Abstract
Spikelet number per panicle is a determinative factor of rice yield. DNA repair epigenetically alters the DNA accessibility, which can eventually regulate the transcription of the target genes. However, what and how DNA repair genes are related to rice spikelet development remains unknown. Here, we report the map-based cloning of a novel spikelet number gene DES4 encoding a tetratricopeptide domain-containing protein. DES4 is a close ortholog of Arabidopsis BRU1, which is functionally related to axillary meristem development. A single base pair deletion in the last exon of DES4 caused a premature stop of the resulting protein. The des4 mutant exhibited dwarf, reduced tiller, and spikelet numbers phenotypes, as well as hypersensitivity to genotoxic stresses, suggesting its essential role in DNA repair. DES4 is predominantly expressed in young panicles and axillary meristems, and DES4 protein is localized in nucleus. A set of DNA repair genes such as cyclins, KUs (KD subunits) and recombinases were differentially regulated in des4. Meanwhile, rice spikelet number genes LAX1, LAX2, and MOC1 were significantly down-regulated in des4. In morphology, des4 showed more severe reduction of spikelet numbers than lax1, lax2, and moc1, suggesting that DES4 may work upstream of the three genes.
1. Introduction
Rice (Oryza sativa L.) is the major source of calories for over half of the world’s population. Meanwhile, rice is also used as a model species for plant molecular biology research due to its completed genome sequencing, mature genetic transformation technology, and genome co-linearity with other crops [1]. Rice yield is a complex agronomic trait determined by three main component traits: number of panicles per plant, number of grains per panicle, and grain weight [2]. Based on morphological dynamic changes, Ikeda et al. (2014) tentatively divided rice panicle development into nine successive stages, spanning from establishment of rachis meristem (RM) to mature panicle [3]. At the onset of panicle differentiation, RM initiates from the shoot apex meristem (SAM), and subsequently produces a number of lateral meristems, which can develop into primary branches. Similarly, second or higher order branches may arise from lateral meristems on each of the primary branches, and spikelet meristems on the secondary branches finally develop into spikelets [2,3,4,5]. In general, spikelet numbers per panicle are mainly determined by the number of primary and secondary branches on the panicle [2,6]. Numerous spikelet number genes and quantitative trait loci have been identified and extensively reviewed in several publications [2,3,5,6,7]. These include Frizzle Panicle (FZP), LAX Panicle 1 (LAX1), LAX2, Small Panicle (SPA), and Aberrant Panicle Organization 1 (APO1), which are implicated in the initiation of RM or conversion of RM into spikelet meristem (SM) [4,8,9,10]; and Grain Number 1a (Gn1a), Dense and Erect Panicle 1 (DEP1), DEP2, and DEP3, which are related to the rate of spikelet differentiation [11,12,13,14]. Regardless of the progress achieved, our understanding of the regulation of spikelet number in rice remains poor, largely due to the fragmented information from a limited number of identified genes. Here, we report the positional cloning of a novel rice spikelet number gene Decreased Spikelet 4 (DES4), which is functionally involved in both DNA damage repair and spikelet number determination. DES4 is a nuclear-localized, tetratricopeptide repeat domain-containing protein similar to BRU1 in Arabidopsis. A set of DNA damage repair genes and spikelet number genes are differentially regulated in the des4 mutant. This work represents a substantial advance toward the understanding of how DNA repair genes are involved in rice reproductive tissue differentiation and development.
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
The recessive mutant des4 was isolated from a γ irradiation-induced mutant population in the background of Zhonghua 11 (ZH11) (O. sativa, ssp. japonica). Genetic mapping population was derived from a cross between des4 and 9311 (O. sativa, ssp. indica). From 2014 to 2018, plants were cultivated in the experimental field of China national rice research institute (May to October) or in Sanya, China (November to March of the following year). Under natural growing conditions, morphological analysis was done on plants in the harvesting stage with fully matured grains.
2.2. Map-Based Cloning
Simple sequence repeat (SSR) and sequence-tagged site markers were derived from (http://www.gramene.org/microsat/ssr.html; Table S1), and screened for polymorphisms in the population. A DNA pool of twelve des4 mutants were used for the draft mapping of the DES4 locus by screening over 120 polymorphic SSR markers. To fine map DES4, about 1002 recessive individuals with the mutant phenotype were selected and screened by newly developed SSR and Indel markers. Primer sequences of the polymorphous markers for fine mapping can be found in Table S1.
2.3. Plasmid Construction and Plant Transformation
For complementation of the des4 mutant, full coding sequence (CDS) of the candidate gene was amplified from the ZH11 panicle cDNA and cloned into expression vector pCambia1300-221-myc, in which the gene is fused with myc peptide tag sequence and driven by a 35S constitutive promoter. The plasmid was then introduced into des4 embryonic calli by Agrobacterium tumefaciens-mediated transformation according to a previous report [15]. Similarly, full CDS of DES4 was cloned into pCambia1300-YFP using the SalI and SmaI sites to fuse with the yellow fluorescent protein (YFP) ORF on the N terminus. pCambia1300-YFP-DES4 and the negative control pCambia1300-YFP plasmids were transiently transformed into tobacco leaf epidermal cells as previously described [16]. Fluorescence was observed on confocal microscopy (Zeiss LSM710, Carl Zeiss, Jena, Germany) at 72 h after infiltration.
2.4. RNA Preparation, RT-PCR Analysis, and mRNA in-Situ Hybridization
Total RNA from various tissues was isolated by using TRIeasyTM Total RNA Extraction Reagent (Yeasen, Shanghai, China). First strand cDNA was transcribed from DNase I-treated RNA using M-MLV reverse transcriptase and oligo (dT) primers (Takara, Dalian, China). qRT-PCR was carried out on a CFX96 touch real-time PCR detection system (Bio-Rad, Hercules, CA, USA) by following a previous publication [16]. Actin gene LOC_Os03g61970 was used as an internal control. Expression was assessed by evaluating threshold cycle (CT) values and calculated by the 2−ΔΔCT method. The experiment was performed in biological triplicates. The mRNA in situ hybridization was conducted as described by Zhang et al. (2010) [17]. Primer sequences are listed in Table S1.
2.5. Genotoxic Stress
One DAG (day after germination) seeds were hydroponically cultured on Hoagland solution with the addition of methyl methanesulfonate (MMS) and zeocin in different concentrations, in a growth chamber (28 °C, 60% humidity, 12 h light/12 h dark). The Hoagland solution was changed every three days to maintain a stable solution pH. The plant heights were manually measured after two weeks of growth. Twenty biological sample repeats were measured for each treatment.
3. Results and Discussion
3.1. des4 Has Reduced Spikelet Number
In an effort to clone rice spikelet number genes, our lab identified a set of des mutants (designated as des1 to des5) by screening a γ irradiation-induced mutant population in the background of ZH11 (O. sativa ssp. japonica). Among the mutants, des4 displayed pleiotropic phenotypes, including dwarf, less tillers, and narrower leaf blades (Figure 1A,C,D). More importantly, the primary and secondary branch numbers of des4 were severely decreased when compared with wild type (WT), which finally led to a significant reduction of spikelet numbers per panicle from 125.4 ± 19.91 (WT) to 38.4 ± 10.38 (des4) (n = 15, p < 0.01) (Figure 1B,D).
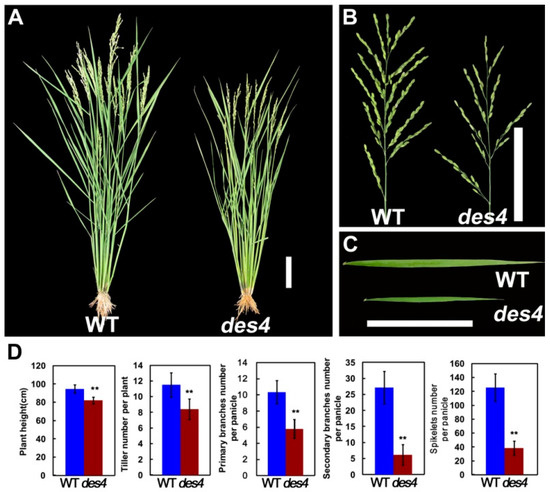
Figure 1.
Phenotypical characterization of the des4 and wild type (WT). (A) Plant architecture, (B) panicle structure, and (C) leaf shape of des4 and WT, bar = 10 cm. (D) Quantification of the plant height, tiller number, primary branches, secondary branches, and spikelet numbers per panicle. Numerical values are expressed as the mean; error bars denote one standard deviation of the mean; asterisk denotes significant difference between mutant and WT within each treatment (** p < 0.01; Student’s t-test).
3.2. Positional Cloning of des4
To clone the gene responsible for des4 phenotypes, we adopted a map-based cloning strategy by using an F2 population derived from the cross between des4 and 9311 (O. sativa spp. indica). We phenotypically characterized 92 F2 lines; the normal and des4 plants followed a 3:1 ratio, suggesting that the des4 phenotype is controlled by a single recessive gene (normal: des4 = 68:24, χ2 = 0.0579 < χ20.05,1 = 3.84). Approximately 4000 individual F2 plants, including 1002 des4 plants, were used for the genetic mapping. We gradually mapped des4 to a 100 kb region between marker M137 and M128 on the long arm of chromosome 2 (Figure 2A). As shown in the rice annotation project database (RAP, https://rapdb.dna.affrc.go.jp/), a total of 18 candidate genes, including 2 transposon genes were located in this region. Subsequently, we Sanger sequenced the 16 non-transposon genes in both WT and des4. None of the candidate genes were mutated, except a cytimidine deletion that occurred on the 3-prime ends of the CDS of Os02g0782800, which should have shifted the open reading frame and caused premature stop of the resulting protein (Figure 2B, Data S1). To the best of our knowledge, no publications regarding the functions of Os02g0782800 are available.
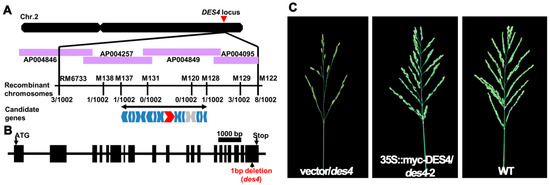
Figure 2.
Genetic mapping and complementation analysis of DES4. (A) Map-based cloning of DES4. The markers for PCR-based mapping are listed in Table S1. “Recombinant” indicates recombination between maker and phenotype; the numbers indicate the numbers of recombinant lines out of 1002 mutant type lines analyzed. Red shape indicates Os02g0782800 candidate gene; blue shapes indicate the 16 sequenced candidate genes; and grey shape indicates the transposon genes. (B) Schematic presentation of the gene structure of DES4. Boxes represent the exons and the line indicates the intron. One base pair deletion occurred on the last exon of DES4. The mutated sequence is presented in Data S1. (C) Panicle architectures of negative control (empty vector) line, genetically complemented line, and WT.
In terms of structure, Os02g0782800 is around 16 kb in length with 20 exons and 19 introns, and the encoded protein is comprised of 1338 amino acids (Figure 2B). Due to the huge size of Os02g0782800, we failed to make a complementation vector carrying the whole genomic sequence of the gene with its native promoter (over 18 kb). Alternatively, we constructed the full CDS of Os02g0782800 into pCambia1300-221-myc, in which the gene is fused with myc-peptide tag sequence under the driving of 35S constitutive promoter. The constructs were genetically transformed into the des4 background. As expected, all the T-DNA positive plants (n = 4) were rescued as WT. Thus, we concluded that Os02g0782800 is DES4 (Figure 2C, Figure S1).
3.3. DES4 Encodes a Novel Tetratricopeptide Repeat Domain-Containing Protein
Conserved domain search on the DES4 sequence identified seven tetratricopeptide repeat (TPR) domains on the N terminus, eight leucine-rich repeat (LRR) domains on the C terminus, and two nuclear localization signal (NLS) sequences in the middle (Figure 3A). As a commonly identified motif in various proteins, TPR is usually comprised of 3–16 tandem repeats of a degenerate 34 amino acid sequence motif unit, which may serve as scaffolds for protein–protein interactions and protein complex assembly. TPR domain-containing proteins have been found to be extensively involved in cell cycling, gene transcription regulation, and protein transportation, as well as protein folding [18,19]. Meanwhile, LRRs are composed of repeating 20–30 amino acid stretches that are rich in hydrophobic leucines. Similar to TPRs, LRRs are also functionally related to the formation of protein–protein complexes [20,21]. Using the DES4 protein sequence as a query, we identified DES4 orthologs in 19 other plant species with a single copy in each one. Phylogenetic analysis revealed that DES4 exhibited the highest similarities with orthologs in Brachypodium distachyon, Triticum urartu and Aegilops tauschii, while all the members in grass were clustered in a major clade, suggesting that those proteins may have conserved functions (Figure 3B). In Arabidopsis thaliana, DES4 ortholog AT3G18730 has been reported as BRU1/Tonsoku/Mgoun3 participating in meristem cellular organization and maintenance. In bru1, the plant became dwarf with distorted phyllotaxy, brush-like branches, and defected inflorescence architectures, which mimicked the phenotypes observed on des4, suggesting that DES4 may have conserved functions as BRU1 [22,23,24].
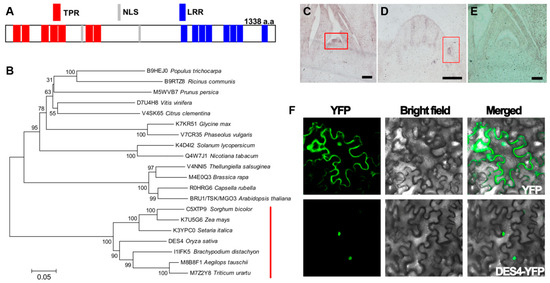
Figure 3.
Protein structure, phylogenetic, and expression analysis of DES4. (A) Conserved domain analysis of the DES4 amino acid sequence. (B) Phylogenetic tree of DES4 and orthologs from other species. Numbers are the bootstrap values. Red line indicates the clade of grasses. (C–E) mRNA in situ hybridization analysis of DES4 in differentiating panicles meristems (C) and tiller meristems (D). Boxes highlight the tissue parts with strong expression signals. Bar = 10 μm. (E) Negative control using sense probes. (F) Subcellular localization of DES4 in tobacco leaf epidermal cells.
We examined the mRNA level of DES4 in various tissues, including root, leaf blade, internodes, and young panicle. DES4 was constitutively transcribed in all the tested tissues, while the highest levels were detected in axillary buds and young panicles (Figure S2). Employing mRNA in situ hybridization technique, we further gained an in-depth view of DES4 expression pattern in differentiating tillers and panicles. Strong signals were detected on the lateral meristems where tiller buds or panicle primary branches initiated, which is in agreement with the decreased tillers and spikelet numbers observed in des4 (Figure 3C,E). Moreover, the NLS on DES4 invited further investigation of its subcellular localization. As shown in Figure 3F, transiently expressed DES4-YFP proteins were detected in the nucleus of tobacco leaf epidermal cells, whereas the empty vector control showed fluorescence in various cellular compartments as expected. This observation is also consistent with the subcellular localization of BRU1 in Arabidopsis.
3.4. des4 Is Hypersensitive to Genotoxic Stress
Previous study indicated that BRU1 is crucial for the recognition and repair of DNA damage [24]. To test the potential role of DES4 in DNA repair, we treated young seedlings of des4 and WT with MMS and zeocin in a series of concentrations. MMS and zeocin have been found to alkylate DNA, which mimics DNA double strand break (DSB) damages [25]. As MMS concentration increased, WT seedlings displayed retarded growth (Figure 4A,B). Interestingly, des4 seedlings were hypersensitive to the MMS-induced genotoxic stress. When <15 ppm MMS treatment was applied, the height of des4 plants was only half that of the WT. We obtained a very similar result from the zeocin treatment (Figure 4B). Therefore, we suggest that DES4 may play an essential role in the DNA repair of DSB damage. DNA repair has been linked with transcriptional gene regulation, as both processes are essentially involved in chromatin compaction and DNA accessibility under very common mechanisms [26]. A few examples are transcription factor IIH (TFIIH) which is required for initiation of RNA transcription, efficient repair of DNA by nucleotide excision in yeast [27]; and BRU1, which has dual roles in DNA damage repair and epigenetic gene silencing in Arabidopsis [24]. Given the high homologies between BRU1 and DES4, as well as similar genotoxic hypersensitivities observed in both mutants, we speculate that DES4 may also have conserved functions similar to BRU1 in gene transcriptional regulation.
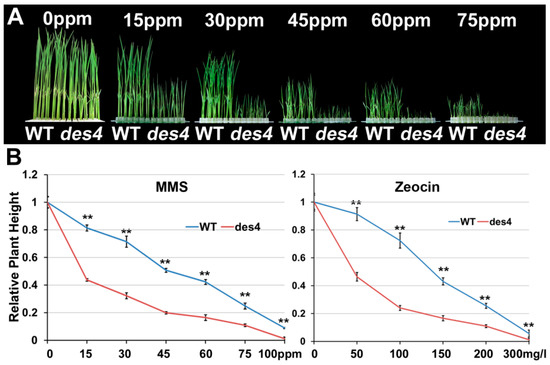
Figure 4.
Genotoxic stresses on des4 and WT seedlings. (A) Growth of des4 and WT seedlings under different methyl methanesulfonate (MMS) concentrations. (B) Quantification of the seedling heights under different concentrations of MMS and zeocin. ** p < 0.01.
3.5. DSB Repair Genes and Spikelet Number Genes Are Differentially Regulated in des4
To investigate the potential downstream genes regulated by DES4, we performed qRT-PCR experiments using 14-day-old seedling cDNAs of WT and des4 as templates. The candidate genes included seven cyclin genes, two KU homolog genes, and six recombinase genes which are potentially involved in DSB repair (Figure 5). Severe DNA damage could directly influence cell cycle and proliferation. Culligan et al. (2004) found that Arabidopsis seedling cells were blocked outside of M phase at G2 and/or at G1/S transition when plants were irradiated with a high dosage [28]. In response to DNA damage, some key cell-cycle-related genes such as CDKB1;2, CDKB2;1, and CyclinB1;2 were down-regulated, whereas CyclinB1;1 was up-regulated in the first few hours and then started to decline after 24 h of treatment [29,30]. In des4, we also found that the transcription of four cyclin genes (CyclinA1;1, CyclinA1;K, CyclinA3;1, and CyclinB2;K) were significantly reduced, while CyclinD3;1 and CyclinD4;1 exhibited higher expression level when compared with the WT. To repair the DSB, eukaryotes developed a nonhomologous end joining (NHEJ) mechanism, which is heavily modulated by DNA-dependent protein kinase complex (DNA-PKcs) containing Ku70/Ku80 heterodimer and DNA ligase4 [31]. Ku70/Ku80 recognizes and binds to the DSB ends, while ligase4 is responsible for the repair of DNA breaks [32]. In our case, des4 showed elevated levels of Ku70 and Ku80. The suggestion is that DNA-PKcs may work in parallel with DES4 as an alternative pathway to compensate the loss of function of DES4, when DNA repair is required. Meanwhile, a list of recombinase genes including RAD50, RAD51A2, and RAD51C were significantly up-regulated in des4. RAD51s have been known as essential proteins in homologous recombination and recombinational repair of DSB. Both RAD51A1 and RAD51A2 possess homologous pairing activity, though RAD51A2 has much higher activities than RAD51A1 [33]. RAD51C was also implicated in DNA repair in somatic cells [34]. The up-regulation of the RAD51s in des4 imply that, in addition to the NHEJ, homologous repair may also take place to compensate the impaired function of DES4. Interestingly, we found that the transcription of RAD17 was down-regulated in des4, possibly as a compensation for these up-regulated RAD sibling genes. It should be noted that the altered expression of these DSB repair-related genes may be an indirect consequence of the loss of function of DES4, and their relationship with DES4 needs to be further explored with more direct evidence, such as multiple gene mutations and genetic complementation. Given the decreased spikelet numbers of des4, we also examined the transcriptional levels of three reported spikelet number genes—LAX1, LAX2, and MOC1—in young panicles. The results showed that all three genes were dramatically decreased in the mutant. We compared the extent of spikelet number decreases in lax1, lax2, moc1, and des4. It seems that DES4 is an up-stream regulator of the other three genes, because it showed the strongest phenotype (Figure S3). As discussed above, the TPR domain-containing protein DES4 may regulate target gene transcription by epigenetically modulating the chromatin configuration and DNA accessibility. We believe that more spikelet number genes are subject to the regulation of DES4.
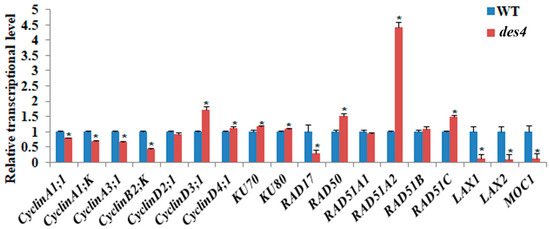
Figure 5.
qRT-PCR analysis of double-strand break (DSB) repair genes and spikelet number genes. qRT-PCR analysis of transcript levels of DSB repair genes and spikelet number genes in WT and des4. Rice actin (LOC_Os03g61970) was used as an internal control. The locus ID for each gene can be found in Table S1. Values are means ± SE. Asterisks indicate significant differences between WT and des4 (* p < 0.05; Student’s t-test).
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/10/3/214/s1, Data S1: Sequence of the mutated DES4. Table S1: Sequences of primers used in this study. Figure S1: The spikelet numbers of genetically complemented des4 lines. Figure S2: qRT-PCR analysis of DES4 in various rice tissues. Figure S3: Panicle morphologies of lax1, lax2, moc1, and des4.
Author Contributions
Z.L., S.N., J.Y. and H.C. performed the experiments and analyzed the data, J.Z. and H.C. conceived of the project, designed and coordinated the experiments, J.Z. wrote the manuscript. All the authors read and approved the final manuscript.
Funding
This work was funded by Zhejiang provincial Natural Science Foundation (Grant No. LY15C130006), and Rice molecular designing breeding project (Grant No. 2016YFD0101801).
Acknowledgments
The authors are grateful to Da Luo for initiating and supervising the project.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Zhang, J.; Guo, D.; Chang, Y.; You, C.; Li, X.; Dai, X.; Weng, Q.; Zhang, J.; Chen, G.; Li, X.; et al. Non-random distribution of T-DNA insertions at various levels of the genome hierarchy as revealed by analyzing 13 804 T-DNA flanking sequences from an enhancer-trap mutant library. Plant J. Cell Mol. Biol. 2007, 49, 947–959. [Google Scholar] [CrossRef] [PubMed]
- Xing, Y.; Zhang, Q. Genetic and molecular bases of rice yield. Annu. Rev. Plant Biol. 2010, 61, 421–442. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, K.; Sunohara, H.; Nagato, Y. Developmental course of inflorescence and spikelet in rice. Jpn. J. Breed. 2004, 54, 147–156. [Google Scholar] [CrossRef]
- Bai, X.; Huang, Y.; Hu, Y.; Liu, H.; Zhang, B.; Smaczniak, C.; Hu, G.; Han, Z.; Xing, Y. Duplication of an upstream silencer of FZP increases grain yield in rice. Nat. Plants 2017, 3, 885. [Google Scholar] [CrossRef]
- Zhang, D.; Yuan, Z. Molecular control of grass inflorescence development. Annu. Rev. Plant Biol. 2014, 65, 553–578. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, J. Branching in rice. Curr. Opin. Plant Biol. 2011, 14, 94–99. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, X.; Fu, D.; Wu, C. Panicle Morphology Mutant 1 (PMM1) determines the inflorescence architecture of rice by controlling brassinosteroid biosynthesis. BMC Plant Biol. 2018, 18, 348. [Google Scholar] [CrossRef]
- Komatsu, M.; Maekawa, M.; Shimamoto, K.; Kyozuka, J. The LAX1 and FRIZZY PANICLE 2 genes determine the inflorescence architecture of rice by controlling rachis-branch and spikelet development. Dev. Biol. 2001, 231, 364–373. [Google Scholar] [CrossRef] [PubMed]
- Keishi, K.; Masahiko, M.; Shin, U.; Yuzuki, S.; Ikuyo, F.; Hironobu, O.; Ko, S.; Junko, K. LAX and SPA: Major regulators of shoot branching in rice. Proc. Natl. Acad. Sci. USA 2003, 100, 11765–11770. [Google Scholar]
- Tabuchi, H.; Zhang, Y.; Hattori, S.; Omae, M.; Shimizusato, S.; Oikawa, T.; Qian, Q.; Nishimura, M.; Kitano, H.; Xie, H. LAX PANICLE2 of rice encodes a novel nuclear protein and regulates the formation of axillary meristems. Plant Cell 2011, 23, 3276–3287. [Google Scholar] [CrossRef]
- Ashikari, M.; Sakakibara, H.; Lin, S.; Yamamoto, T.; Takashi, T.; Nishimura, A.; Angeles, E.R.; Qian, Q.; Kitano, H.; Matsuoka, M. Cytokinin oxidase regulates rice grain production. Science 2005, 309, 741–745. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Qian, Q.Z.; Sun, H.; He, S.; Luo, D.; Xia, G.; Chu, C.; Li, J.; Fu, X. Natural variation at the DEP1 locus enhances grain yield in rice. Nat. Genet. 2009, 41, 494–497. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Liu, W.; Tang, J.; Chen, J.; Tong, H.; Hu, B.; Li, C.; Fang, J.; Chen, M.; Chu, C. Rice DENSE AND ERECT PANICLE 2 is essential for determining panicle outgrowth and elongation. Cell Res. 2010, 20, 838–849. [Google Scholar] [CrossRef]
- Sun, H.Y.; Qian, Q.; Wu, K.; Wang, W.; Luo, J.J.; Wang, S.S.; Zhang, C.W.; Ma, Y.F.; Liu, Q.; Huang, X.Z. Heterotrimeric G proteins regulate nitrogen-use efficiency in rice. Sci. Found. China 2015, 46, 652. [Google Scholar] [CrossRef]
- Hiei, Y.; Komari, T.; Kubo, T. Transformation of rice mediated by Agrobacterium tumefaciens. Plant Mol. Biol. 1997, 35, 205–218. [Google Scholar] [CrossRef] [PubMed]
- Bello, B.K.; Hou, Y.; Zhao, J.; Jiao, G.; Wu, Y.; Li, Z.; Wang, Y.; Tong, X.; Wang, W.; Yuan, W.; et al. NF-YB1-YC12-bHLH144 complex directly activates Wx to regulate grain quality in rice (Oryza sativa L.). Plant Biotechnol. J. 2018. [Google Scholar] [CrossRef]
- Zhang, J.; Nallamilli, B.R.; Mujahid, H.; Peng, Z. OsMADS6 plays an essential role in endosperm nutrient accumulation and is subject to epigenetic regulation in rice (Oryza sativa). Plant J. Cell Mol. Biol. 2010, 64, 604–617. [Google Scholar] [CrossRef]
- Sikorski, R.S.; Boguski, M.S.; Goebl, M.; Hieter, P. A repeating amino acid motif in CDC23 defines a family of proteins and a new relationship among genes required for mitosis and RNA synthesis. Cell 1990, 60, 307–317. [Google Scholar] [CrossRef]
- Goebl, M.; Yanagida, M. The TPR snap helix: A novel protein repeat motif from mitosis to transcription. Trends Biochem. Sci. 1991, 16, 173. [Google Scholar] [CrossRef]
- Kobe, B.; Deisenhofer, J. Crystal structure of porcine ribonuclease inhibitor, a protein with leucine-rich repeats. Nature 1993, 366, 751–756. [Google Scholar] [CrossRef]
- Kobe, B.; Deisenhofer, J. The leucine-rich repeat: A versatile binding motif. Trends Biochem. Sci. 1994, 19, 415–421. [Google Scholar] [CrossRef]
- Guyomarc’H, S.; Vernoux, T.; Traas, J.; Zhou, D.X.; Delarue, M. MGOUN3, an Arabidopsis gene with TetratricoPeptide-Repeat-related motifs, regulates meristem cellular organization. J. Exp. Bot. 2004, 55, 673–684. [Google Scholar] [CrossRef]
- Suzuki, T.; Inagaki, S.; Nakajima, S.; Akashi, T.; Ohto, M.A.; Kobayashi, M.; Seki, M.; Shinozaki, K.; Kato, T.; Tabata, S. A novel Arabidopsis gene TONSOKU is required for proper cell arrangement in root and shoot apical meristems. Plant J. 2010, 38, 673–684. [Google Scholar] [CrossRef] [PubMed]
- Takeda, S.; Tadele, Z.; Hofmann, I.; Aline, V.P.; Angelis, K.J.; Hidetaka, K.; Takashi, A.; Tesfaye, M.; Ortrun, M.S.; et al. BRU1, a novel link between responses to DNA damage and epigenetic gene silencing in Arabidopsis. Genes Dev. 2004, 18, 782–793. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, J.L. Monofunctional alkylating agent-induced S-phase-dependent DNA damage. Mutat. Res. 1989, 216, 111–118. [Google Scholar] [CrossRef]
- Green, C.M.; Almouzni, G. When repair meets chromatin. EMBO Rep. 2002, 3, 28–33. [Google Scholar] [CrossRef] [PubMed]
- Feaver, W.J.; Svejstrup, J.Q.; Bardwell, L.; Bardwell, A.J.; Buratowski, S.; Gulyas, K.D.; Donahue, T.F.; Friedberg, E.C.; Kornberg, R.D. Dual roles of a multiprotein complex from S. cerevisiae in transcription and DNA repair. Cell 1993, 75, 1379. [Google Scholar] [CrossRef]
- Culligan, K.; Tissier, A.; Britt, A. ATR regulates a G2-phase cell-cycle checkpoint in Arabidopsis thaliana. Plant Cell 2004, 16, 1091. [Google Scholar] [CrossRef] [PubMed]
- Ricaud, L.; Proux, C.; Renou, J.-P.; Pichon, O.; Fochesato, S.; Ortet, P.; Montané, M.-H. ATM-mediated transcriptional and developmental responses to γ-rays in Arabidopsis. PLoS ONE 2007, 2, e430. [Google Scholar] [CrossRef] [PubMed]
- Culligan, K.M.; Robertson, C.E.; Julia, F.; Peter, D.; Britt, A.B. ATR and ATM play both distinct and additive roles in response to ionizing radiation. Plant J. 2010, 48, 947–961. [Google Scholar] [CrossRef] [PubMed]
- Lieber, M.R.; Karanjawala, Z.E. Ageing, repetitive genomes and DNA damage. Nat. Rev. Mol. Cell Biol. 2004, 5, 69–75. [Google Scholar] [CrossRef]
- Dynan, W.S.; Yoo, S. Interaction of Ku protein and DNA-dependent protein kinase catalytic subunit with nucleic acids. Nucleic Acids Res. 1998, 26, 1551–1559. [Google Scholar] [CrossRef]
- Morozumi, Y.; Ino, R.; Ikawa, S.; Mimida, N.; Shimizu, T.; Toki, S.; Ichikawa, H.; Shibata, T.; Kurumizaka, H. Homologous pairing activities of two rice RAD51 proteins, RAD51A1 and RAD51A2. PLoS ONE 2013, 8, e75451. [Google Scholar] [CrossRef]
- Kou, Y.; Chang, Y.; Li, X.; Xiao, J.; Wang, S. The rice RAD51C gene is required for the meiosis of both female and male gametocytes and the DNA repair of somatic cells. J. Exp. Bot. 2012, 63, 5323–5335. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).