Modulation and Evolution of Animal Development through microRNA Regulation of Gene Expression
Abstract
:1. Introduction
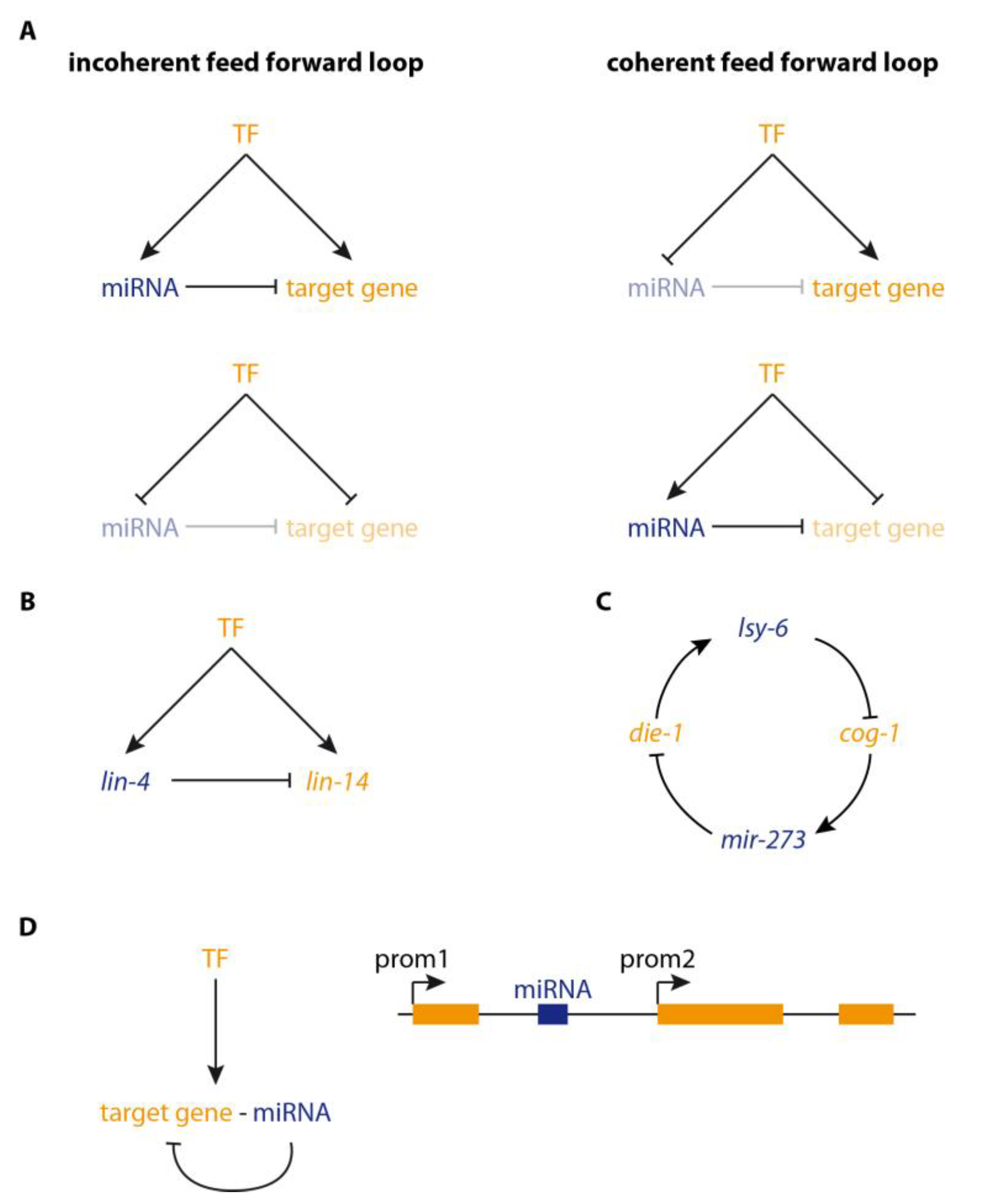
2. microRNAs in Regulatory Loops
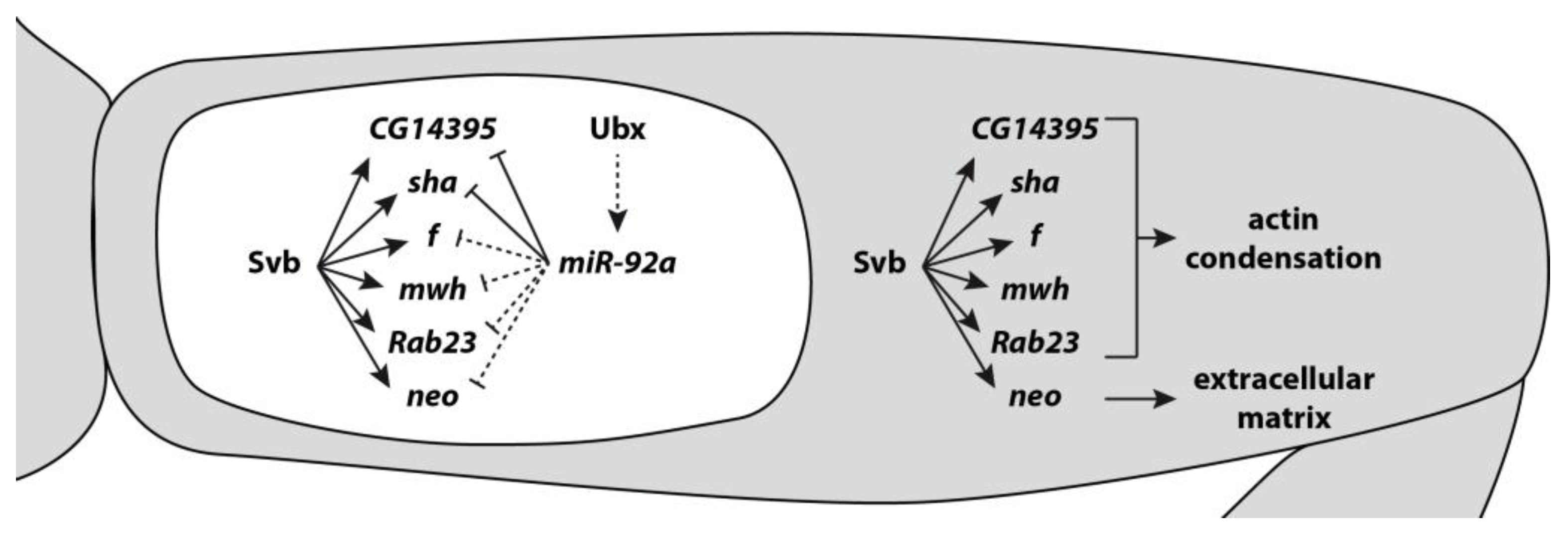
3. microRNA Targets in GRNs
4. Evolution of microRNAs and Targets Leading to Phenotypic Change
Acknowledgments
Conflicts of Interest
References
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef]
- Mukherji, S.; Ebert, M.S.; Zheng, G.X.; Tsang, J.S.; Sharp, P.A.; van Oudenaarden, A. MicroRNAs can generate thresholds in target gene expression. Nat. Genet. 2011, 43, 854–859. [Google Scholar] [CrossRef]
- Posadas, D.M.; Carthew, R.W. MicroRNAs and their roles in developmental canalization. Curr. Opin. Genet. Dev. 2014, 27, 1–6. [Google Scholar] [CrossRef]
- Agarwal, V.; Subtelny, A.O.; Thiru, P.; Ulitsky, I.; Bartel, D.P. Predicting microRNA targeting efficacy in Drosophila. Genome Biol. 2018, 19, 152. [Google Scholar] [CrossRef]
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef]
- Jan, C.H.; Friedman, R.C.; Ruby, J.G.; Bartel, D.P. Formation, regulation and evolution of Caenorhabditis elegans 3’UTRs. Nature 2011, 469, 97–101. [Google Scholar] [CrossRef]
- Chen, Y.W.; Song, S.; Weng, R.; Verma, P.; Kugler, J.M.; Buescher, M.; Rouam, S.; Cohen, S.M. Systematic study of Drosophila microRNA functions using a collection of targeted knockout mutations. Dev. Cell 2014, 31, 784–800. [Google Scholar] [CrossRef]
- Berezikov, E. Evolution of microRNA diversity and regulation in animals. Nat. Rev. Genet. 2011, 12, 846–860. [Google Scholar] [CrossRef]
- Niwa, R.; Slack, F.J. The evolution of animal microRNA function. Curr. Opin. Genet. Dev. 2007, 17, 145–150. [Google Scholar] [CrossRef]
- Alberti, C.; Cochella, L. A framework for understanding the roles of miRNAs in animal development. Development 2017, 144, 2548–2559. [Google Scholar] [CrossRef]
- Liu, N.; Okamura, K.; Tyler, D.M.; Phillips, M.D.; Chung, W.J.; Lai, E.C. The evolution and functional diversification of animal microRNA genes. Cell Res. 2008, 18, 985–996. [Google Scholar] [CrossRef]
- Bartel, D.P.; Chen, C.Z. Micromanagers of gene expression: The potentially widespread influence of metazoan microRNAs. Nat. Rev. Genet. 2004, 5, 396–400. [Google Scholar] [CrossRef]
- Cora, D.; Re, A.; Caselle, M.; Bussolino, F. MicroRNA-mediated regulatory circuits: Outlook and perspectives. Phys. Biol. 2017, 14, 045001. [Google Scholar] [CrossRef]
- Tsang, J.; Zhu, J.; van Oudenaarden, A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol. Cell 2007, 26, 753–767. [Google Scholar] [CrossRef]
- Osella, M.; Bosia, C.; Cora, D.; Caselle, M. The role of incoherent microRNA-mediated feedforward loops in noise buffering. PLoS Comput. Biol. 2011, 7, e1001101. [Google Scholar] [CrossRef]
- Kim, D.; Grun, D.; van Oudenaarden, A. Dampening of expression oscillations by synchronous regulation of a microRNA and its target. Nat. Genet. 2013, 45, 1337–1344. [Google Scholar] [CrossRef]
- Chang, S.; Johnston, R.J., Jr.; Frokjaer-Jensen, C.; Lockery, S.; Hobert, O. MicroRNAs act sequentially and asymmetrically to control chemosensory laterality in the nematode. Nature 2004, 430, 785–789. [Google Scholar] [CrossRef]
- Johnston, R.J.; Hobert, O. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature 2003, 426, 845–849. [Google Scholar] [CrossRef]
- Johnston, R.J., Jr.; Chang, S.; Etchberger, J.F.; Ortiz, C.O.; Hobert, O. MicroRNAs acting in a double-negative feedback loop to control a neuronal cell fate decision. Proc. Natl. Acad. Sci. USA 2005, 102, 12449–12454. [Google Scholar] [CrossRef]
- Hinske, L.C.; Galante, P.A.; Kuo, W.P.; Ohno-Machado, L. A potential role for intragenic miRNAs on their hosts’ interactome. BMC Genom. 2010, 11, 533. [Google Scholar] [CrossRef]
- Hinske, L.C.; Franca, G.S.; Torres, H.A.; Ohara, D.T.; Lopes-Ramos, C.M.; Heyn, J.; Reis, L.F.; Ohno-Machado, L.; Kreth, S.; Galante, P.A. miRIAD-integrating microRNA inter- and intragenic data. Database J. Biol. Databases Curation 2014, 2014. [Google Scholar] [CrossRef]
- Baskerville, S.; Bartel, D.P. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA 2005, 11, 241–247. [Google Scholar] [CrossRef]
- Rodriguez, A.; Griffiths-Jones, S.; Ashurst, J.L.; Bradley, A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004, 14, 1902–1910. [Google Scholar] [CrossRef]
- Gennarino, V.A.; Sardiello, M.; Avellino, R.; Meola, N.; Maselli, V.; Anand, S.; Cutillo, L.; Ballabio, A.; Banfi, S. MicroRNA target prediction by expression analysis of host genes. Genome Res. 2009, 19, 481–490. [Google Scholar] [CrossRef]
- Liang, Y.; Ridzon, D.; Wong, L.; Chen, C. Characterization of microRNA expression profiles in normal human tissues. BMC Genom. 2007, 8, 166. [Google Scholar] [CrossRef]
- Ozsolak, F.; Poling, L.L.; Wang, Z.; Liu, H.; Liu, X.S.; Roeder, R.G.; Zhang, X.; Song, J.S.; Fisher, D.E. Chromatin structure analyses identify miRNA promoters. Genes Dev. 2008, 22, 3172–3183. [Google Scholar] [CrossRef]
- Monteys, A.M.; Spengler, R.M.; Wan, J.; Tecedor, L.; Lennox, K.A.; Xing, Y.; Davidson, B.L. Structure and activity of putative intronic miRNA promoters. RNA 2010, 16, 495–505. [Google Scholar] [CrossRef]
- Punnamoottil, B.; Rinkwitz, S.; Giacomotto, J.; Svahn, A.J.; Becker, T.S. Motor neuron-expressed microRNAs 218 and their enhancers are nested within introns of Slit2/3 genes. Genesis 2015, 53, 321–328. [Google Scholar] [CrossRef]
- Paraboschi, E.M.; Cardamone, G.; Rimoldi, V.; Duga, S.; Solda, G.; Asselta, R. miR-634 is a Pol III-dependent intronic microRNA regulating alternative-polyadenylated isoforms of its host gene PRKCA. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 1046–1056. [Google Scholar] [CrossRef]
- Yuva-Aydemir, Y.; Xu, X.L.; Aydemir, O.; Gascon, E.; Sayin, S.; Zhou, W.; Hong, Y.; Gao, F.B. Downregulation of the Host Gene jigr1 by miR-92 Is Essential for Neuroblast Self-Renewal in Drosophila. PLoS Genet. 2015, 11, e1005264. [Google Scholar] [CrossRef]
- Herranz, H.; Cohen, S.M. MicroRNAs and gene regulatory networks: Managing the impact of noise in biological systems. Genes Dev. 2010, 24, 1339–1344. [Google Scholar] [CrossRef]
- Belles, X. MicroRNAs and the Evolution of Insect Metamorphosis. Annu. Rev. Entomol. 2017, 62, 111–125. [Google Scholar] [CrossRef]
- Lozano, J.; Montanez, R.; Belles, X. MiR-2 family regulates insect metamorphosis by controlling the juvenile hormone signaling pathway. Proc. Natl. Acad. Sci. USA 2015, 112, 3740–3745. [Google Scholar] [CrossRef]
- Zhao, Z.; Boyle, T.J.; Liu, Z.; Murray, J.I.; Wood, W.B.; Waterston, R.H. A negative regulatory loop between microRNA and Hox gene controls posterior identities in Caenorhabditis elegans. PLoS Genet. 2010, 6, e1001089. [Google Scholar] [CrossRef]
- Mok, G.F.; Lozano-Velasco, E.; Maniou, E.; Viaut, C.; Moxon, S.; Wheeler, G.; Munsterberg, A. miR-133-mediated regulation of the Hedgehog pathway orchestrates embryo myogenesis. Development 2018, 145. [Google Scholar] [CrossRef]
- Shalgi, R.; Lieber, D.; Oren, M.; Pilpel, Y. Global and local architecture of the mammalian microRNA-transcription factor regulatory network. PLoS Comput. Biol. 2007, 3, e131. [Google Scholar] [CrossRef]
- Wu, S.; Huang, S.; Ding, J.; Zhao, Y.; Liang, L.; Liu, T.; Zhan, R.; He, X. Multiple microRNAs modulate p21Cip1/Waf1 expression by directly targeting its 3’ untranslated region. Oncogene 2010, 29, 2302–2308. [Google Scholar] [CrossRef]
- Lai, X.; Schmitz, U.; Gupta, S.K.; Bhattacharya, A.; Kunz, M.; Wolkenhauer, O.; Vera, J. Computational analysis of target hub gene repression regulated by multiple and cooperative miRNAs. Nucleic Acids Res. 2012, 40, 8818–8834. [Google Scholar] [CrossRef]
- Wong, T.S.; Liu, X.B.; Wong, B.Y.; Ng, R.W.; Yuen, A.P.; Wei, W.I. Mature miR-184 as Potential Oncogenic microRNA of Squamous Cell Carcinoma of Tongue. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2008, 14, 2588–2592. [Google Scholar] [CrossRef]
- Jongen-Lavrencic, M.; Sun, S.M.; Dijkstra, M.K.; Valk, P.J.; Lowenberg, B. MicroRNA expression profiling in relation to the genetic heterogeneity of acute myeloid leukemia. Blood 2008, 111, 5078–5085. [Google Scholar] [CrossRef]
- Lionetti, M.; Biasiolo, M.; Agnelli, L.; Todoerti, K.; Mosca, L.; Fabris, S.; Sales, G.; Deliliers, G.L.; Bicciato, S.; Lombardi, L.; et al. Identification of microRNA expression patterns and definition of a microRNA/mRNA regulatory network in distinct molecular groups of multiple myeloma. Blood 2009, 114, e20–e26. [Google Scholar] [CrossRef] [PubMed]
- Voorhoeve, P.M.; le Sage, C.; Schrier, M.; Gillis, A.J.; Stoop, H.; Nagel, R.; Liu, Y.P.; van Duijse, J.; Drost, J.; Griekspoor, A.; et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell 2006, 124, 1169–1181. [Google Scholar] [CrossRef]
- Guled, M.; Lahti, L.; Lindholm, P.M.; Salmenkivi, K.; Bagwan, I.; Nicholson, A.G.; Knuutila, S. CDKN2A, NF2, and JUN are dysregulated among other genes by miRNAs in malignant mesothelioma-A miRNA microarray analysis. Genes Chromosom. Cancer 2009, 48, 615–623. [Google Scholar] [CrossRef]
- Gottardo, F.; Liu, C.G.; Ferracin, M.; Calin, G.A.; Fassan, M.; Bassi, P.; Sevignani, C.; Byrne, D.; Negrini, M.; Pagano, F.; et al. Micro-RNA profiling in kidney and bladder cancers. Urol. Oncol. 2007, 25, 387–392. [Google Scholar] [CrossRef]
- Cursons, J.; Pillman, K.A.; Scheer, K.G.; Gregory, P.A.; Foroutan, M.; Hediyeh-Zadeh, S.; Toubia, J.; Crampin, E.J.; Goodall, G.J.; Bracken, C.P.; et al. Combinatorial Targeting by MicroRNAs Co-ordinates Post-transcriptional Control of EMT. Cell Syst. 2018, 7, 77–91. [Google Scholar] [CrossRef]
- Pinzon, N.; Li, B.; Martinez, L.; Sergeeva, A.; Presumey, J.; Apparailly, F.; Seitz, H. microRNA target prediction programs predict many false positives. Genome Res. 2017, 27, 234–245. [Google Scholar] [CrossRef] [PubMed]
- Cassidy, J.J.; Jha, A.R.; Posadas, D.M.; Giri, R.; Venken, K.J.; Ji, J.; Jiang, H.; Bellen, H.J.; White, K.P.; Carthew, R.W. miR-9a minimizes the phenotypic impact of genomic diversity by buffering a transcription factor. Cell 2013, 155, 1556–1567. [Google Scholar] [CrossRef] [PubMed]
- Cassidy, J.J.; Straughan, A.J.; Carthew, R.W. Differential Masking of Natural Genetic Variation by miR-9a in Drosophila. Genetics 2016, 202, 675–687. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Cassidy, J.J.; Reinke, C.A.; Fischboeck, S.; Carthew, R.W. A microRNA imparts robustness against environmental fluctuation during development. Cell 2009, 137, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Coolen, M.; Katz, S.; Bally-Cuif, L. miR-9: A versatile regulator of neurogenesis. Front. Cell. Neurosci. 2013, 7. [Google Scholar] [CrossRef]
- Bonev, B.; Pisco, A.; Papalopulu, N. MicroRNA-9 reveals regional diversity of neural progenitors along the anterior-posterior axis. Dev. Cell 2011, 20, 19–32. [Google Scholar] [CrossRef]
- Coolen, M.; Thieffry, D.; Drivenes, O.; Becker, T.S.; Bally-Cuif, L. miR-9 controls the timing of neurogenesis through the direct inhibition of antagonistic factors. Dev. Cell 2012, 22, 1052–1064. [Google Scholar] [CrossRef]
- Bonev, B.; Stanley, P.; Papalopulu, N. MicroRNA-9 Modulates Hes1 ultradian oscillations by forming a double-negative feedback loop. Cell Rep. 2012, 2, 10–18. [Google Scholar] [CrossRef]
- Tan, S.L.; Ohtsuka, T.; Gonzalez, A.; Kageyama, R. MicroRNA9 regulates neural stem cell differentiation by controlling Hes1 expression dynamics in the developing brain. Genes Cells Devot. Mol. Cell. Mech. 2012, 17, 952–961. [Google Scholar] [CrossRef]
- Shibata, M.; Kurokawa, D.; Nakao, H.; Ohmura, T.; Aizawa, S. MicroRNA-9 modulates Cajal-Retzius cell differentiation by suppressing Foxg1 expression in mouse medial pallium. J. Neurosci. Off. J. Soc. Neurosci. 2008, 28, 10415–10421. [Google Scholar] [CrossRef]
- Zhao, C.; Sun, G.; Li, S.; Shi, Y. A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat. Struct. Mol. Biol. 2009, 16, 365–371. [Google Scholar] [CrossRef]
- Packer, A.N.; Xing, Y.; Harper, S.Q.; Jones, L.; Davidson, B.L. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington’s disease. J. Neurosci. Off. J. Soc. Neurosci. 2008, 28, 14341–14346. [Google Scholar] [CrossRef]
- Li, Y.; Wang, F.; Lee, J.A.; Gao, F.B. MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes Dev. 2006, 20, 2793–2805. [Google Scholar] [CrossRef]
- Biryukova, I.; Asmar, J.; Abdesselem, H.; Heitzler, P. Drosophila mir-9a regulates wing development via fine-tuning expression of the LIM only factor, dLMO. Dev. Biol. 2009, 327, 487–496. [Google Scholar] [CrossRef]
- Bejarano, F.; Smibert, P.; Lai, E.C. miR-9a prevents apoptosis during wing development by repressing Drosophila LIM-only. Dev. Biol. 2010, 338, 63–73. [Google Scholar] [CrossRef]
- Arif, S.; Murat, S.; Almudi, I.; Nunes, M.D.; Bortolamiol-Becet, D.; McGregor, N.S.; Currie, J.M.; Hughes, H.; Ronshaugen, M.; Sucena, E.; et al. Evolution of mir-92a Underlies Natural Morphological Variation in Drosophila melanogaster. Curr. Biol. 2013, 23, 523–528. [Google Scholar] [CrossRef]
- Chen, X.; Rosbash, M. MicroRNA-92a is a circadian modulator of neuronal excitability in Drosophila. Nat. Commun. 2017, 8, 14707. [Google Scholar] [CrossRef]
- Schertel, C.; Rutishauser, T.; Forstemann, K.; Basler, K. Functional Characterization of Drosophila microRNAs by a Novel in vivo Library. Genetics 2012, 192, 1543–1552. [Google Scholar] [CrossRef]
- Stern, D.L. A role of Ultrabithorax in morphological differences between Drosophila species. Nature 1998, 396, 463–466. [Google Scholar] [CrossRef]
- Kittelmann, S.; Buffry, A.D.; Franke, F.A.; Almudi, I.; Yoth, M.; Sabaris, G.; Couso, J.P.; Nunes, M.D.S.; Frankel, N.; Gomez-Skarmeta, J.L.; et al. Gene regulatory network architecture in different developmental contexts influences the genetic basis of morphological evolution. PLoS Genet. 2018, 14, e1007375. [Google Scholar] [CrossRef]
- Chanut-Delalande, H.; Fernandes, I.; Roch, F.; Payre, F.; Plaza, S. Shavenbaby couples patterning to epidermal cell shape control. PLoS Biol. 2006, 4, e290. [Google Scholar] [CrossRef]
- Menoret, D.; Santolini, M.; Fernandes, I.; Spokony, R.; Zanet, J.; Gonzalez, I.; Latapie, Y.; Ferrer, P.; Rouault, H.; White, K.P.; et al. Genome-wide analyses of Shavenbaby target genes reveals distinct features of enhancer organization. Genome Biol. 2013, 14, R86. [Google Scholar] [CrossRef]
- Ronshaugen, M.; Biemar, F.; Piel, J.; Levine, M.; Lai, E.C. The Drosophila microRNA iab-4 causes a dominant homeotic transformation of halteres to wings. Genes Dev. 2005, 19, 2947–2952. [Google Scholar] [CrossRef]
- Tyler, D.M.; Okamura, K.; Chung, W.J.; Hagen, J.W.; Berezikov, E.; Hannon, G.J.; Lai, E.C. Functionally distinct regulatory RNAs generated by bidirectional transcription and processing of microRNA loci. Genes Dev. 2008, 22, 26–36. [Google Scholar] [CrossRef]
- Stark, A.; Bushati, N.; Jan, C.H.; Kheradpour, P.; Hodges, E.; Brennecke, J.; Bartel, D.P.; Cohen, S.M.; Kellis, M. A single Hox locus in Drosophila produces functional microRNAs from opposite DNA strands. Genes Dev. 2008, 22, 8–13. [Google Scholar] [CrossRef]
- Kaschula, R.; Pinho, S.; Alonso, C.R. MicroRNA-dependent regulation of Hox gene expression sculpts fine-grain morphological patterns in a Drosophila appendage. Development 2018, 145. [Google Scholar] [CrossRef]
- Franchini, P.; Xiong, P.; Fruciano, C.; Meyer, A. The Role of microRNAs in the Repeated Parallel Diversification of Lineages of Midas Cichlid Fish from Nicaragua. Genome Biol. Evolut. 2016, 8, 1543–1555. [Google Scholar] [CrossRef]
- Quah, S.; Hui, J.H.; Holland, P.W. A Burst of miRNA Innovation in the Early Evolution of Butterflies and Moths. Mol. Biol. Evolut. 2015, 32, 1161–1174. [Google Scholar] [CrossRef]
- Li, J.; Zhang, Z. miRNA regulatory variation in human evolution. Trends Genet. 2013, 29, 116–124. [Google Scholar] [CrossRef]
- Somel, M.; Liu, X.; Tang, L.; Yan, Z.; Hu, H.; Guo, S.; Jiang, X.; Zhang, X.; Xu, G.; Xie, G.; et al. MicroRNA-driven developmental remodeling in the brain distinguishes humans from other primates. PLoS Biol. 2011, 9, e1001214. [Google Scholar] [CrossRef]
- Martin, A.; Orgogozo, V. The Loci of repeated evolution: A catalog of genetic hotspots of phenotypic variation. Evolution 2013, 67, 1235–1250. [Google Scholar] [CrossRef] [PubMed]
- Carroll, S.B. Evo-devo and an expanding evolutionary synthesis: A genetic theory of morphological evolution. Cell 2008, 134, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Selbach, M.; Schwanhausser, B.; Thierfelder, N.; Fang, Z.; Khanin, R.; Rajewsky, N. Widespread changes in protein synthesis induced by microRNAs. Nature 2008, 455, 58–63. [Google Scholar] [CrossRef]
- Baek, D.; Villen, J.; Shin, C.; Camargo, F.D.; Gygi, S.P.; Bartel, D.P. The impact of microRNAs on protein output. Nature 2008, 455, 64–71. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kittelmann, S.; McGregor, A.P. Modulation and Evolution of Animal Development through microRNA Regulation of Gene Expression. Genes 2019, 10, 321. https://doi.org/10.3390/genes10040321
Kittelmann S, McGregor AP. Modulation and Evolution of Animal Development through microRNA Regulation of Gene Expression. Genes. 2019; 10(4):321. https://doi.org/10.3390/genes10040321
Chicago/Turabian StyleKittelmann, Sebastian, and Alistair P. McGregor. 2019. "Modulation and Evolution of Animal Development through microRNA Regulation of Gene Expression" Genes 10, no. 4: 321. https://doi.org/10.3390/genes10040321
APA StyleKittelmann, S., & McGregor, A. P. (2019). Modulation and Evolution of Animal Development through microRNA Regulation of Gene Expression. Genes, 10(4), 321. https://doi.org/10.3390/genes10040321




