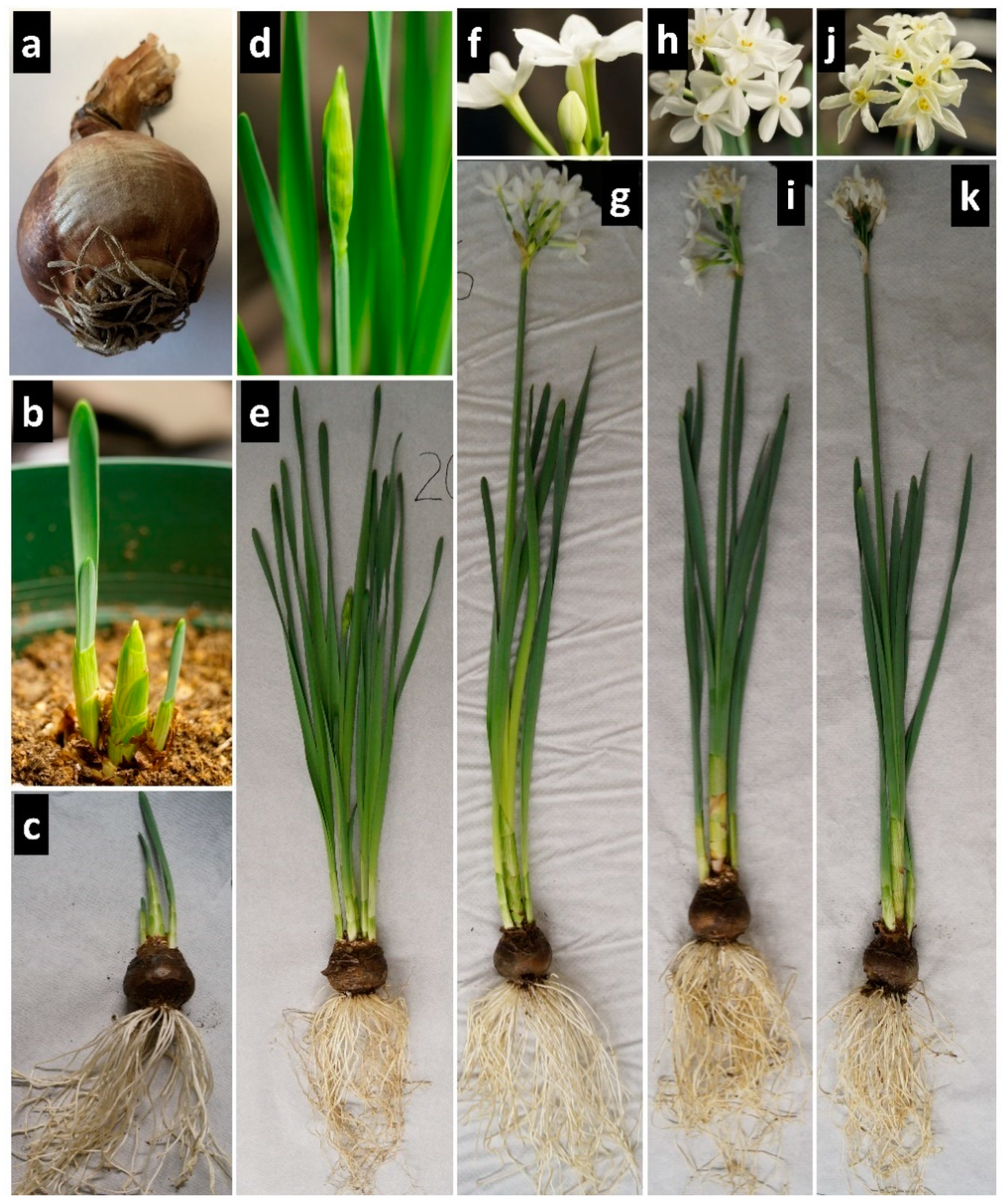
3.1. RNA-Seq of Narcissus papyraceus Bulb
To study gene expression in non-model plants such as Amaryllidaceae, for which genomic information is lacking, a transcriptome must be de novo assembled. To achieve this, RNA-Seq of the
N. papyraceus bulb was first performed. The bulb was chosen because of its high alkaloid content. For example, Viladomat et al. (1986) [
34] found that the bulb of
N. assoanus accumulated a large amount of alkaloids, while Kreh (2002) [
32] reported that the concentration of galanthamine was relatively high in the bulb of
N. pseudonarcissus (L.) cv. Carlton. In a more recent study by Shawky et al. (2015) [
33], in which nine different samples (different parts at different developmental stages) of
N. papyraceus were analyzed, alkaloids were found to be most abundant in the bulb.
RNA was extracted from
N. papyraceus bulbs and screened for sufficient quality and quantity prior to cDNA library creation and deep sequencing. A total of 70,409,091 raw paired reads were obtained, which were trimmed to give 64,038,268 surviving paired reads that corresponds to 91% of the initial raw reads (
Table 1;
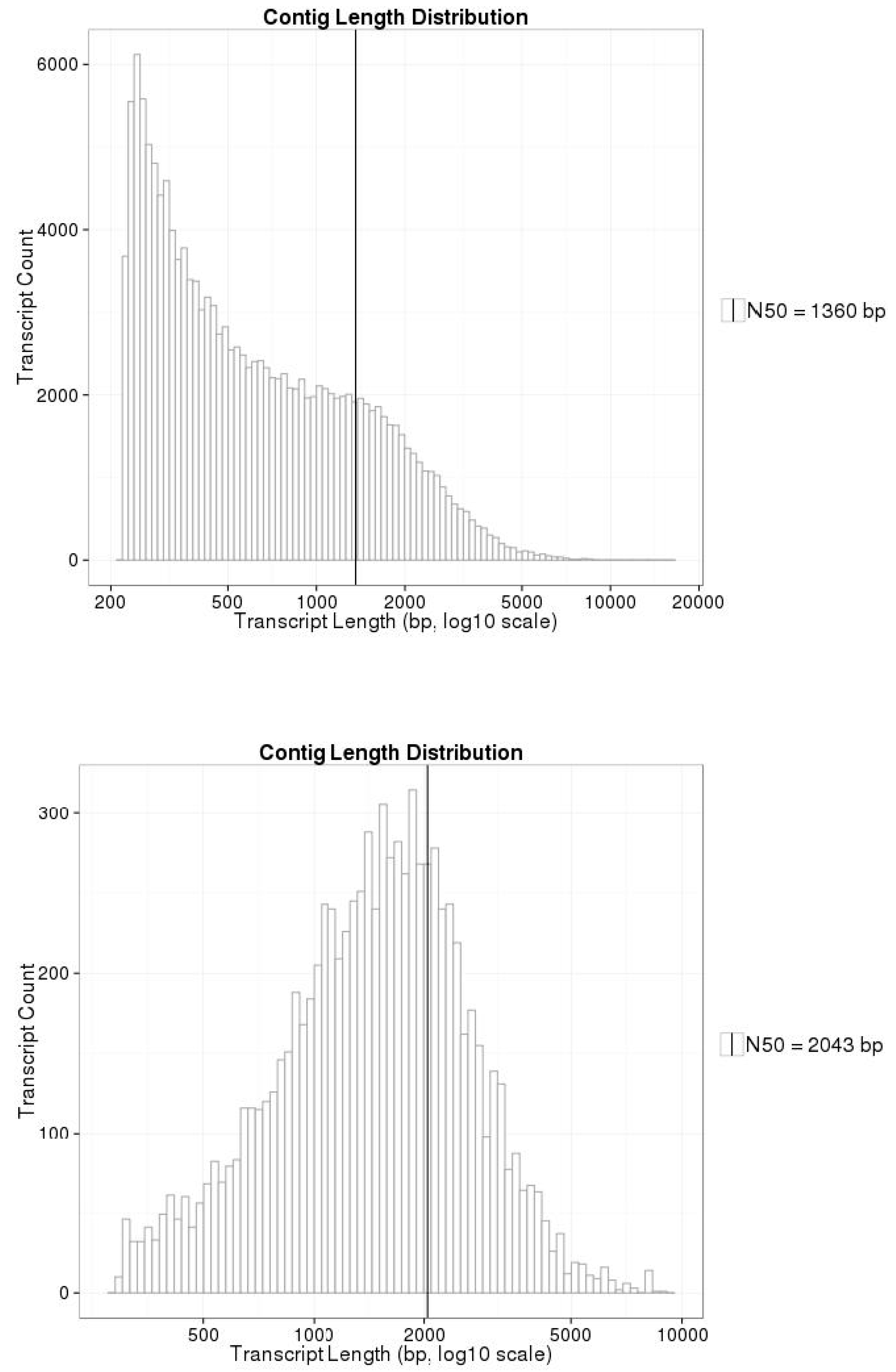
Table A5). A total of 8,945,044 paired reads were obtained after the normalization step, corresponding to 14% of the initial raw reads. The normalized reads were used to assemble the transcriptome generating 148,563 transcripts (or unigenes) with a length distribution N50 of 1360 bp (
Figure A2). For identification of the transcripts, they were aligned against the uniprot_sprot.trinotate_v2.0.pep protein database. BLAST annotation yielded 8866 transcripts with an average length of 2043 bp (
Table 1;
Table A5). Thus, the remaining 139,697 transcripts represented a large number of transcript sequences showing no similarity to known genes. They appeared to represent transcripts of uncharacterized genes or sequences specific to
N. papyraceus. Altogether, we concluded that a good quality transcriptome was developed with a high number of surviving paired reads and full-length assembled transcripts.
To identify specific transcripts encoding enzymes involved in the AA biosynthesis in our assembled transcriptome, we performed local BLASTx analyses (
Table 2). Several transcript variants of orthologous genes were identified. In a study conducted by Singh and Desgagné-Penix (2017) [
25], two
TYDC transcript variants were identified in
N. pseudonarcissus. We observed that
N. papyraceus’s TYDC1 and TYDC2 together share 57% homology of the amino acid sequence with
N. pseudonarcissus’s TYDC1 and TYDC2;
N. papyraceus TYDC1 is 90% homologous to
N. pseudonarcissus TYDC1 and
N. papyraceus TYDC2 is 94% homologous to
N. pseudonarcissus TYDC2. The BLASTx analysis further reported that
N. papyraceus TYDC1 shared 70% homology with opium poppy’s (
Papaver somniferum) TYDC2, which is involved in the synthesis of benzylisoquinoline alkaloids, and that
N. papyraceus TYDC2 was 74% homologous to rice’s (
Oryza sativa) TYDC1 (
Table 2).
PAL genes have also been reported in various plant species including the Amaryllidaceae
Lycoris radiata and
N. pseudonarcissus in which two transcript variants were identified in each species [
25,
31,
45]. BLASTx searches for
PAL genes in the assembled
N. papyraceus bulb transcriptome led to the identification of three full-length transcript variants (
Table 2). Similarly, several transcript variants were also identified for
C4H,
4CL and
HCT (
Table 2). For gene transcripts “specific” to AA biosynthesis, we identified one transcript variant of
NBS, one of
N4OMT, two of
CYP96T and three of
NorRed (
Table 2). Altogether, we were able to identify full-length transcripts, and several variants, with expect values ≤ 0, corresponding to genes encoding biosynthetic enzymes involved in AA production.
Comparative FPKM digital expression of the
N. papyraceus bulb transcriptome indicated that the “AA-specific” gene transcripts, such as those of
N4OMT,
CYP96T and
NorRed, were more abundantly expressed than transcripts encoding enzymes involved in the “AA-precursor” biochemical reactions including those of
TYDC,
PAL and others (
Table 2). In addition,
TYDC and
PAL had similar levels of expression (
Table 2). They may thus be coordinately regulated since both these genes encode enzymes catalyzing the first reactions in the AA biosynthetic pathway (
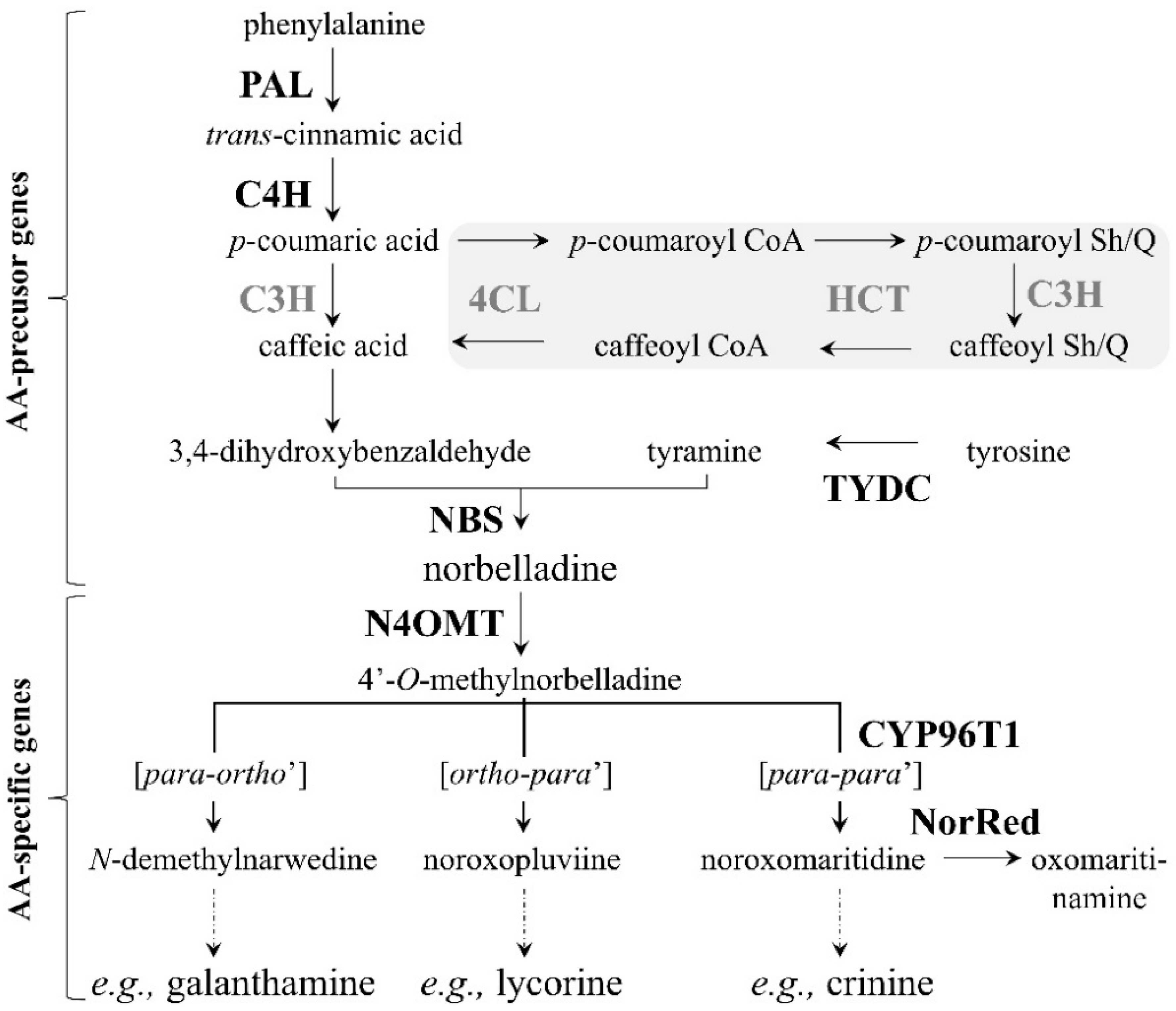
Figure 1). Furthermore, the digital expression profile of the
N. papyraceus bulb is comparable to that reported for the
N. pseudonarcissus bulb [
25].
3.2. Gene Expression Analysis with RT-qPCR
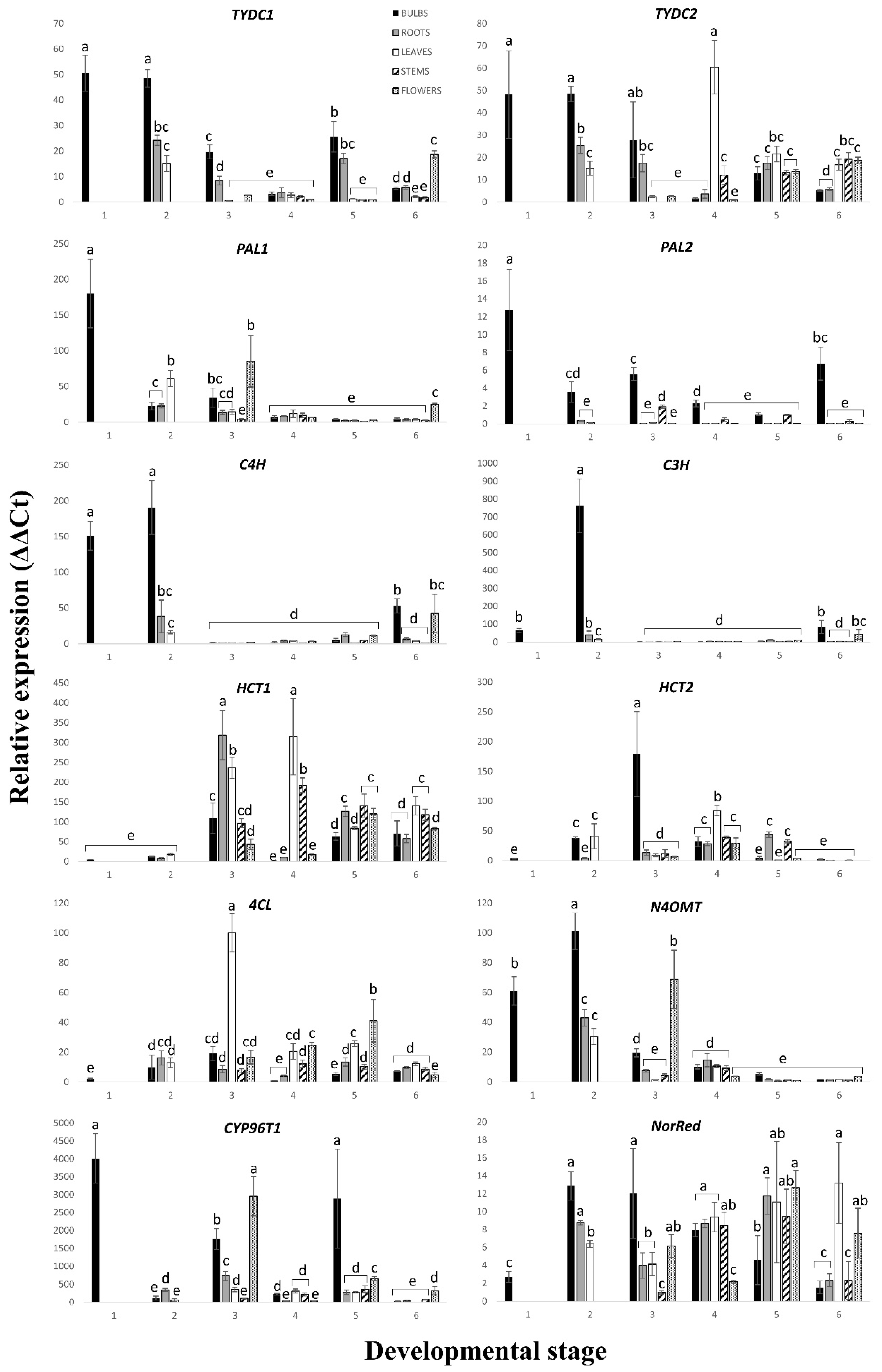
The relative expression levels of the genes analyzed are shown in
Figure 3. The results indicate that the transcripts exhibited different expression kinetics that can be described as early (stage 1), mid (stages 2–4) and late (stages 5–6) responses. The fold-expression varied from relatively low (<2-fold) to high (>4000-fold), compared to each other. Looking at the expression levels of any gene in all the 24
N. papyraceus samples, it is obvious that its expression differs among the various parts and changes as the plant grows and develops. For example, the physiological conditions of an organism, as well as environmental stimuli, have long been known to affect the transcriptional regulation of PAL [
46]. The same may, most likely, be true for all the other AA biosynthetic genes, which can explain the observed spatial and temporal differences in gene expression. Obviously, as time goes on, a plant adapts to its changing needs and responds to changes in its environment, to varying extents, in the various parts of the plant.
The biosynthetic genes
PAL,
C4H,
4CL,
HCT and
C3H (
Figure 1) are involved in phenylpropanoid biosynthesis. In other words, 3,4-DHBA is a product of the phenylpropanoid biosynthetic pathway [
25]. This pathway is responsible for the production of a large number of specialized metabolites including coumarins, hydroxycinnamates and lignins [
47,
48]. These compounds play essential roles in plant growth and development and plant–environment interactions [
46]. Being one of the first biosynthetic genes of the pathway, PAL is responsible for converting the amino acid phenylalanine, a product of primary metabolism, to trans-cinnamic acid (
Figure 1). Thus, PAL resides at a metabolically important position, linking primary metabolism to specialized metabolism [
49]. The same is true for the biosynthetic gene
TYDC, which encodes TYDC that is responsible for transforming the amino acid tyrosine (primary metabolite) to tyramine. In addition to AAs, TYDC is involved in the biosynthesis of several other types of specialized metabolites [
27] including benzylisoquinoline alkaloids [
50] and hydroxycinnamic acid amides of tyramine (HCAATs) [
51]. Morphine and codeine are the best-known examples of benzylisoquinoline alkaloids produced by the opium poppy. The production of HCAATs has been associated with plant stress responses. Following oxidative polymerization, HCAATs are believed to reinforce cell walls rendering them less susceptible to penetration by pathogens. Since
PAL and
TYDC are genes encoding the first biosynthetic enzymes involved in the production of many types of specialized metabolites, they have attracted considerable attention and are targets for metabolic engineering [
52].
Several transcript variants of
PAL exist since PAL is encoded by a multi-gene family in plants [
53]. For example, there are four variants of
PAL in Arabidopsis, five in poplar and nine in rice [
54]. We analyzed the expression of two transcript variants of
PAL in our study of
N. papyraceus (
Figure 3). At each developmental stage,
PAL1 was expressed at generally similar levels in all parts.
PAL2, for its part, was expressed the highest in the bulb, at almost all stages, followed by the stem, indicating that PAL2 is more specifically involved in the synthesis of phenylpropanoids in these two parts. This profile of
PAL2 expression has also been reported for
PAL2 of
N. pseudonarcissus ‘King Alfred’ [
25]. The fact that the two transcript variants are expressed at different levels in the various
N. papyraceus plant samples suggests that they are involved in the phenylpropanoid pathway to different degrees for the production of various specialized metabolites in the plant.
Such differential spatial and/or temporal expression of variants of
PAL, associated with specific metabolic activities, has also been reported for other plants species. For example, in a study conducted on
PAL in another Amaryllidaceae species,
Lycoris radiata, Jiang et al. (2013) [
31] identified two transcript variants of
PAL, which differed in their expression in different parts of the flower at the blooming stage of the plant; the expression of
LrPAL2 was always much higher than that of
LrPAL1. Singh and Desgagné-Penix (2017) [
25] also noted different expression profiles for two
PAL variants in five parts of
N. pseudonarcissus ‘King Alfred’ at the flowering stage. In both studies, the authors suggest that the
PAL variants encode enzymes that may have distinct functions in different branches of the phenylpropanoid pathway. In an analysis of PAL in quaking aspen (
Populus tremuloides Michx.), Kao et al. (2002) [
55] observed that
PtPAL1 is implicated in the biosynthesis of non-lignin metabolites while
PtPAL2 is involved in lignin formation. Lignins are phenylpropanoid-derived compounds involved in cell wall formation.
Regarding the biosynthetic gene
TYDC, similar to
PAL, it is encoded by multiple genes. For example, eight copies of the
TYDC gene have been detected in the opium poppy [
57], whereas four similar genes encode
TYDC in parsley [
58]. In our study of
N. papyraceus, when comparing the expression profiles of
TYDC1 and
TYDC2, the most interesting feature is that they are practically the same in the first three developmental stages (
Figure 3). This may be a sign that these two transcript variants are regulated in a similar manner early in the life of the plant. However, differences in gene expression levels between
TYDC1 and
TYDC2 appear in the later stages of 4–6.
Different expression profiles for different
TYDC transcript variants have also been observed in the mature opium poppy [
59]. They noted that
TYDC1-like genes were predominantly expressed in the roots and, to a much lesser extent, in the stem. The opposite was true for
TYDC2-like genes. In the opium poppy,
TYDC is involved in the synthesis of benzylisoquinoline alkaloids. Facchini and De Luca (1995) [
59] analyzed the benzylisoquinoline alkaloids sanguinarine and morphine, each of which is synthesized by different branch pathways further downstream in the benzylisoquinoline alkaloid biosynthetic pathway [
50]. The presence of high quantities of sanguinarine in the roots, where
TYDC1-like genes were highly expressed, and the lack of detectable quantities of sanguinarine in the aerial parts, where
TYDC1-like genes were lowly expressed, led the authors to believe that
TYDC1-like genes were particularly involved in the production of sanguinarine and other related alkaloids. On the other hand, the abundance of morphine in the aerial parts together with the high levels of
TYDC2-like transcripts in the stem, led the authors to suggest that
TYDC2-like genes may be implicated in the synthesis of morphine and other closely related alkaloids. Based on these observations, the authors propose that
TYDC1-like genes may be coordinately regulated with genes present in the branch pathway involving sanguinarine biosynthesis, whereas
TYDC2-like genes may be coordinately regulated with genes present in the branch pathway involving morphine biosynthesis. Such coordinated regulation of expression between different genes involved upstream and downstream in a biosynthetic pathway may be occurring in the AA pathway as well. Further elucidation of the AA pathway will eventually allow the investigation of whether such a phenomenon is present in the AA biosynthetic pathway.
HCT is another gene for which we analyzed the expression levels of two transcript variants (
Figure 3). Like for
PAL and
TYDC, there are considerable differences between the expression level profiles of the
HCT variants. We assume that, like for
PAL, the different variants of
HCT are involved to varying extents in the phenylpropanoid pathway.
HCT has been reported to be quite active in lignin formation [
60,
61]. Therefore, maybe the relatively higher expression of
HCT1 during stages 3–6 could be due to a possibly greater production of lignins during these stages and that
HCT1 in
N. papyraceus is perhaps more involved in lignin formation than
HCT2.
C4H and
C3H are two biosynthetic genes in the phenylpropanoid pathway, which code for cytochrome P450 enzymes. Curiously, both genes display a similar expression pattern for stages 2–6 (
Figure 3). It, thus, seems that these two genes are being coordinately regulated as the plant is growing and developing. Regarding
C4H, Fock-Bastide et al. (2014) [
62] reported that its expression level in
Vanilla planifolia pods was highest during the earlier stages of its maturation, but declined during the later stages. This expression profile resembles that observed by us for
N. papyraceus in that
C4H expression is high during the 1st and 2nd developmental stages (
Figure 3). During stages 3–5, the level of expression of
C4H remains constant even though the
N. papyraceus plant continues to grow and develop during these stages. Phimchan et al. (2014) [
63] also noted that
C4H activity remained constant in various cultivars of
Capsicum regardless of fluctuations in capsaicinoid accumulation in these cultivars.
Unlike the genes discussed so far,
N4OMT is specific to AA biosynthesis. N4OMT is responsible for the methylation of norbelladine, the compound from which all AAs are derived (
Figure 1). Kilgore et al. (2014) [
30] and Singh and Desgagné-Penix (2017) [
25] analyzed the expression of
N4OMT in the
Narcissus species at the flowering stage in different parts: Bulb, leaves, flowers and/or roots and stem. Kilgore et al. (2014) [
30] noted that the highest expression was in the bulb, followed by the flowers in close second, while it was barely expressed in the leaves. Singh and Desgagné-Penix’s (2017) [
25] results demonstrated that the highest expression was, once again, in the bulb but very low in all the other parts. In our results of
N4OMT’s expression in
N. papyraceus at developmental stage 5 (flowering stage;
Figure 3), we observed that the expression in the bulb was slightly higher than that in the other tissues, and the lowest expression was in the leaves. Our results abound in the same way as those presented above with smaller differences. One reason for the differences in the results of
N4OMT expression between our and the other two studies may be that the same
Narcissus species was not analyzed in each study.
3.3. Alkaloid Profiling
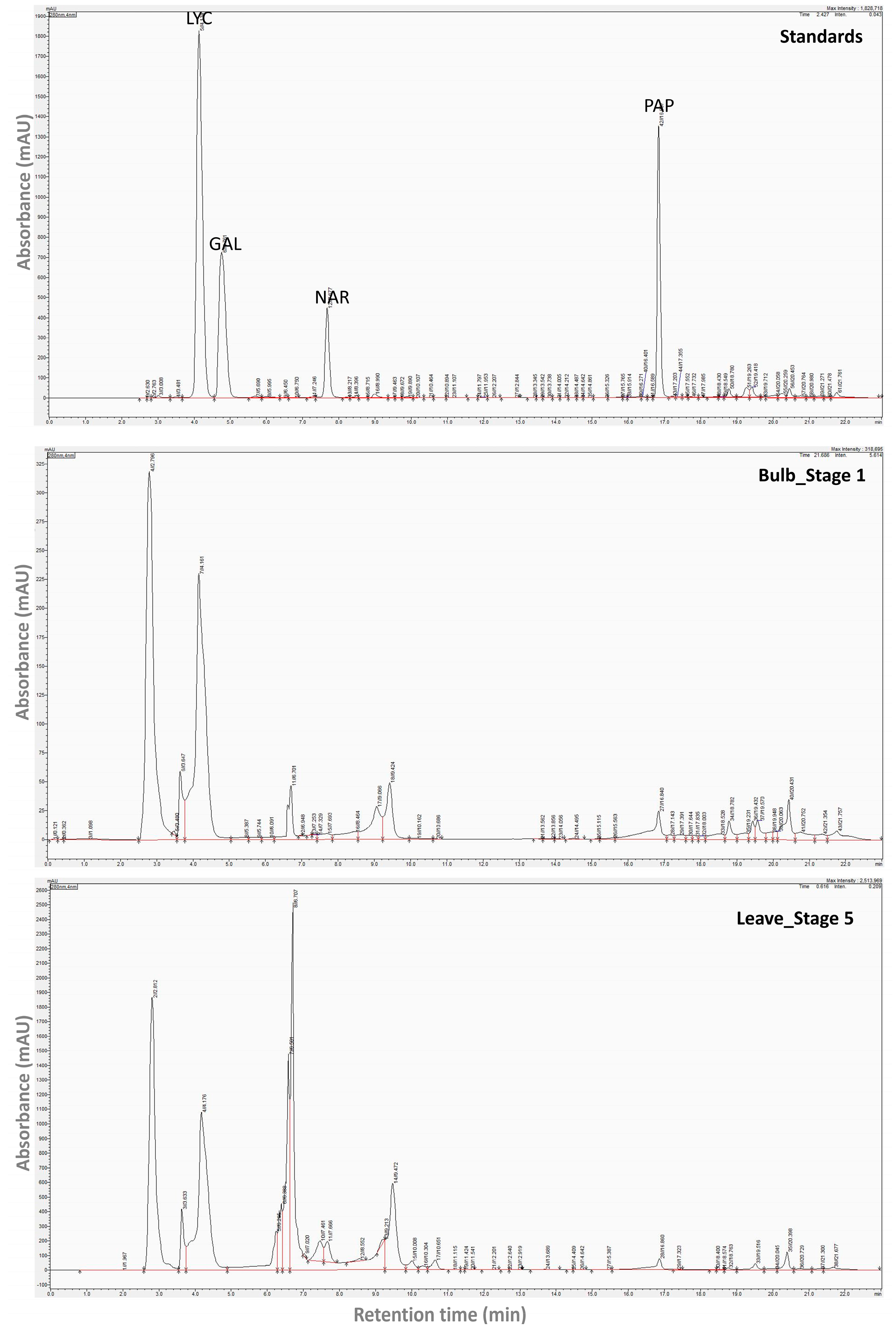
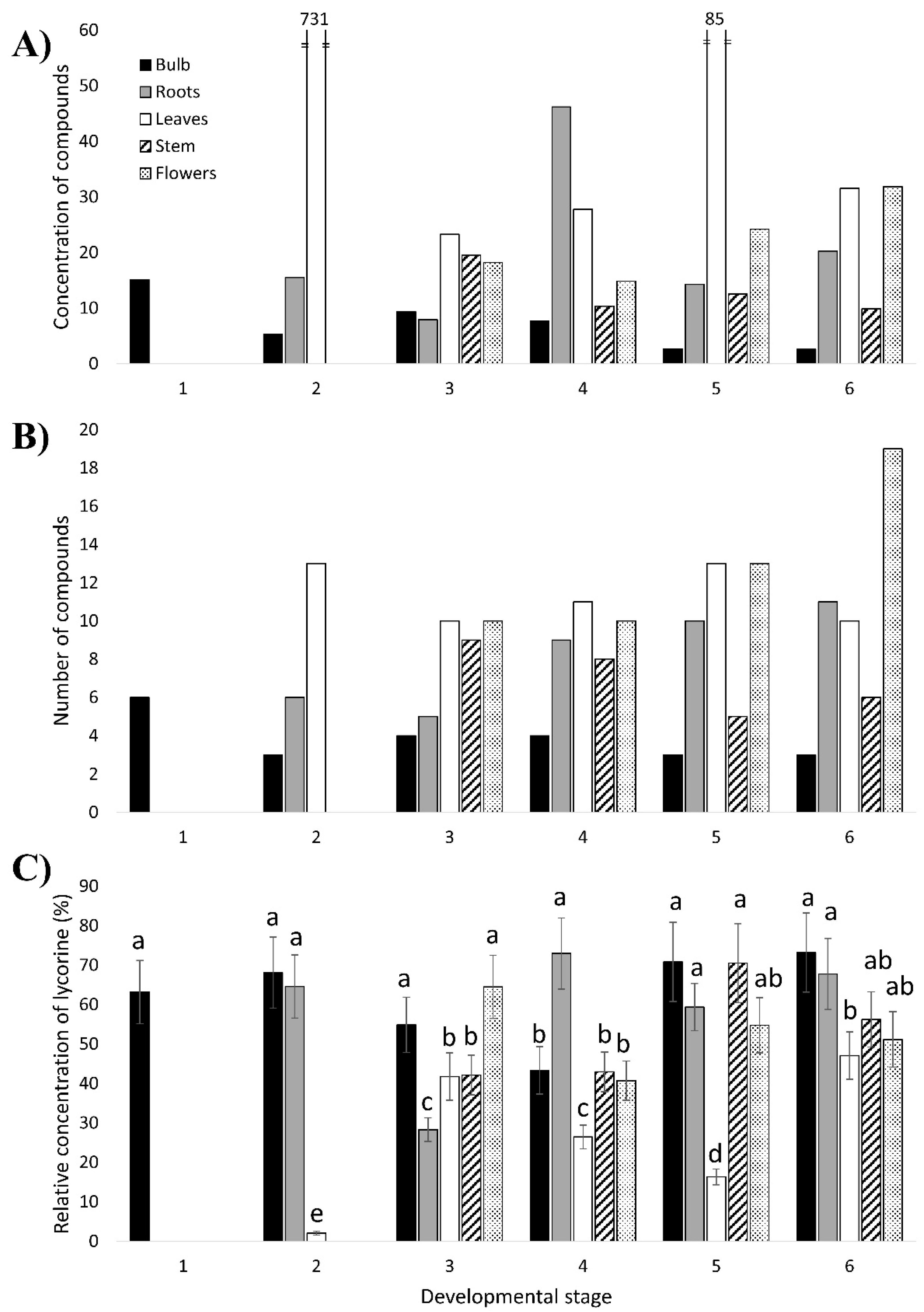
Representative chromatograms of the alkaloid extract of certain
N. papyraceus samples are shown in
Figure A3. Among the various compounds detected in all the
N. papyraceus samples, 28 presented interesting features and were selected for analysis (
Table A4). Some of these were detected in all or almost all the samples, while others were present in particular parts throughout the life of the plant. The most striking findings are the extremely high and very high concentration of compounds in the leaves of stages 2 and 5, respectively (
Figure 4A). This is specifically due to the six compounds with an average Rt ranging from 6.43 to 7.20, which are present at surprisingly high concentrations (
Table A4). Looking at the distribution of the compounds among the different parts of the plant, the leaves and flowers had the highest abundance of compounds, whereas the bulb, the lowest (
Figure 4A,B). A similar finding was reported in a recent study of AA biosynthesis in
N. pseudonarcissus ‘King Alfred’ using LC–MS in that leaves had the highest concentration of AAs [
25].
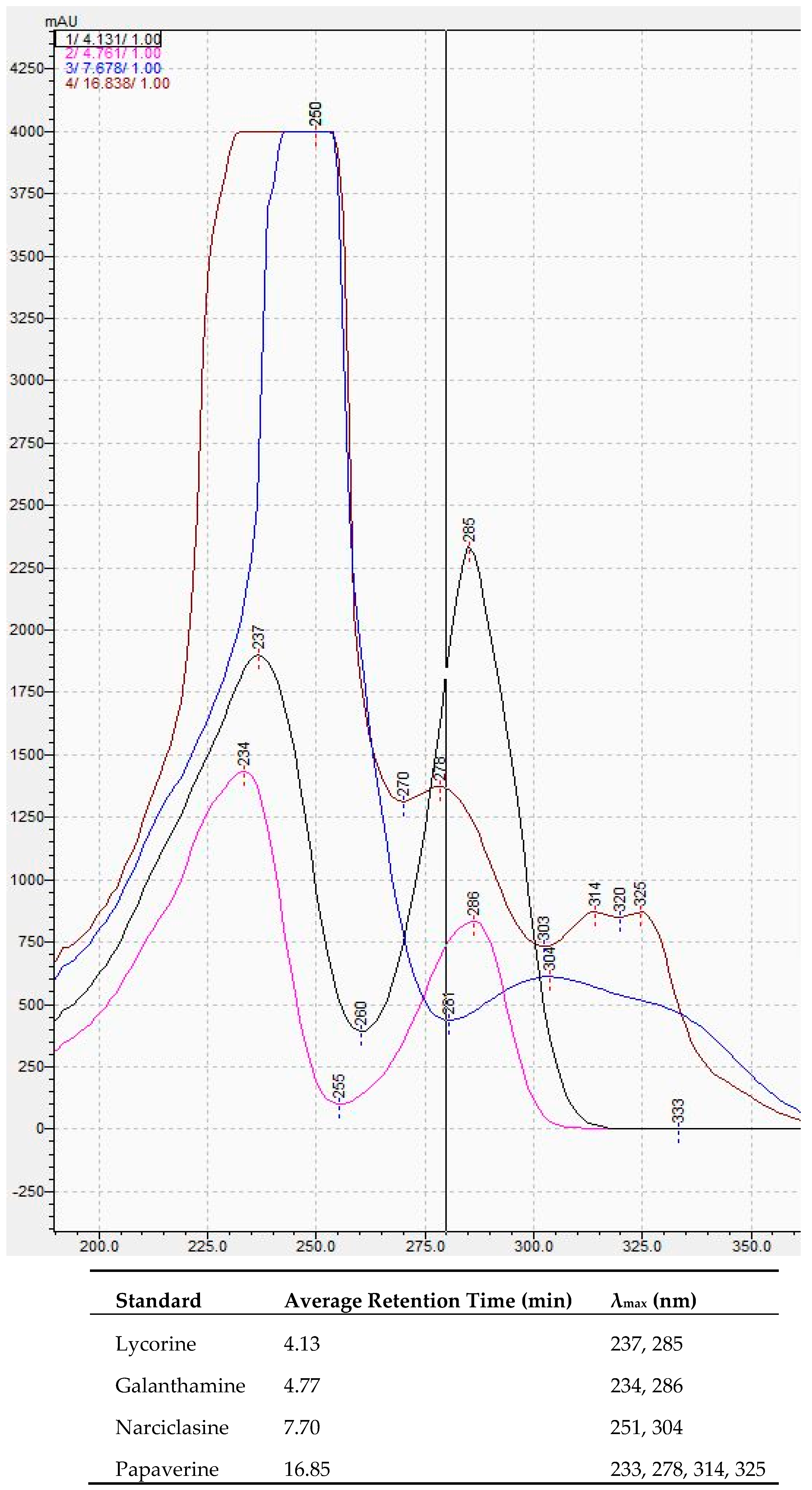
By comparing the 28 selected compounds (
Table A4) with the results obtained for the AA standards (
Figure A1), the following conclusions can be made. The compound with an average Rt of 4.22 min (average λ
max of 221 and 285 nm, not shown) corresponds to lycorine since these values are in agreement with those of the lycorine standard. Contrary to lycorine, galanthamine was not detected in any of the samples. Narciclasine also does not seem to be present even though there was a compound detected in almost all of the samples with an average Rt of 7.69 min, which is very close to that obtained for narciclasine (7.70 min). However, the mean λ
max values (221 and 283 nm, not shown) of this compound differ considerably from those of narciclasine (251 and 304 nm). Therefore, among the three AA standards, only lycorine was found to be detected in
N. papyraceus.
Lycorine was detected in each plant sample and had a high concentration in most samples (
Figure 4C;
Table A4). This is in accordance with the findings reported by Shawky et al. (2015) [
33] and confirmed by Tarakemeh et al. (2019) [
64] who found that lycorine was the predominant alkaloid in
N. papyraceus with no/low galanthamine, and with the fact that lycorine is one of the most abundant AAs in the
Narcissus genus [
65]. A reason for this is that lycorine is possibly, phylogenetically, among the oldest AAs in
Narcissus [
66]. Furthermore, lycorine was present at higher concentrations in the leaf samples compared to the bulb samples (
Table A4). This is in agreement with gas chromatography-mass spectrometry (GC–MS) results of six ornamental species of
Narcissus [
65]. It is also interesting to note that the fraction of the concentration of lycorine out of the total concentration of heterocyclic compounds in each plant part remains more or less constant as the plant ages (
Figure 4C;
Table A4). This suggests that a somewhat constant level of production of lycorine occurs in each part of the plant throughout its growth and development.
3.4. Comparison of Gene Expression and Compound Content Profiles
To better understand the involvement of the proposed AA biosynthetic genes in the production of the AAs at each developmental stage of
N. papyraceus, we compared the expression profiles (
Figure 3) of the AA biosynthetic genes with the heterocyclic compound content profiles (
Figure 4A,B). It should be noted that HPLC is a limited analytical method for identification of AA thus correlations about spatial and temporal pattern of AA accumulation and expression of biosynthetic pathway are observations. Some of the noteworthy observations are described below.
At the 2nd stage, relative to the other stages,
TYDC1,
TYDC2,
PAL2,
C4H,
C3H and
N4OMT all exhibit very high expression in the bulb, considerably lower in the roots, and lowest in the leaves. Therefore, at this stage, during which the plant begins its growth from the 1st stage bulb, the expression of most of the genes remains high in the bulb compared to the roots and leaves. According to this, a great amount of alkaloids would be expected to be produced in the bulb and least in the leaves. However, the opposite is observed in
Figure 4A,B where the bulb has the lowest amount of compounds and the leaves have the highest. A plausible explanation for this may be that the biosynthetic enzymes and/or alkaloids produced in the bulb are being transported to other parts of the plant, leading to a decreased amount of alkaloids in the bulb and higher amounts in the roots and leaves. Such transport has been previously described in the opium poppy [
67,
68,
69].
At the 3rd developmental stage, the greater amount of compounds being accumulated in the leaves and flowers is in accordance with the high, or very high, expression of PAL1, 4CL, HCT1 and N4OMT in the leaves or flowers. The surprisingly high content of compounds in the stem, where almost all genes are expressed at very low levels, points, once again, to a possible transport of enzymes and/or alkaloids from, possibly, the bulb and roots to the stem.
At stage 4, the expression of
PAL2 is highest in the flowers,
4CL is higher in all aerial parts,
HCT1 is highest in the leaves and stem and
TYDC2 is highest in the leaves. This is consistent with the high accumulation of compounds in the aerial parts. There is also an increase of compounds in the roots at this stage, particularly the concentration of lycorine (
Figure 4A,C;
Table A4). The high concentration of lycorine in the stage 4 roots may be due to high expression levels of many other, yet unknown, genes implicated further downstream in the pathway, more precisely, in the ortho-para’ phenol coupling subgroup, which is specific to lycorine synthesis (
Figure 1).
Therefore, as the plant grows and develops, it tends to store the compounds in certain parts rather than others regardless of the level of gene expression. This is possibly due, most likely, to a transport of enzymes and AAs within the plant, as mentioned for the 2nd and 3rd stages. When the plant is young, i.e., at stages 2 and 3, it is clear that there is a greater accumulation of compounds in all aerial parts (leaves, stem and un-blossomed flowers;
Figure 4A,B). At stage 4, the leaves and flowers continue to accumulate large amounts of compounds. The presence of a high amount of compounds in the aerial parts is in line with the theory that alkaloids act as defense molecules for a plant [
1] as the aerial parts, besides being vulnerable to various pathogens, are also visible and easily accessible to herbivores. Additionally, leaving aside the high concentration of lycorine in the stage 4 roots, the amount of compounds in the roots increased as of stage 4 (
Figure 4A,B). This may be because roots can be attacked by pathogens present in the soil [
59].