Genome-Wide Analysis of Artificial Mutations Induced by Ethyl Methanesulfonate in the Eggplant (Solanum melongena L.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Illumina Sequencing Analysis
2.3. SNPs’/Indels’ Identification and Annotation
2.4. GO and KEGG Pathway
2.5. Expression Analysis
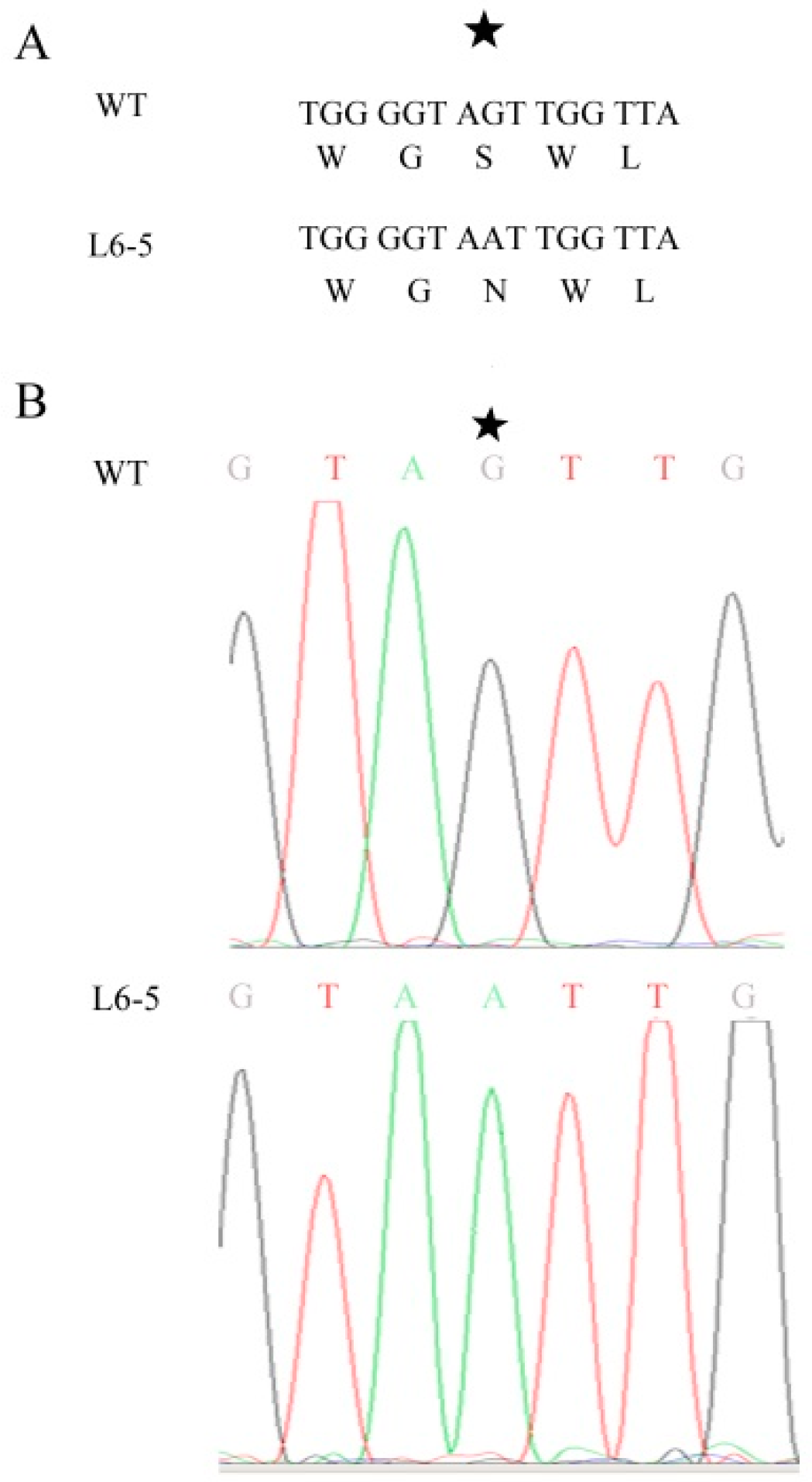
2.6. Sanger Sequencing
3. Results
3.1. Whole-Genome Re-Sequencing of Five Eggplant Lines
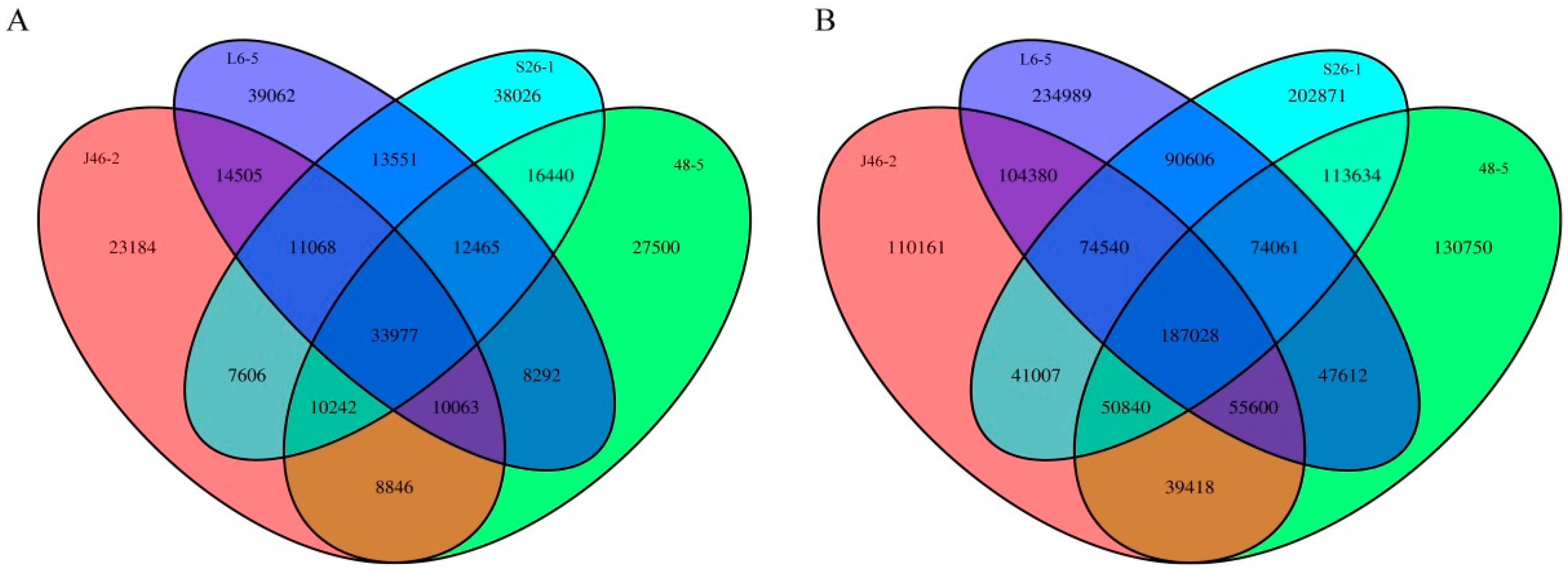
3.2. Identification of Single Nucleotide Substitutions and Indels
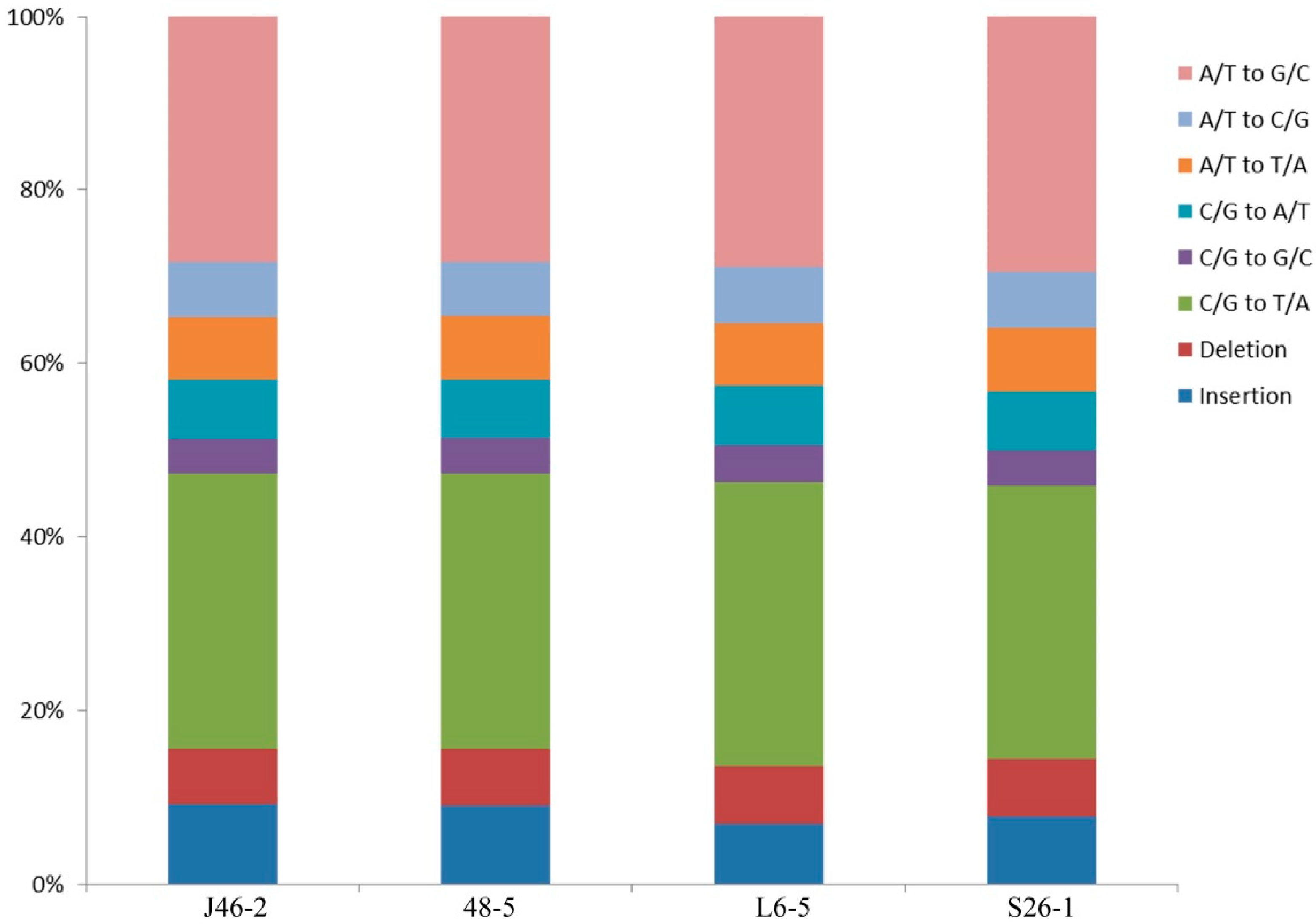
3.3. Characterization of the SNPs and Indels
3.4. Effects of Mutations on Gene Function
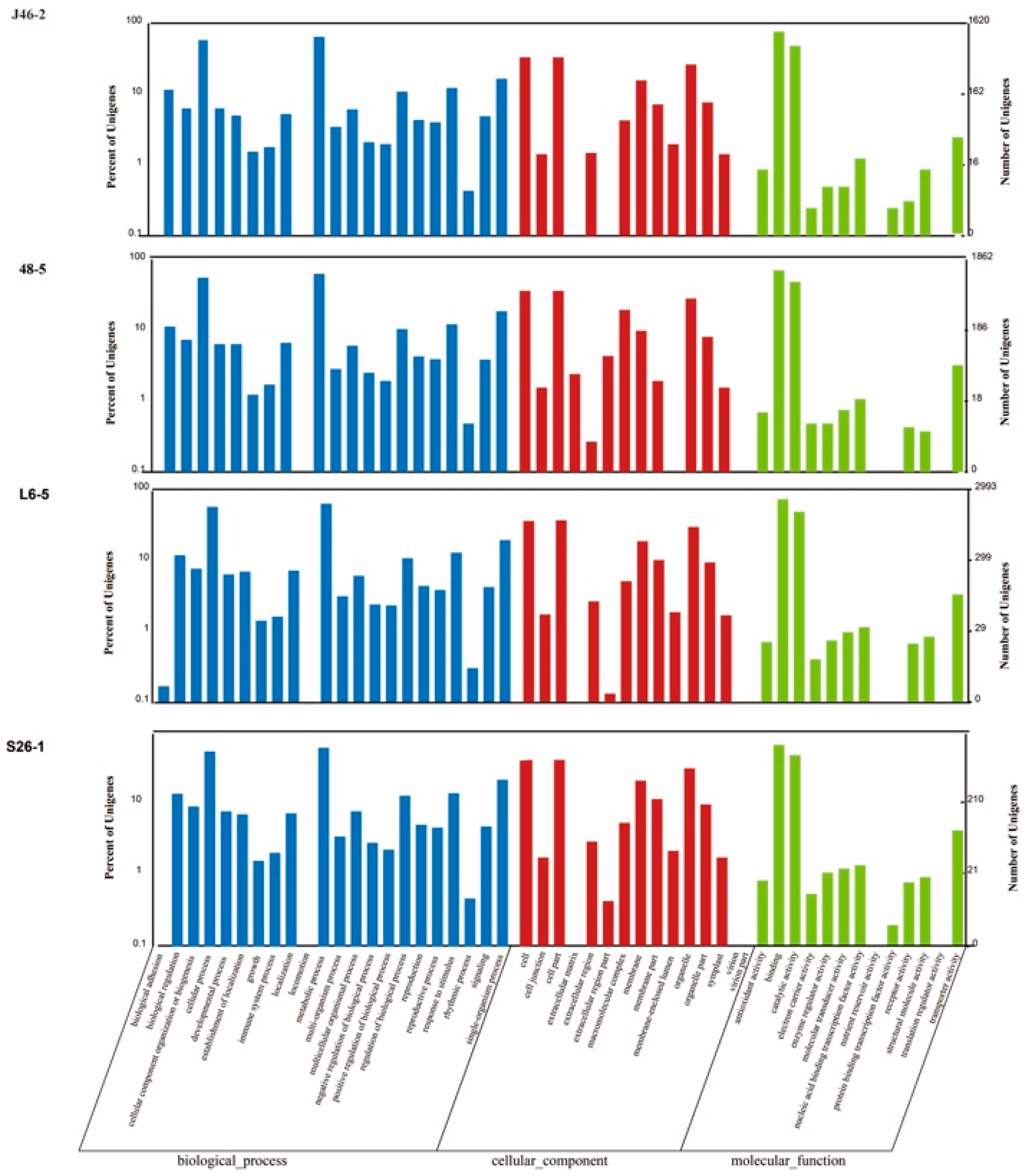
3.5. Gene Ontology (GO) Annotation
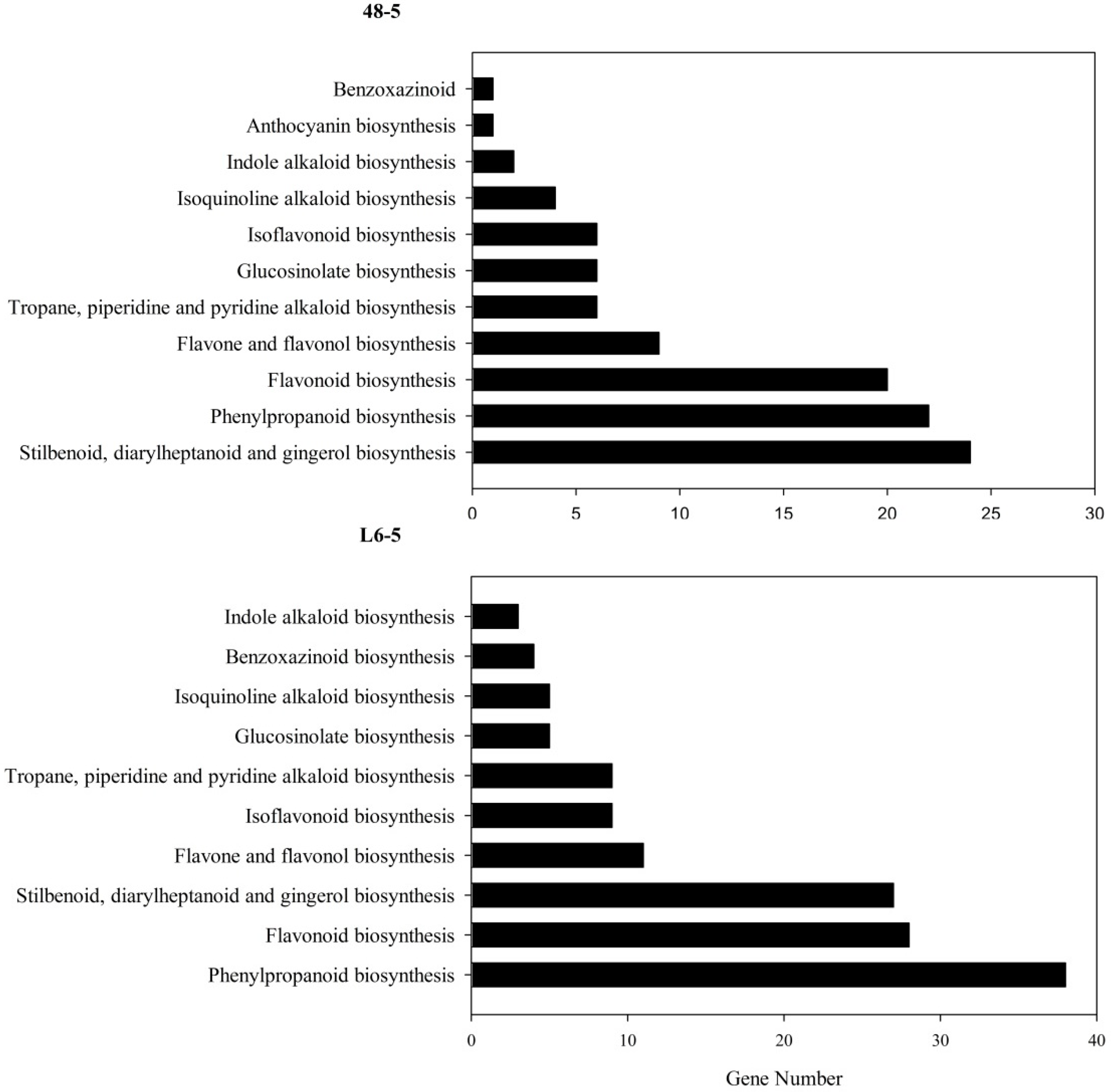
3.6. KEGG Pathway Mapping
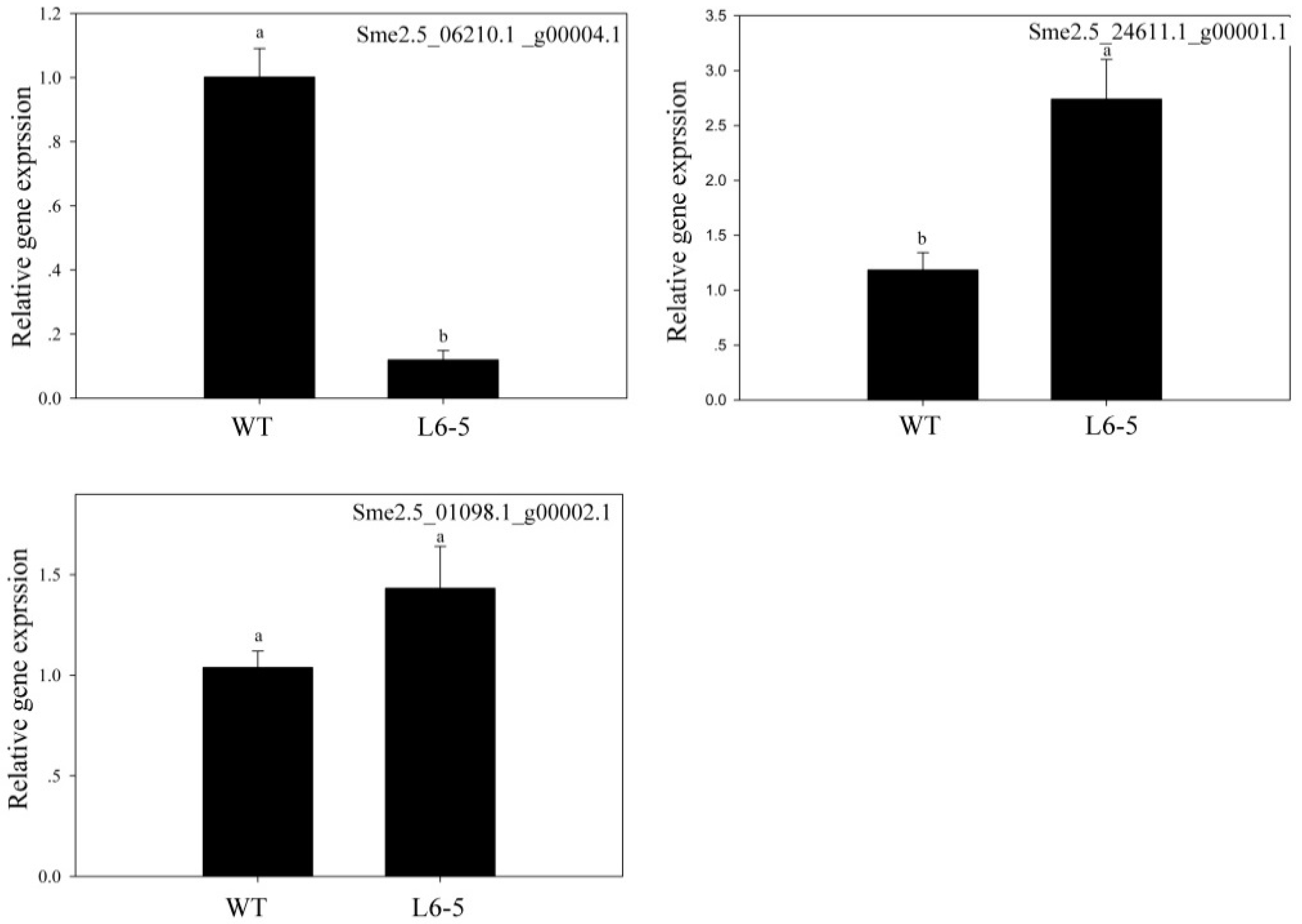
3.7. The Expression of Sme2.5_06210.1_g00004.1 Decreased Significantly in the L6-5 Mutant
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Plazas, M.; Andujar, I.; Vilanova, S.; Hurtado, M.; Gramazio, P.; Herraiz, F.J.; Prohens, J. Breeding for chlorogenic acid content in eggplant: Interest and prospects. Not. Bot. Horti Agrobot. Cluj-Napoca 2013, 41, 26. [Google Scholar] [CrossRef]
- Meyer, R.S.; Karol, K.G.; Little, D.P.; Nee, M.H.; Litt, A. Phylogeographic relationships among asian eggplants and new perspectives on eggplant domestication. Mol. Phylogenet. Evol. 2012, 63, 685–701. [Google Scholar] [CrossRef] [PubMed]
- Saito, T.; Ariizumi, T.; Okabe, Y.; Asamizu, E.; Hiwasa-Tanase, K.; Fukuda, N.; Mizoguchi, T.; Yamazaki, Y.; Aoki, K.; Ezura, H. TOMATOMA: A novel tomato mutant database distributing Micro-Tom mutant collections. Plant Cell Physiol. 2011, 52, 283–296. [Google Scholar] [CrossRef] [PubMed]
- Shikata, M.; Hoshikawa, K.; Ariizumi, T.; Fukuda, N.; Yamazaki, Y.; Ezura, H. TOMATOMA update: Phenotypic and metabolite information in the Micro-Tom mutant resource. Plant Cell Physiol 2016, 57, e11. [Google Scholar] [CrossRef] [PubMed]
- Arisha, M.H.; Shah, S.N.; Gong, Z.H.; Jing, H.; Li, C.; Zhang, H.X. Ethyl methane sulfonate induced mutations in M2 generation and physiological variations in M1 generation of peppers (Capsicum annuum L.). Front. Plant Sci. 2015, 6, 399. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, B.; Li, J.; Yang, X.; Ren, Z. Ethyl methanesulfonate (EMS)-mediated mutagenesis of cucumber (Cucumis sativus L.). Agric. Sci. 2014, 5, 716–721. [Google Scholar]
- Xi-Ou, X.; Wenqiu, L.; Wei, L.; Xiaoming, G.; Lingling, L.; Feiyue, M.; Yuge, L. The analysis of physiological variations in m2 generation of solanum melongena l. Mutagenized by ethyl methane sulfonate. Front. Plant Sci. 2017, 8, 17. [Google Scholar] [CrossRef] [PubMed]
- Martin, B.; Ramiro, M.; Martinez-Zapater, J.M.; Alonso-Blanco, C. A high-density collection of ems-induced mutations for tilling in landsberg erecta genetic background of arabidopsis. BMC Plant Biol. 2009, 9, 147. [Google Scholar] [CrossRef] [PubMed]
- Uchida, N.; Sakamoto, T.; Kurata, T.; Tasaka, M. Identification of EMS-induced causal mutations in a non-reference Arabidopsis thaliana accession by whole genome sequencing. Plant Cell Physiol. 2011, 52, 716–722. [Google Scholar] [CrossRef] [PubMed]
- Tsuda, M.; Kaga, A.; Anai, T.; Shimizu, T.; Sayama, T.; Takagi, K.; Machita, K.; Watanabe, S.; Nishimura, M.; Yamada, N.; et al. Construction of a high-density mutant library in soybean and development of a mutant retrieval method using amplicon sequencing. BMC Genom. 2015, 16, 1014. [Google Scholar] [CrossRef]
- Henry, I.M.; Nagalakshmi, U.; Lieberman, M.C.; Ngo, K.J.; Krasileva, K.V.; Vasquez-Gross, H.; Akhunova, A.; Akhunov, E.; Dubcovsky, J.; Tai, T.H.; et al. Efficient genome-wide detection and cataloging of EMS-induced mutations using exome capture and next-generation sequencing. Plant Cell 2014, 26, 1382–1397. [Google Scholar] [CrossRef]
- Shirasawa, K.; Hirakawa, H.; Nunome, T.; Tabata, S.; Isobe, S. Genome-wide survey of artificial mutations induced by ethyl methanesulfonate and gamma rays in tomato. Plant Biotechnol. J. 2016, 14, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Abe, A.; Kosugi, S.; Yoshida, K.; Natsume, S.; Takagi, H.; Kanzaki, H.; Matsumura, H.; Yoshida, K.; Mitsuoka, C.; Tamiru, M.; et al. Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 2012, 30, 174–178. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Wang, S.; Zhang, X.; Dong, Z.; Chen, F.; Cui, D. Transcriptome analysis of the Chinese bread wheat cultivar Yunong 201 and its ethyl methanesulfonate mutant line. Gene 2016, 575, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Feuillet, C.; Travella, S.; Stein, N.; Albar, L.; Nublat, A.; Keller, B. Map-based isolation of the leaf rust disease resistance gene Lr10 from the hexaploid wheat (Triticum aestivum L.) genome. Proc. Natl. Acad. Sci. USA 2003, 100, 15253–15258. [Google Scholar] [CrossRef] [PubMed]
- Peters, J.L.; Cnudde, F.; Gerats, T. Forward genetics and map-based cloning approaches. Trends Plant Sci. 2003, 8, 484–491. [Google Scholar] [CrossRef] [PubMed]
- Takagi, H.; Uemura, A.; Yaegashi, H.; Tamiru, M.; Abe, A.; Mitsuoka, C.; Utsushi, H.; Natsume, S.; Kanzaki, H.; Matsumura, H.; et al. MutMap-Gap: Whole-genome resequencing of mutant F2 progeny bulk combined with de novo assembly of gap regions identifies the rice blast resistance gene Pii. New Phytol. 2013, 200, 276–283. [Google Scholar] [CrossRef] [PubMed]
- McCallum, C.M.; Comai, L.; Greene, E.A.; Henikoff, S. Targeting induced local lesions in genomes (TILLING) for plant functional genomics. Plant Physiol. 2000, 123, 439–442. [Google Scholar] [CrossRef]
- Sevanthi, A.M.V.; Kandwal, P.; Kale, P.B.; Prakash, C.; Ramkumar, M.K.; Yadav, N.; Mahato, A.K.; Sureshkumar, V.; Behera, M.; Deshmukh, R.K.; et al. Whole genome characterization of a few EMS-induced mutants of upland rice variety Nagina 22 reveals a staggeringly high frequency of SNPs which show high phenotypic plasticity towards the wild-type. Front. Plant Sci. 2018, 9, 1179. [Google Scholar] [CrossRef]
- Kanehisa, M. From genomics to chemical genomics: New developments in KEGG. Nucleic Acids Res. 2006, 34, D354–D357. [Google Scholar] [CrossRef]
- Zhang, Y.; Hu, Z.; Chu, G.; Huang, C.; Tian, S.; Zhao, Z.; Chen, G. Anthocyanin accumulation and molecular analysis of anthocyanin biosynthesis-associated genes in eggplant (Solanum melongena L.). J. Agric. Food Chem. 2014, 62, 2906–2912. [Google Scholar] [CrossRef]
- Li, J.; Ren, L.; Gao, Z.; Jiang, M.; Liu, Y.; Zhou, L.; He, Y.; Chen, H. Combined transcriptomic and proteomic analysis constructs a new model for light-induced anthocyanin biosynthesis in eggplant (Solanum melongena L.). Plant Cell Environ. 2017, 40, 3069–3087. [Google Scholar] [CrossRef]
- Mengnan, X.; Shenhao, W.; Shu, Z.; Qingzhi, C.; Dongli, G.; Huiming, C.; Sanwen, H. A new gene conferring the glabrous trait in cucumber identified using MutMap. Hortic. Plant J. 2015, 1, 29–34. [Google Scholar]
- Greene, E.A.; Codomo, C.A.; Taylor, N.E.; Henikoff, J.G.; Till, B.J.; Reynolds, S.H.; Enns, L.C.; Burtner, C.; Johnson, J.E.; Odden, A.R.; et al. Spectrum of chemically induced mutations from a large-scale reverse-genetic screen in Arabidopsis. Genetics 2003, 164, 731. [Google Scholar] [PubMed]
- Zhou, Q.; Wang, S.; Hu, B.; Chen, H.; Zhang, Z.; Huang, S. An accumulation and replication of chloroplasts 5 gene mutation confers light green peel in cucumber. J. Integr. Plant Biol. 2015, 57, 936–942. [Google Scholar] [CrossRef]
- Kodym, A.; Afza, R. Physical and chemical mutagenesis. In Plant Functional Genomics; Grotewold, E., Ed.; Humana Press: Totowa, NJ, USA, 2003; pp. 189–203. [Google Scholar]
- Noda, Y.; Kaneyuki, T.; Igarashi, K.; Mori, A.; Packer, L. Antioxidant activity of nasunin, an anthocyanin in eggplant peels. Toxicology 2000, 148, 119–123. [Google Scholar] [CrossRef]
- Hiromoto, T.; Honjo, E.; Noda, N.; Tamada, T.; Kazuma, K.; Suzuki, M.; Blaber, M.; Kuroki, R. Structural basis for acceptor-substrate recognition of UDP-glucose: Anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea. Protein Sci. Publ. Protein Soc. 2015, 24, 395–407. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Gong, Q.; Ni, X.; Zhou, Y.; Gao, Z. UFGT: The key enzyme associated with the petals variegation in Japanese apricot. Front. Plant Sci. 2017, 8, 108. [Google Scholar] [CrossRef]
- Li, J.; He, Y.J.; Zhou, L.; Liu, Y.; Jiang, M.; Ren, L.; Chen, H. Transcriptome profiling of genes related to light-induced anthocyanin biosynthesis in eggplant (Solanum melongena L.) before purple color becomes evident. BMC Genom. 2018, 19, 201. [Google Scholar] [CrossRef]

| Line | Fruit Shape | Fruit Color | Sepal Color | Apex | Fruit Diameter | Fruit Length | Florescence |
|---|---|---|---|---|---|---|---|
| WT | Long | Aubergine | Purple | Concave | 5 cm | 30 cm | |
| J46-2 | Long | Aubergine | Purple | Raised | 5 cm | 30 cm | |
| 48-5 | Oval | White | Green | Concave | 10 cm | 25 cm | |
| L6-5 | Long | White | Green | Concave | 5 cm | 30 cm | |
| S26-1 | Long | Purple black | Purple | Concave | 5 cm | 30 cm | Early |
| Items | WT | J46-2 | 48-5 | L6-5 | S26-1 |
|---|---|---|---|---|---|
| Clean base (bp) | 56619013585 | 31640172900 | 30425850600 | 33955170600 | 33049549200 |
| Average depth | 58.2× | 30.98× | 29.98× | 33.85× | 31.89× |
| Genome Coverage | 94.52% | 98.70% | 98.70% | 98.67% | 98.68% |
| Item | Line | Variat_Number | Average (bp) | Max (bp) | Min (bp) |
|---|---|---|---|---|---|
| SNP | J46-2 | 294,966 | 2690 | 477,958 | 1 |
| 48-5 | 331,414 | 2394 | 546892 | 1 | |
| L6-5 | 477,587 | 1661 | 562,258 | 1 | |
| S26-1 | 448,118 | 1770 | 568,513 | 1 | |
| Indel | J46-2 | 54,109 | 14,665 | 515,567 | 1 |
| 48-5 | 61,014 | 13,005 | 538,563 | 1 | |
| L6-5 | 75,273 | 10,542 | 550,336 | 1 | |
| S26-1 | 75,500 | 10,510 | 547,033 | 1 |
| Intem | J46-2 | 48-5 | L6-5 | S26-1 | |
|---|---|---|---|---|---|
| High-impact | nonsense mutations | 18,638 | 20,737 | 28,489 | 27,220 |
| frameshift mutations | 2323 | 2371 | 2285 | 2288 | |
| Modifier-impact | Intron mutation | 96,836 | 108,394 | 159,089 | 147,403 |
| intergenic mutations | 15,360 | 167,494 | 232,069 | 220,473 | |
| Low-impact | synonymous mutations | 8767 | 10,276 | 13,236 | 13,071 |
| Mutatant Line | Gene ID | Mutation | Mutation Type | Description |
|---|---|---|---|---|
| Sme2.5_00384.1_g00003.1 | nonsynonymous | G-T | F3’H, VvF3’h4; flavonoid 3′ hydroxylase | |
| Sme2.5_00928.1_g00014.1 | nonsynonymous | C-T | flavonol-3-O-glycoside-7-O-glucosyltransferase 1 | |
| Sme2.5_01087.1_g00007.1 | nonsynonymous | A-G | F3H1; flavonoid 3′-hydroxylase | |
| Sme2.5_03930.1_g00006.1 | nonsynonymous | G-C | flavonol-3-O-glycoside-7-O-glucosyltransferase 1 | |
| Sme2.5_05662.1_g00003.1 | nonsynonymous | C-T | flavonol-3-O-glycoside-7-O-glucosyltransferase 1 | |
| 48-5 | Sme2.5_13126.1_g00003.1 | nonsynonymous | G-A | flavonol-3-O-glycoside-7-O-glucosyltransferase 1 |
| Sme2.5_13996.1_g00001.1 | nonsynonymous | T-G | Flavonoid 3′ 5′-hydroxylase | |
| Sme2.5_24902.1_g00001.1 | nonsynonymous | C-T | Flavonoid 3′,5′-hydroxylase 2-like | |
| Sme2.5_26073.1_g00001.1 | nonsynonymous | T-C | COMT1; caffeic acid 3-O-methyltransferase | |
| Sme2.5_03583.1_g00011.1 | nonsynonymous | T-C | anthocyanin 5-O-glucosyltransferase | |
| Sme2.5_00081.1_g00002.1 | frameshift_deletion | GC-C | COMT2; caffeic acid 3-O-methyltransferase | |
| Sme2.5_01098.1_g00002.1 | nonsynonymous | G-A | Anthocyanidin 3-O-glucosyltransferase 5-like | |
| Sme2.5_01191.1_g00004.1 | nonsynonymous | T-G | Flavonoid 3′ 5′-hydroxylase | |
| Sme2.5_02030.1_g00005.1 | nonsynonymous | A-G | Flavonol 3-O-methyltransferase | |
| Sme2.5_02274.1_g00006.1 | nonsynonymous | T-C | flavonol-3-O-glycoside-7-O-glucosyltransferase 1 | |
| L6-5 | Sme2.5_06210.1_g00004.1 | nonsynonymous | C-T | flavonol-3-O-glycoside-7-O-glucosyltransferase 1 |
| Sme2.5_07919.1_g00001.1 | nonsynonymous | G-A | UDP-glucosyl transferase family protein | |
| Sme2.5_12449.1_g00001.1 | nonsynonymous | G-A | flavonoid 3′ hydroxylase | |
| Sme2.5_13126.1_g00003.1 | nonsynonymous | G-A | flavonol-3-O-glycoside-7-O-glucosyltransferase 1 | |
| Sme2.5_24611.1_g00001.1 | nonsynonymous | G-A | anthocyanidin 3-O-glucosyltransferase 5-like | |
| Sme2.5_26073.1_g00001.1 | nonsynonymous | T-C | COMT1; caffeic acid 3-O-methyltransferase | |
| Sme2.5_00081.1_g00002.1 | frameshift_deletion | GC-C | COMT2; caffeic acid 3-O-methyltransferase |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiao, X.-o.; Lin, W.; Li, K.; Feng, X.; Jin, H.; Zou, H. Genome-Wide Analysis of Artificial Mutations Induced by Ethyl Methanesulfonate in the Eggplant (Solanum melongena L.). Genes 2019, 10, 595. https://doi.org/10.3390/genes10080595
Xiao X-o, Lin W, Li K, Feng X, Jin H, Zou H. Genome-Wide Analysis of Artificial Mutations Induced by Ethyl Methanesulfonate in the Eggplant (Solanum melongena L.). Genes. 2019; 10(8):595. https://doi.org/10.3390/genes10080595
Chicago/Turabian StyleXiao, Xi-ou, Wenqiu Lin, Ke Li, Xuefeng Feng, Hui Jin, and Huafeng Zou. 2019. "Genome-Wide Analysis of Artificial Mutations Induced by Ethyl Methanesulfonate in the Eggplant (Solanum melongena L.)" Genes 10, no. 8: 595. https://doi.org/10.3390/genes10080595
APA StyleXiao, X.-o., Lin, W., Li, K., Feng, X., Jin, H., & Zou, H. (2019). Genome-Wide Analysis of Artificial Mutations Induced by Ethyl Methanesulfonate in the Eggplant (Solanum melongena L.). Genes, 10(8), 595. https://doi.org/10.3390/genes10080595




