisms | Features |
|---|---|---|---|
| circBase | http://www.circbase.org | Human Mouse Fly Worm Fish Planaria | Most updated catalogue of predicted circRNAs. Beside human and mouse, it also collects data from several other organisms |
| circInteractome | https://circinteractome.nia.nih.gov | Human | Enables the prediction and mapping of binding sites for RNA binding proteins and miRNA on known circRNAs. It includes also a module for siRNA design for knock-down experiments and primer design for PCR |
| circNet | http://syslab5.nchu.edu.tw/CircNet/ | Human | Provides tissue-specific expression patterns, integrated miRNA-circRNA-mRNA networks, circRNA isoform expression and genomic annotation |
| ENCORI StarBase v2 | http://starbase.sysu.edu.cn/index.php http://starbase.sysu.edu.cn/starbase2/index.php | Human Mouse Worm | Designed for investigating interaction networks of lncRNAs, miRNAs, ceRNA, RNA binding proteins and mRNAs from public CLIP-Seq data. It also allows to browse for circRNA-miRNA interactions. |
| circ2Traits | http://gyanxet-beta.com/circdb/ | Human | Link of circRNA with disease inferred by miRNA-disease associations |
| Tool | Website | Organisms | Browse by Sequenc/Gene ID | Standalone Version |
|---|---|---|---|---|
| STarMir | http://sfold.wadsworth.org/cgi-bin/starmirtest2.pl | Human Mouse Worm Other | Sequence/ Gene ID | no |
| STarMirDB | http://sfold.wadsworth.org/starmirDB.php | Human Mouse Worm | Gene ID | no |
| PITA | https://genie.weizmann.ac.il/pubs/mir07/index.html | Human Mouse Fly Worm | Gene ID | yes |
| miRanda/ mirSVR | http://www.microrna.org/microrna/home.do | Human Mouse Rat Fly Worm | Gene ID | yes |
| TargetScan | http://www.targetscan.org/vert_72/ | Human Mouse Fly Worm Zebrafish | Gene ID | yes |
| RNAhybrid | https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid/ | Any | Sequence | yes |
| TarBase v8 | http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php | Human Mouse Rat Chicken Zebrafish Fly Worm Chimpanzees Macaque Soy Maize Barrelclover Grape wine Earthmoss Epstein–Barr virus KSHV | Gene ID | no |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dori, M.; Bicciato, S. Integration of Bioinformatic Predictions and Experimental Data to Identify circRNA-miRNA Associations. Genes 2019, 10, 642. https://doi.org/10.3390/genes10090642
Dori M, Bicciato S. Integration of Bioinformatic Predictions and Experimental Data to Identify circRNA-miRNA Associations. Genes. 2019; 10(9):642. https://doi.org/10.3390/genes10090642
Chicago/Turabian StyleDori, Martina, and Silvio Bicciato. 2019. "Integration of Bioinformatic Predictions and Experimental Data to Identify circRNA-miRNA Associations" Genes 10, no. 9: 642. https://doi.org/10.3390/genes10090642
APA StyleDori, M., & Bicciato, S. (2019). Integration of Bioinformatic Predictions and Experimental Data to Identify circRNA-miRNA Associations. Genes, 10(9), 642. https://doi.org/10.3390/genes10090642





