Control of Eukaryotic DNA Replication Initiation—Mechanisms to Ensure Smooth Transitions
Abstract
1. Introduction
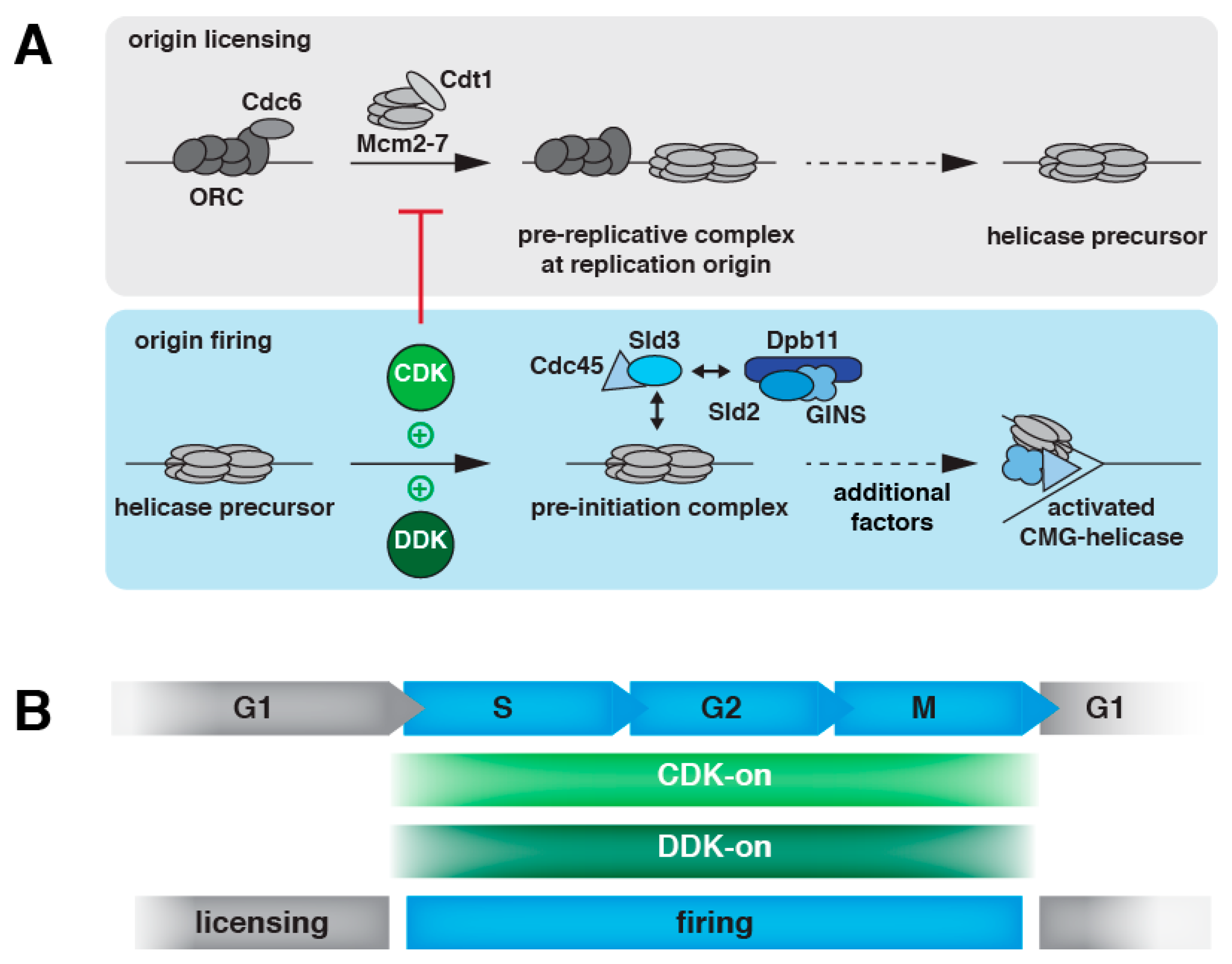
2. DNA Replication Initiation in Eukaryotes
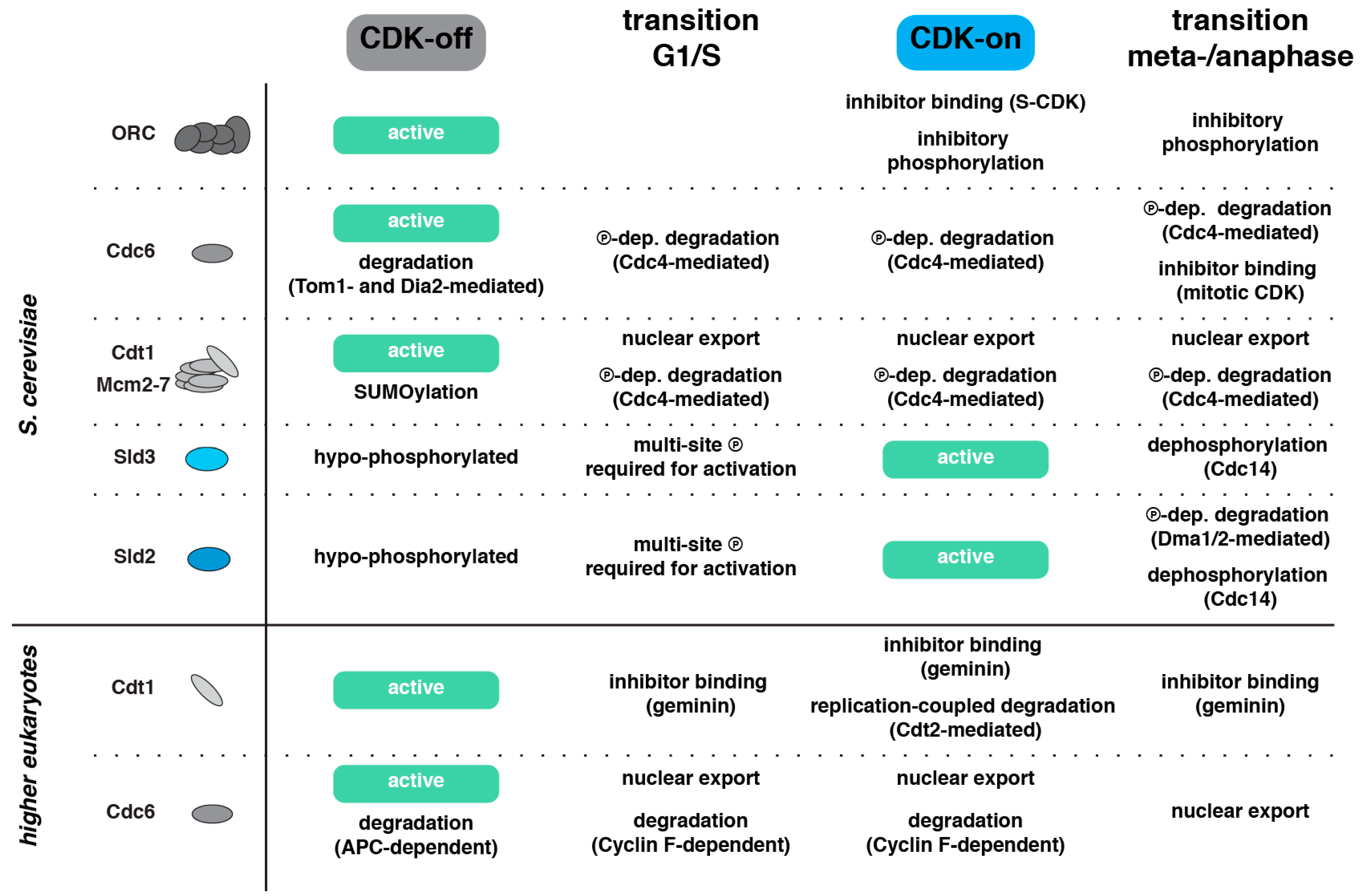
2.1. DNA Replication Initiation Control in Budding Yeast
2.2. Additional DNA Replication Initiation Control Mechanisms in Metazoa
2.3. Deregulation of DNA Replication Initiation—Over-Replication and Genome Instability
2.4. Partial Deregulation of DNA Replication Initiation—Sporadic Over-Replication
3. DNA Replication Control at Cell Cycle Transitions
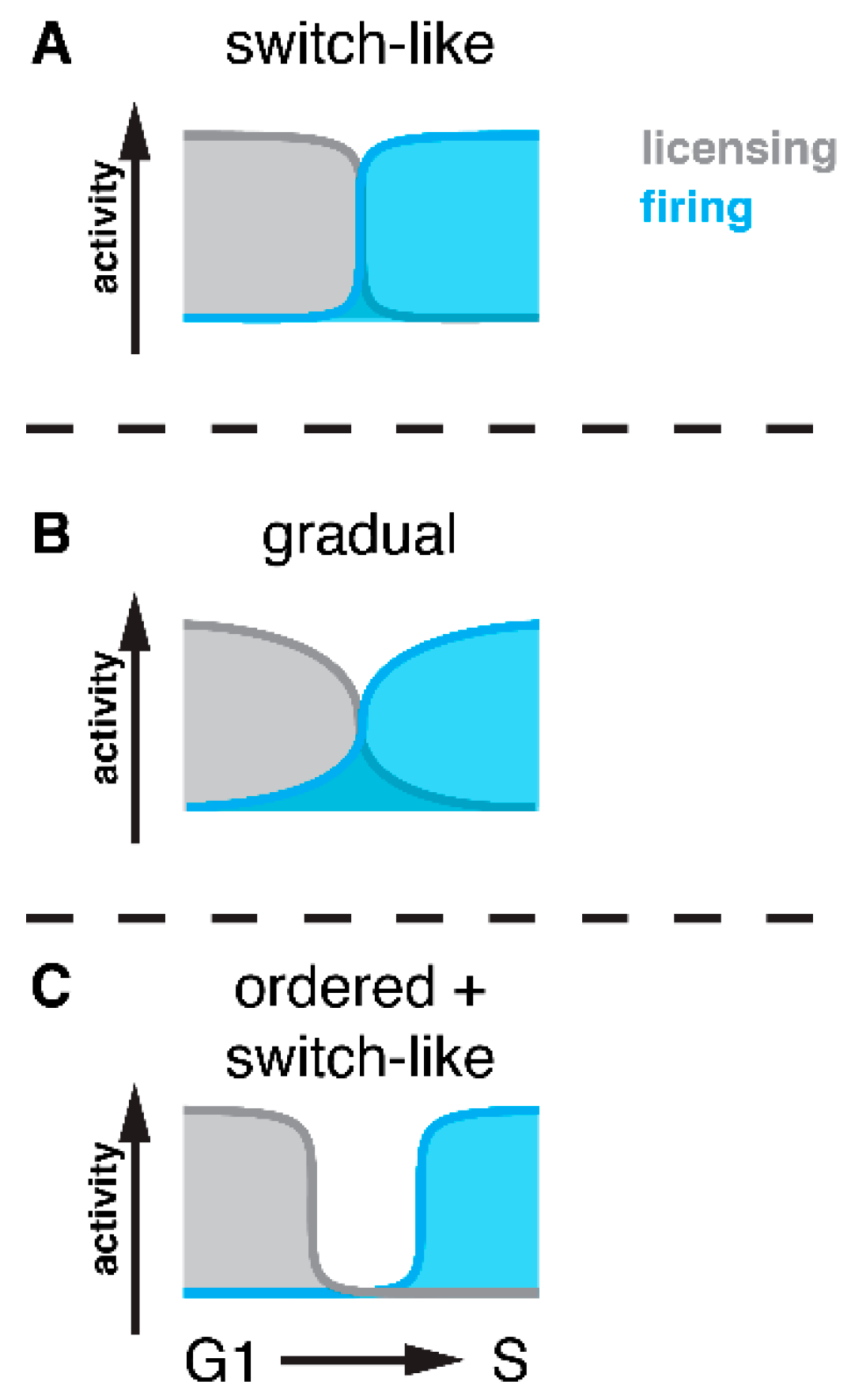
3.1. Bistable Switches—The Fundament of DNA Replication Control at Cell Cycle Transitions
3.2. Temporal Order of Licensing/Firing Activation/Inactivation at Cell Cycle Transitions
3.3. Intrinsic Temporal Order by CDK–Substrate Interactions
3.4. Temporal Order by Phosphatase–Substrate Interactions
3.5. Temporal Order by Degradation
3.6. Temporal Order by a Two-Kinase System
4. Conclusions and Outlook
Funding
Acknowledgments
Conflicts of Interest
References
- Bianconi, E.; Piovesan, A.; Facchin, F.; Beraudi, A.; Casadei, R.; Frabetti, F.; Vitale, L.; Pelleri, M.C.; Tassani, S.; Piva, F.; et al. An estimation of the number of cells in the human body. Ann. Hum. Biol. 2013, 40, 463–471. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, K.; On, K.F.; Diffley, J.F.X. Regulating DNA Replication in Eukarya. Cold Spring Harb. Perspect. Biol. 2013, 5, a012930. [Google Scholar] [CrossRef] [PubMed]
- Blow, J.J.; Dutta, A. Preventing re-replication of chromosomal DNA. Nat. Rev. Mol. Cell Biol. 2005, 6, 476–486. [Google Scholar] [CrossRef] [PubMed]
- Arias, E.E.; Walter, J.C. Strength in numbers: Preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev. 2007, 21, 497–518. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.Q.; Co, C.; Li, J.J. Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms. Nature 2001, 411, 1068–1073. [Google Scholar] [CrossRef]
- Green, B.M.; Finn, K.J.; Li, J.J. Loss of DNA Replication Control Is a Potent Inducer of Gene Amplification. Science 2010, 329, 943–946. [Google Scholar] [CrossRef] [PubMed]
- Alexander, J.L.; Barrasa, M.I.; Orr-Weaver, T.L. Replication fork progression during re-replication requires the DNA damage checkpoint and double-strand break repair. Curr. Biol. 2015, 25, 1654–1660. [Google Scholar] [CrossRef] [PubMed]
- Green, B.M.; Li, J.J. Loss of rereplication control in Saccharomyces cerevisiae results in extensive DNA damage. Mol. Biol. Cell 2005, 16, 421–432. [Google Scholar] [CrossRef] [PubMed]
- Halazonetis, T.D.; Gorgoulis, V.G.; Bartek, J. An Oncogene-Induced DNA Damage Model for Cancer Development. Science 2008, 319, 1352–1355. [Google Scholar] [CrossRef] [PubMed]
- Macheret, M.; Halazonetis, T.D. Intragenic origins due to short G1 phases underlie oncogene-induced DNA replication stress. Nature 2018, 555, 112. [Google Scholar] [CrossRef] [PubMed]
- Macheret, M.; Halazonetis, T.D. DNA replication stress as a hallmark of cancer. Annu. Rev. Pathol. 2015, 10, 425–448. [Google Scholar] [CrossRef] [PubMed]
- Kotsantis, P.; Petermann, E.; Boulton, S.J. Mechanisms of Oncogene-Induced Replication Stress: Jigsaw Falling into Place. Cancer Discov. 2018, 8, 537–555. [Google Scholar] [CrossRef] [PubMed]
- Hills, S.A.; Diffley, J.F.X. DNA replication and oncogene-induced replicative stress. Curr. Biol. 2014, 24, R435–R444. [Google Scholar] [CrossRef] [PubMed]
- Sherr, C.J.; McCormick, F. The RB and p53 pathways in cancer. Cancer Cell 2002, 2, 103–112. [Google Scholar] [CrossRef]
- Meloche, S.; Pouysségur, J. The ERK1/2 mitogen-activated protein kinase pathway as a master regulator of the G1- to S-phase transition. Oncogene 2007, 26, 3227–3239. [Google Scholar] [CrossRef] [PubMed]
- Wong, K.M.; Hudson, T.J.; McPherson, J.D. Unraveling the genetics of cancer: Genome sequencing and beyond. Annu. Rev. Genom. Hum. Genet. 2011, 12, 407–430. [Google Scholar] [CrossRef] [PubMed]
- Bleichert, F.; Botchan, M.R.; Berger, J.M. Mechanisms for initiating cellular DNA replication. Science 2017, 355, eaah6317. [Google Scholar] [CrossRef] [PubMed]
- Bell, S.P.; Labib, K. Chromosome Duplication in Saccharomyces cerevisiae. Genetics 2016, 203, 1027–1067. [Google Scholar] [CrossRef] [PubMed]
- Burgers, P.M.J.; Kunkel, T.A. Eukaryotic DNA Replication Fork. Annu. Rev. Biochem. 2017, 86, 417–438. [Google Scholar] [CrossRef] [PubMed]
- Dewar, J.M.; Walter, J.C. Mechanisms of DNA replication termination. Nat. Rev. Mol. Cell Biol. 2017, 18, 507–516. [Google Scholar] [CrossRef]
- Gambus, A. Termination of Eukaryotic Replication Forks. Adv. Exp. Med. Biol. 2017, 1042, 163–187. [Google Scholar]
- Bhat, K.P.; Cortez, D. RPA and RAD51: Fork reversal, fork protection, and genome stability. Nat. Struct. Mol. Biol. 2018, 25, 446–453. [Google Scholar] [CrossRef] [PubMed]
- Neelsen, K.J.; Lopes, M. Replication fork reversal in eukaryotes: From dead end to dynamic response. Nat. Rev. Mol. Cell Biol. 2015, 16, 207–220. [Google Scholar] [CrossRef] [PubMed]
- Branzei, D.; Szakal, B. Building up and breaking down: Mechanisms controlling recombination during replication. Crit. Rev. Biochem. Mol. Biol. 2017, 52, 381–394. [Google Scholar] [CrossRef] [PubMed]
- Remus, D.; Beuron, F.; Tolun, G.; Griffith, J.D.; Morris, E.P.; Diffley, J.F.X. Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell 2009, 139, 719–730. [Google Scholar] [CrossRef] [PubMed]
- Evrin, C.; Clarke, P.; Zech, J.; Lurz, R.; Sun, J.; Uhle, S.; Li, H.; Stillman, B.; Speck, C. A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc. Natl. Acad. Sci. USA 2009, 106, 20240–20245. [Google Scholar] [CrossRef]
- Donovan, S.; Harwood, J.; Drury, L.S.; Diffley, J.F. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc. Natl. Acad. Sci. USA 1997, 94, 5611–5616. [Google Scholar] [CrossRef]
- Seki, T.; Diffley, J.F. Stepwise assembly of initiation proteins at budding yeast replication origins in vitro. Proc. Natl. Acad. Sci. USA 2000, 97, 14115–14120. [Google Scholar] [CrossRef]
- Rowles, A.; Tada, S.; Blow, J.J. Changes in association of the Xenopus origin recognition complex with chromatin on licensing of replication origins. J. Cell Sci. 1999, 112 Pt 12, 2011–2018. [Google Scholar]
- Yeeles, J.T.P.; Janska, A.; Early, A.; Diffley, J.F.X. How the Eukaryotic Replisome Achieves Rapid and Efficient DNA Replication. Mol. Cell 2017, 65, 105–116. [Google Scholar] [CrossRef]
- Heller, R.C.; Kang, S.; Lam, W.M.; Chen, S.; Chan, C.S.; Bell, S.P. Eukaryotic Origin-Dependent DNA Replication In Vitro Reveals Sequential Action of DDK and S-CDK Kinases. Cell 2011, 146, 80–91. [Google Scholar] [CrossRef]
- Ilves, I.; Petojevic, T.; Pesavento, J.J.; Botchan, M.R. Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol. Cell 2010, 37, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Costa, A.; Ilves, I.; Tamberg, N.; Petojevic, T.; Nogales, E.; Botchan, M.R.; Berger, J.M. The structural basis for MCM2-7 helicase activation by GINS and Cdc45. Nat. Struct. Mol. Biol. 2011, 18, 471–477. [Google Scholar] [CrossRef] [PubMed]
- Moyer, S.E.; Lewis, P.W.; Botchan, M.R. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc. Natl. Acad. Sci. USA 2006, 103, 10236–10241. [Google Scholar] [CrossRef] [PubMed]
- Aparicio, T.; Guillou, E.; Coloma, J.; Montoya, G.; Mendez, J. The human GINS complex associates with Cdc45 and MCM and is essential for DNA replication. Nucleic Acids Res. 2009, 37, 2087–2095. [Google Scholar] [CrossRef] [PubMed]
- Gambus, A.; Jones, R.C.; Sanchez-Diaz, A.; Kanemaki, M.; van Deursen, F.; Edmondson, R.D.; Labib, K. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat. Cell Biol. 2006, 8, 358–366. [Google Scholar] [CrossRef] [PubMed]
- Yeeles, J.T.P.; Deegan, T.D.; Janska, A.; Early, A.; Diffley, J.F.X. Regulated eukaryotic DNA replication origin firing with purified proteins. Nature 2015, 519, 431–435. [Google Scholar] [CrossRef] [PubMed]
- Kurat, C.F.; Yeeles, J.T.P.; Patel, H.; Early, A.; Diffley, J.F.X. Chromatin Controls DNA Replication Origin Selection, Lagging-Strand Synthesis, and Replication Fork Rates. Mol. Cell 2017, 65, 117–130. [Google Scholar] [CrossRef]
- Devbhandari, S.; Jiang, J.; Kumar, C.; Whitehouse, I.; Remus, D. Chromatin Constrains the Initiation and Elongation of DNA Replication. Mol. Cell 2017, 65, 131–141. [Google Scholar] [CrossRef]
- Azmi, I.F.; Watanabe, S.; Maloney, M.F.; Kang, S.; Belsky, J.A.; MacAlpine, D.M.; Peterson, C.L.; Bell, S.P. Nucleosomes influence multiple steps during replication initiation. eLife 2017, 6, e22512. [Google Scholar] [CrossRef]
- Coster, G.; Diffley, J.F.X. Bidirectional eukaryotic DNA replication is established by quasi-symmetrical helicase loading. Science 2017, 357, 314–318. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, S.; Diffley, J.F.X. Interdependent nuclear accumulation of budding yeast Cdt1 and Mcm2-7 during G1 phase. Nat. Cell Biol. 2002, 4, 198–207. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, J.F.; Beach, D. cdt1 is an essential target of the Cdc10/Sct1 transcription factor: Requirement for DNA replication and inhibition of mitosis. EMBO J. 1994, 13, 425–434. [Google Scholar] [CrossRef] [PubMed]
- Bell, S.P.; Stillman, B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature 1992, 357, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Weinreich, M.; Stillman, B. ORC and Cdc6p interact and determine the frequency of initiation of DNA replication in the genome. Cell 1995, 81, 667–676. [Google Scholar] [CrossRef]
- Bueno, A.; Russell, P. Dual functions of CDC6: A yeast protein required for DNA replication also inhibits nuclear division. EMBO J. 1992, 11, 2167–2176. [Google Scholar] [CrossRef] [PubMed]
- Douglas, M.E.; Ali, F.A.; Costa, A.; Diffley, J.F.X. The mechanism of eukaryotic CMG helicase activation. Nature 2018, 555, 265–268. [Google Scholar] [CrossRef] [PubMed]
- Zegerman, P.; Diffley, J.F.X. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature 2007, 445, 281–285. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, S.; Umemori, T.; Hirai, K.; Muramatsu, S.; Kamimura, Y.; Araki, H. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature 2007, 445, 328–332. [Google Scholar] [CrossRef]
- Masumoto, H.; Sugino, A.; Araki, H. Dpb11 controls the association between DNA polymerases alpha and epsilon and the autonomously replicating sequence region of budding yeast. Mol. Cell. Biol. 2000, 20, 2809–2817. [Google Scholar] [CrossRef]
- Masumoto, H.; Muramatsu, S.; Kamimura, Y.; Araki, H. S-Cdk-dependent phosphorylation of Sld2 essential for chromosomal DNA replication in budding yeast. Nature 2002, 415, 651–655. [Google Scholar] [CrossRef] [PubMed]
- Douglas, M.E.; Diffley, J.F.X. Recruitment of Mcm10 to Sites of Replication Initiation Requires Direct Binding to the MCM Complex. J. Biol. Chem. 2016, 291, 5879–5888. [Google Scholar] [CrossRef] [PubMed]
- Homesley, L.; Lei, M.; Kawasaki, Y.; Sawyer, S.; Christensen, T.; Tye, B.K. Mcm10 and the MCM2-7 complex interact to initiate DNA synthesis and to release replication factors from origins. Genes Dev. 2000, 14, 913–926. [Google Scholar] [PubMed]
- Pacek, M.; Tutter, A.V.; Kubota, Y.; Takisawa, H.; Walter, J.C. Localization of MCM2-7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol. Cell 2006, 21, 581–587. [Google Scholar] [CrossRef] [PubMed]
- Miyazawa-Onami, M.; Araki, H.; Tanaka, S. Pre-initiation complex assembly functions as a molecular switch that splits the Mcm2-7 double hexamer. EMBO Rep. 2017, 18, 1752–1761. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, S.; van Deursen, F.; De Piccoli, G.; Labib, K. Dpb2 integrates the leading-strand DNA polymerase into the eukaryotic replisome. Curr. Biol. 2013, 23, 543–552. [Google Scholar] [CrossRef] [PubMed]
- Aves, S.J.; Liu, Y.; Richards, T.A. Evolutionary diversification of eukaryotic DNA replication machinery. Subcell. Biochem. 2012, 62, 19–35. [Google Scholar] [PubMed]
- Prioleau, M.-N.; MacAlpine, D.M. DNA replication origins-where do we begin? Genes Dev. 2016, 30, 1683–1697. [Google Scholar] [CrossRef]
- Gros, J.; Kumar, C.; Lynch, G.; Yadav, T.; Whitehouse, I.; Remus, D. Post-licensing Specification of Eukaryotic Replication Origins by Facilitated Mcm2-7 Sliding along DNA. Mol. Cell 2015, 60, 797–807. [Google Scholar] [CrossRef]
- Santocanale, C.; Diffley, J.F. ORC- and Cdc6-dependent complexes at active and inactive chromosomal replication origins in Saccharomyces cerevisiae. EMBO J. 1996, 15, 6671–6679. [Google Scholar] [CrossRef]
- Diffley, J.F.; Cocker, J.H.; Dowell, S.J.; Rowley, A. Two steps in the assembly of complexes at yeast replication origins in vivo. Cell 1994, 78, 303–316. [Google Scholar] [CrossRef]
- Dahmann, C.; Diffley, J.F.; Nasmyth, K.A. S-phase-promoting cyclin-dependent kinases prevent re-replication by inhibiting the transition of replication origins to a pre-replicative state. Curr. Biol. 1995, 5, 1257–1269. [Google Scholar] [CrossRef]
- Cheng, L.; Collyer, T.; Hardy, C.F. Cell cycle regulation of DNA replication initiator factor Dbf4p. Mol. Cell. Biol. 1999, 19, 4270–4278. [Google Scholar] [CrossRef]
- Oshiro, G.; Owens, J.C.; Shellman, Y.; Sclafani, R.A.; Li, J.J. Cell cycle control of Cdc7p kinase activity through regulation of Dbf4p stability. Mol. Cell. Biol. 1999, 19, 4888–4896. [Google Scholar] [CrossRef] [PubMed]
- Pasero, P.; Duncker, B.P.; Schwob, E.; Gasser, S.M. A role for the Cdc7 kinase regulatory subunit Dbf4p in the formation of initiation-competent origins of replication. Genes Dev. 1999, 13, 2159–2176. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, M.F.; Santocanale, C.; Drury, L.S.; Diffley, J.F. Dbf4p, an essential S phase-promoting factor, is targeted for degradation by the anaphase-promoting complex. Mol. Cell. Biol. 2000, 20, 242–248. [Google Scholar] [CrossRef] [PubMed]
- Surana, U.; Robitsch, H.; Price, C.; Schuster, T.; Fitch, I.; Futcher, A.B.; Nasmyth, K. The role of CDC28 and cyclins during mitosis in the budding yeast S. cerevisiae. Cell 1991, 65, 145–161. [Google Scholar] [CrossRef]
- Hadwiger, J.A.; Wittenberg, C.; Richardson, H.E.; de Barros Lopes, M.; Reed, S.I. A family of cyclin homologs that control the G1 phase in yeast. Proc. Natl. Acad. Sci. USA 1989, 86, 6255–6259. [Google Scholar] [CrossRef]
- Fitch, I.; Dahmann, C.; Surana, U.; Amon, A.; Nasmyth, K.; Goetsch, L.; Byers, B.; Futcher, B. Characterization of four B-type cyclin genes of the budding yeast Saccharomyces cerevisiae. Mol. Biol. Cell 1992, 3, 805–818. [Google Scholar] [CrossRef]
- Epstein, C.B.; Cross, F.R. CLB5: A novel B cyclin from budding yeast with a role in S phase. Genes Dev. 1992, 6, 1695–1706. [Google Scholar] [CrossRef]
- Schwob, E.; Nasmyth, K. CLB5 and CLB6, a new pair of B cyclins involved in DNA replication in Saccharomyces cerevisiae. Genes Dev. 1993, 7, 1160–1175. [Google Scholar] [CrossRef] [PubMed]
- Kitada, K.; Johnston, L.H.; Sugino, T.; Sugino, A. Temperature-sensitive cdc7 mutations of Saccharomyces cerevisiae are suppressed by the DBF4 gene, which is required for the G1/S cell cycle transition. Genetics 1992, 131, 21–29. [Google Scholar] [PubMed]
- Bishop, A.C.; Ubersax, J.A.; Petsch, D.T.; Matheos, D.P.; Gray, N.S.; Blethrow, J.; Shimizu, E.; Tsien, J.Z.; Schultz, P.G.; Rose, M.D.; et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature 2000, 407, 395–401. [Google Scholar] [CrossRef] [PubMed]
- Donaldson, A.D.; Fangman, W.L.; Brewer, B.J. Cdc7 is required throughout the yeast S phase to activate replication origins. Genes Dev. 1998, 12, 491–501. [Google Scholar] [CrossRef] [PubMed]
- Ubersax, J.A.; Woodbury, E.L.; Quang, P.N.; Paraz, M.; Blethrow, J.D.; Shah, K.; Shokat, K.M.; Morgan, D.O. Targets of the cyclin-dependent kinase Cdk1. Nature 2003, 425, 859–864. [Google Scholar] [CrossRef] [PubMed]
- Holt, L.J.; Tuch, B.B.; Villén, J.; Johnson, A.D.; Gygi, S.P.; Morgan, D.O. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science 2009, 325, 1682–1686. [Google Scholar] [CrossRef]
- Randell, J.C.W.; Fan, A.; Chan, C.; Francis, L.I.; Heller, R.C.; Galani, K.; Bell, S.P. Mec1 Is One of Multiple Kinases that Prime the Mcm2-7 Helicase for Phosphorylation by Cdc7. Mol. Cell 2010, 40, 353–363. [Google Scholar] [CrossRef]
- Sheu, Y.-J.; Stillman, B. Cdc7-Dbf4 phosphorylates MCM proteins via a docking site-mediated mechanism to promote S phase progression. Mol. Cell 2006, 24, 101–113. [Google Scholar] [CrossRef]
- Devault, A.; Gueydon, E.; Schwob, E. Interplay between S-cyclin-dependent kinase and Dbf4-dependent kinase in controlling DNA replication through phosphorylation of yeast Mcm4 N-terminal domain. Mol. Biol. Cell 2008, 19, 2267–2277. [Google Scholar] [CrossRef]
- Cho, W.-H.; Lee, Y.-J.; Kong, S.-I.; Hurwitz, J.; Lee, J.-K. CDC7 kinase phosphorylates serine residues adjacent to acidic amino acids in the minichromosome maintenance 2 protein. Proc. Natl. Acad. Sci. USA 2006, 103, 11521–11526. [Google Scholar] [CrossRef]
- Hardy, C.F.; Dryga, O.; Seematter, S.; Pahl, P.M.; Sclafani, R.A. mcm5/cdc46-bob1 bypasses the requirement for the S phase activator Cdc7p. Proc. Natl. Acad. Sci. USA 1997, 94, 3151–3155. [Google Scholar] [CrossRef] [PubMed]
- Sheu, Y.-J.; Stillman, B. The Dbf4-Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4. Nature 2010, 463, 113–117. [Google Scholar] [CrossRef] [PubMed]
- Jackson, A.L.; Pahl, P.M.; Harrison, K.; Rosamond, J.; Sclafani, R.A. Cell cycle regulation of the yeast Cdc7 protein kinase by association with the Dbf4 protein. Mol. Cell. Biol. 1993, 13, 2899–2908. [Google Scholar] [CrossRef] [PubMed]
- Muramatsu, S.; Hirai, K.; Tak, Y.-S.; Kamimura, Y.; Araki, H. CDK-dependent complex formation between replication proteins Dpb11, Sld2, Pol epsilon, and GINS in budding yeast. Genes Dev. 2010, 24, 602–612. [Google Scholar] [CrossRef] [PubMed]
- Tak, Y.-S.; Tanaka, Y.; Endo, S.; Kamimura, Y.; Araki, H. A CDK-catalysed regulatory phosphorylation for formation of the DNA replication complex Sld2-Dpb11. EMBO J. 2006, 25, 1987–1996. [Google Scholar] [CrossRef] [PubMed]
- Araki, H.; Leem, S.H.; Phongdara, A.; Sugino, A. Dpb11, which interacts with DNA polymerase II(epsilon) in Saccharomyces cerevisiae, has a dual role in S-phase progression and at a cell cycle checkpoint. Proc. Natl. Acad. Sci. USA 1995, 92, 11791–11795. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Elledge, S.J. Genetic and physical interactions between DPB11 and DDC1 in the yeast DNA damage response pathway. Genetics 2002, 160, 1295–1304. [Google Scholar]
- Garcia, V.; Furuya, K.; Carr, A.M. Identification and functional analysis of TopBP1 and its homologs. DNA Repair 2005, 4, 1227–1239. [Google Scholar] [CrossRef]
- Puddu, F.; Granata, M.; Di Nola, L.; Balestrini, A.; Piergiovanni, G.; Lazzaro, F.; Giannattasio, M.; Plevani, P.; Muzi-Falconi, M. Phosphorylation of the budding yeast 9-1-1 complex is required for Dpb11 function in the full activation of the UV-induced DNA damage checkpoint. Mol. Cell. Biol. 2008, 28, 4782–4793. [Google Scholar] [CrossRef]
- Pfander, B.; Diffley, J.F.X. Dpb11 coordinates Mec1 kinase activation with cell cycle-regulated Rad9 recruitment. EMBO J. 2011, 30, 4897–4907. [Google Scholar] [CrossRef]
- Gritenaite, D.; Princz, L.N.; Szakal, B.; Bantele, S.C.S.; Wendeler, L.; Schilbach, S.; Habermann, B.H.; Matos, J.; Lisby, M.; Branzei, D.; et al. A cell cycle-regulated Slx4-Dpb11 complex promotes the resolution of DNA repair intermediates linked to stalled replication. Genes Dev. 2014, 28, 1604–1619. [Google Scholar] [CrossRef] [PubMed]
- Bantele, S.C.; Ferreira, P.; Gritenaite, D.; Boos, D.; Pfander, B. Targeting of the Fun30 nucleosome remodeller by the Dpb11 scaffold facilitates cell cycle-regulated DNA end resection. eLife 2017, 6, e21687. [Google Scholar] [CrossRef] [PubMed]
- Ohouo, P.Y.; de Oliveira, F.M.B.; Liu, Y.; Ma, C.J.; Smolka, M.B. DNA-repair scaffolds dampen checkpoint signalling by counteracting the adaptor Rad9. Nature 2012, 493, 120–124. [Google Scholar] [CrossRef] [PubMed]
- Kamimura, Y.; Tak, Y.S.; Sugino, A.; Araki, H. Sld3, which interacts with Cdc45 (Sld4), functions for chromosomal DNA replication in Saccharomyces cerevisiae. EMBO J. 2001, 20, 2097–2107. [Google Scholar] [CrossRef] [PubMed]
- Kanemaki, M.; Labib, K. Distinct roles for Sld3 and GINS during establishment and progression of eukaryotic DNA replication forks. EMBO J. 2006, 25, 1753–1763. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, S.; Nakato, R.; Katou, Y.; Shirahige, K.; Araki, H. Origin Association of Sld3, Sld7, and Cdc45 Proteins Is a Key Step for Determination of Origin-Firing Timing. Curr. Biol. 2011, 21, 2055–2063. [Google Scholar] [CrossRef] [PubMed]
- Deegan, T.D.; Yeeles, J.T.; Diffley, J.F. Phosphopeptide binding by Sld3 links Dbf4-dependent kinase to MCM replicative helicase activation. EMBO J. 2016, 35, 961–973. [Google Scholar] [CrossRef]
- Takayama, Y.; Kamimura, Y.; Okawa, M.; Muramatsu, S.; Sugino, A.; Araki, H. GINS, a novel multiprotein complex required for chromosomal DNA replication in budding yeast. Genes Dev. 2003, 17, 1153–1165. [Google Scholar] [CrossRef]
- Chen, S.; Bell, S.P. CDK prevents Mcm2-7 helicase loading by inhibiting Cdt1 interaction with Orc6. Genes Dev. 2011, 25, 363–372. [Google Scholar] [CrossRef]
- Drury, L.S.; Perkins, G.; Diffley, J.F. The Cdc4/34/53 pathway targets Cdc6p for proteolysis in budding yeast. EMBO J. 1997, 16, 5966–5976. [Google Scholar] [CrossRef]
- Drury, L.S.; Perkins, G.; Diffley, J.F. The cyclin-dependent kinase Cdc28p regulates distinct modes of Cdc6p proteolysis during the budding yeast cell cycle. Curr. Biol. 2000, 10, 231–240. [Google Scholar] [CrossRef]
- Elsasser, S.; Chi, Y.; Yang, P.; Campbell, J.L. Phosphorylation controls timing of Cdc6p destruction: A biochemical analysis. Mol. Biol. Cell 1999, 10, 3263–3277. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Zhang, W.; Koepp, D.M. The Hect-domain E3 ligase Tom1 and the F-box protein Dia2 control Cdc6 degradation in G1. J. Biol. Chem. 2012. [Google Scholar] [CrossRef]
- Labib, K.; Diffley, J.F.; Kearsey, S.E. G1-phase and B-type cyclins exclude the DNA-replication factor Mcm4 from the nucleus. Nat. Cell Biol. 1999, 1, 415–422. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Makino, N.; Nagai, M.; Araki, H.; Ushimaru, T. CDK phosphorylation regulates Mcm3 degradation in budding yeast. Biochem. Biophys. Res. Commun. 2018, 506, 680–684. [Google Scholar] [CrossRef] [PubMed]
- Weinreich, M.; Liang, C.; Chen, H.H.; Stillman, B. Binding of cyclin-dependent kinases to ORC and Cdc6p regulates the chromosome replication cycle. Proc. Natl. Acad. Sci. USA 2001, 98, 11211–11217. [Google Scholar] [CrossRef] [PubMed]
- Wilmes, G.M.; Archambault, V.; Austin, R.J.; Jacobson, M.D.; Bell, S.P.; Cross, F.R. Interaction of the S-phase cyclin Clb5 with an “RXL” docking sequence in the initiator protein Orc6 provides an origin-localized replication control switch. Genes Dev. 2004, 18, 981–991. [Google Scholar] [CrossRef]
- Mimura, S.; Seki, T.; Tanaka, S.; Diffley, J.F.X. Phosphorylation-dependent binding of mitotic cyclins to Cdc6 contributes to DNA replication control. Nature 2004, 431, 1118–1123. [Google Scholar] [CrossRef]
- Drury, L.S.; Diffley, J.F.X. Factors Affecting the Diversity of DNA Replication Licensing Control in Eukaryotes. Curr. Biol. 2009, 19, 530–535. [Google Scholar] [CrossRef]
- Kumagai, A.; Shevchenko, A.; Shevchenko, A.; Dunphy, W.G. Treslin collaborates with TopBP1 in triggering the initiation of DNA replication. Cell 2010, 140, 349–359. [Google Scholar] [CrossRef]
- Boos, D.; Sanchez-Pulido, L.; Rappas, M.; Pearl, L.H.; Oliver, A.W.; Ponting, C.P.; Diffley, J.F.X. Regulation of DNA Replication through Sld3-Dpb11 Interaction Is Conserved from Yeast to Humans. Curr. Biol. 2011, 21, 1152–1157. [Google Scholar] [CrossRef] [PubMed]
- Strausfeld, U.P.; Howell, M.; Rempel, R.; Maller, J.L.; Hunt, T.; Blow, J.J. Cip1 blocks the initiation of DNA replication in Xenopus extracts by inhibition of cyclin-dependent kinases. Curr. Biol. 1994, 4, 876–883. [Google Scholar] [CrossRef]
- Walter, J.C. Evidence for sequential action of cdc7 and cdk2 protein kinases during initiation of DNA replication in Xenopus egg extracts. J. Biol. Chem. 2000, 275, 39773–39778. [Google Scholar] [CrossRef] [PubMed]
- Jares, P.; Blow, J.J. Xenopus cdc7 function is dependent on licensing but not on XORC, XCdc6, or CDK activity and is required for XCdc45 loading. Genes Dev. 2000, 14, 1528–1540. [Google Scholar] [PubMed]
- Wohlschlegel, J.A.; Dwyer, B.T.; Dhar, S.K.; Cvetic, C.; Walter, J.C.; Dutta, A. Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science 2000, 290, 2309–2312. [Google Scholar] [CrossRef] [PubMed]
- Tada, S.; Li, A.; Maiorano, D.; Méchali, M.; Blow, J.J. Repression of origin assembly in metaphase depends on inhibition of RLF-B/Cdt1 by geminin. Nat. Cell Biol. 2001, 3, 107–113. [Google Scholar] [CrossRef] [PubMed]
- McGarry, T.J.; Kirschner, M.W. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell 1998, 93, 1043–1053. [Google Scholar] [CrossRef]
- Quinn, L.M.; Herr, A.; McGarry, T.J.; Richardson, H. The Drosophila Geminin homolog: Roles for Geminin in limiting DNA replication, in anaphase and in neurogenesis. Genes Dev. 2001, 15, 2741–2754. [Google Scholar] [CrossRef]
- Yanagi, K.-I.; Mizuno, T.; Tsuyama, T.; Tada, S.; Iida, Y.; Sugimoto, A.; Eki, T.; Enomoto, T.; Hanaoka, F. Caenorhabditis elegans geminin homologue participates in cell cycle regulation and germ line development. J. Biol. Chem. 2005, 280, 19689–19694. [Google Scholar] [CrossRef]
- Arias, E.E.; Walter, J.C. Replication-dependent destruction of Cdt1 limits DNA replication to a single round per cell cycle in Xenopus egg extracts. Genes Dev. 2005, 19, 114–126. [Google Scholar] [CrossRef]
- Jin, J.; Arias, E.E.; Chen, J.; Harper, J.W.; Walter, J.C. A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol. Cell 2006, 23, 709–721. [Google Scholar] [CrossRef] [PubMed]
- Arias, E.E.; Walter, J.C. PCNA functions as a molecular platform to trigger Cdt1 destruction and prevent re-replication. Nat. Cell Biol. 2006, 8, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Senga, T.; Sivaprasad, U.; Zhu, W.; Park, J.H.; Arias, E.E.; Walter, J.C.; Dutta, A. PCNA is a cofactor for Cdt1 degradation by CUL4/DDB1-mediated N-terminal ubiquitination. J. Biol. Chem. 2006, 281, 6246–6252. [Google Scholar] [CrossRef] [PubMed]
- Higa, L.A.; Banks, D.; Wu, M.; Kobayashi, R.; Sun, H.; Zhang, H. L2DTL/CDT2 interacts with the CUL4/DDB1 complex and PCNA and regulates CDT1 proteolysis in response to DNA damage. Cell Cycle 2006, 5, 1675–1680. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Xiong, Y. An evolutionarily conserved function of proliferating cell nuclear antigen for Cdt1 degradation by the Cul4-Ddb1 ubiquitin ligase in response to DNA damage. J. Biol. Chem. 2006, 281, 3753–3756. [Google Scholar] [CrossRef] [PubMed]
- Ralph, E.; Boye, E.; Kearsey, S.E. DNA damage induces Cdt1 proteolysis in fission yeast through a pathway dependent on Cdt2 and Ddb1. EMBO Rep. 2006, 7, 1134–1139. [Google Scholar] [CrossRef] [PubMed]
- Alexander, J.L.; Orr-Weaver, T.L. Replication fork instability and the consequences of fork collisions from rereplication. Genes Dev. 2016, 30, 2241–2252. [Google Scholar] [CrossRef] [PubMed]
- Neelsen, K.J.; Zanini, I.M.Y.; Mijic, S.; Herrador, R.; Zellweger, R.; Ray Chaudhuri, A.; Creavin, K.D.; Blow, J.J.; Lopes, M. Deregulated origin licensing leads to chromosomal breaks by rereplication of a gapped DNA template. Genes Dev. 2013, 27, 2537–2542. [Google Scholar] [CrossRef] [PubMed]
- Green, B.M.; Morreale, R.J.; Ozaydin, B.; Derisi, J.L.; Li, J.J. Genome-wide mapping of DNA synthesis in Saccharomyces cerevisiae reveals that mechanisms preventing reinitiation of DNA replication are not redundant. Mol. Biol. Cell 2006, 17, 2401–2414. [Google Scholar] [CrossRef] [PubMed]
- Finn, K.J.; Li, J.J. Single-Stranded Annealing Induced by Re-Initiation of Replication Origins Provides a Novel and Efficient Mechanism for Generating Copy Number Expansion via Non-Allelic Homologous Recombination. PLoS Genet. 2013, 9, e1003192. [Google Scholar] [CrossRef] [PubMed]
- Archambault, V.; Ikui, A.E.; Drapkin, B.J.; Cross, F.R. Disruption of mechanisms that prevent rereplication triggers a DNA damage response. Mol. Cell. Biol. 2005, 25, 6707–6721. [Google Scholar] [CrossRef] [PubMed]
- Tanny, R.E.; MacAlpine, D.M.; Blitzblau, H.G.; Bell, S.P. Genome-wide analysis of re-replication reveals inhibitory controls that target multiple stages of replication initiation. Mol. Biol. Cell 2006, 17, 2415–2423. [Google Scholar] [CrossRef] [PubMed]
- Hanlon, S.L.; Li, J.J. Re-replication of a Centromere Induces Chromosomal Instability and Aneuploidy. PLoS Genet. 2015, 11, e1005039. [Google Scholar] [CrossRef] [PubMed]
- Symington, L.S.; Rothstein, R.; Lisby, M. Mechanisms and regulation of mitotic recombination in Saccharomyces cerevisiae. Genetics 2014, 198, 795–835. [Google Scholar] [CrossRef] [PubMed]
- Mehta, A.; Haber, J.E. Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb. Perspect. Biol. 2014, 6, a016428. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Blow, J.J. Cdt1 downregulation by proteolysis and geminin inhibition prevents DNA re-replication in Xenopus. EMBO J. 2005, 24, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Takisawa, H.; Kubota, Y. Intrinsic nuclear import activity of geminin is essential to prevent re-initiation of DNA replication in Xenopus eggs. Genes Cells 2005, 10, 63–73. [Google Scholar] [CrossRef]
- Maiorano, D.; Krasinska, L.; Lutzmann, M.; Mechali, M. Recombinant Cdt1 induces rereplication of G2 nuclei in Xenopus egg extracts. Curr. Biol. 2005, 15, 146–153. [Google Scholar] [CrossRef]
- Melixetian, M.; Ballabeni, A.; Masiero, L.; Gasparini, P.; Zamponi, R.; Bartek, J.; Lukas, J.; Helin, K. Loss of Geminin induces rereplication in the presence of functional p53. J. Cell Biol. 2004, 165, 473–482. [Google Scholar] [CrossRef]
- Nishitani, H.; Lygerou, Z.; Nishimoto, T. Proteolysis of DNA replication licensing factor Cdt1 in S-phase is performed independently of geminin through its N-terminal region. J. Biol. Chem. 2004, 279, 30807–30816. [Google Scholar] [CrossRef]
- Zhu, W.; Chen, Y.; Dutta, A. Rereplication by depletion of geminin is seen regardless of p53 status and activates a G2/M checkpoint. Mol. Cell. Biol. 2004, 24, 7140–7150. [Google Scholar] [CrossRef] [PubMed]
- Machida, Y.J.; Dutta, A. The APC/C inhibitor, Emi1, is essential for prevention of rereplication. Genes Dev. 2007, 21, 184–194. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Dutta, A. An ATR- and BRCA1-mediated Fanconi anemia pathway is required for activating the G2/M checkpoint and DNA damage repair upon rereplication. Mol. Cell. Biol. 2006, 26, 4601–4611. [Google Scholar] [CrossRef] [PubMed]
- Diffley, J.F.X. Quality control in the initiation of eukaryotic DNA replication. Philos. Trans. R. Soc. B Biol. Sci. 2011, 366, 3545–3553. [Google Scholar] [CrossRef] [PubMed]
- Reußwig, K.-U.; Zimmermann, F.; Galanti, L.; Pfander, B. Robust Replication Control Is Generated by Temporal Gaps between Licensing and Firing Phases and Depends on Degradation of Firing Factor Sld2. Cell Rep. 2016, 17, 556–569. [Google Scholar] [CrossRef] [PubMed]
- Ciccia, A.; Elledge, S.J. The DNA Damage Response: Making It Safe to Play with Knives. Mol. Cell 2010, 40, 179–204. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, S.; Araki, H. Multiple Regulatory Mechanisms to Inhibit Untimely Initiation of DNA Replication Are Important for Stable Genome Maintenance. PLoS Genet. 2011, 7, e1002136. [Google Scholar] [CrossRef] [PubMed]
- Tyson, J.J.; Novak, B. Regulation of the eukaryotic cell cycle: Molecular antagonism, hysteresis, and irreversible transitions. J. Biol. 2001, 210, 249–263. [Google Scholar] [CrossRef]
- Chen, K.C.; Calzone, L.; Csikasz-Nagy, A.; Cross, F.R.; Novak, B.; Tyson, J.J. Integrative analysis of cell cycle control in budding yeast. Mol. Biol. Cell 2004, 15, 3841–3862. [Google Scholar] [CrossRef] [PubMed]
- Cross, F.R. Two redundant oscillatory mechanisms in the yeast cell cycle. Dev. Cell 2003, 4, 741–752. [Google Scholar] [CrossRef]
- Santos, S.D.M.; Ferrell, J.E. Systems biology: On the cell cycle and its switches. Nature 2008, 454, 288–289. [Google Scholar] [CrossRef] [PubMed]
- Skotheim, J.M.; Di Talia, S.; Siggia, E.D.; Cross, F.R. Positive feedback of G1 cyclins ensures coherent cell cycle entry. Nature 2008, 454, 291–296. [Google Scholar] [CrossRef] [PubMed]
- Schwob, E.; Böhm, T.; Mendenhall, M.D.; Nasmyth, K. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell 1994, 79, 233–244. [Google Scholar] [CrossRef]
- Verma, R.; Annan, R.S.; Huddleston, M.J.; Carr, S.A.; Reynard, G.; Deshaies, R.J. Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science 1997, 278, 455–460. [Google Scholar] [CrossRef] [PubMed]
- Pomerening, J.R.; Kim, S.Y.; Ferrell, J.E. Systems-level dissection of the cell-cycle oscillator: Bypassing positive feedback produces damped oscillations. Cell 2005, 122, 565–578. [Google Scholar] [CrossRef]
- Cross, F.R.; Archambault, V.; Miller, M.; Klovstad, M. Testing a mathematical model of the yeast cell cycle. Mol. Biol. Cell 2002, 13, 52–70. [Google Scholar] [CrossRef] [PubMed]
- Nash, P.; Tang, X.; Orlicky, S.; Chen, Q.; Gertler, F.B.; Mendenhall, M.D.; Sicheri, F.; Pawson, T.; Tyers, M. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature 2001, 414, 514–521. [Google Scholar] [CrossRef]
- Brümmer, A.; Salazar, C.; Zinzalla, V.; Alberghina, L.; Höfer, T. Mathematical Modelling of DNA Replication Reveals a Trade-off between Coherence of Origin Activation and Robustness against Rereplication. PLoS Comput. Biol. 2010, 6, e1000783. [Google Scholar] [CrossRef]
- Loog, M.; Morgan, D.O. Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature 2005, 434, 104–108. [Google Scholar] [CrossRef]
- Zhai, Y.; Yung, P.Y.K.; Huo, L.; Liang, C. Cdc14p resets the competency of replication licensing by dephosphorylating multiple initiation proteins during mitotic exit in budding yeast. J. Cell Sci. 2010, 123, 3933–3943. [Google Scholar] [CrossRef]
- Bloom, J.; Cross, F.R. Novel role for Cdc14 sequestration: Cdc14 dephosphorylates factors that promote DNA replication. Mol. Cell. Biol. 2007, 27, 842–853. [Google Scholar] [CrossRef] [PubMed]
- Powers, B.L.; Hall, M.C. Re-examining the role of Cdc14 phosphatase in reversal of Cdk phosphorylation during mitotic exit. J. Cell Sci. 2017, 130, 2673–2681. [Google Scholar] [CrossRef] [PubMed]
- Bouchoux, C.; Uhlmann, F. A Quantitative Model for Ordered Cdk Substrate Dephosphorylation during Mitotic Exit. Cell 2011, 147, 803–814. [Google Scholar] [CrossRef] [PubMed]
- Nishitani, H.; Sugimoto, N.; Roukos, V.; Nakanishi, Y.; Saijo, M.; Obuse, C.; Tsurimoto, T.; Nakayama, K.I.; Nakayama, K.; Fujita, M.; et al. Two E3 ubiquitin ligases, SCF-Skp2 and DDB1-Cul4, target human Cdt1 for proteolysis. EMBO J. 2006, 25, 1126–1136. [Google Scholar] [CrossRef] [PubMed]
- Phizicky, D.V.; Berchowitz, L.E.; Bell, S.P. Multiple kinases inhibit origin licensing and helicase activation to ensure reductive cell division during meiosis. eLife 2018, 7, e33309. [Google Scholar] [CrossRef] [PubMed]
- Petersen, B.O.; Wagener, C.; Marinoni, F.; Kramer, E.R.; Melixetian, M.; Lazzerini Denchi, E.; Gieffers, C.; Matteucci, C.; Peters, J.M.; Helin, K. Cell cycle- and cell growth-regulated proteolysis of mammalian CDC6 is dependent on APC-CDH1. Genes Dev. 2000, 14, 2330–2343. [Google Scholar] [CrossRef] [PubMed]
- Mailand, N.; Diffley, J.F.X. CDKs promote DNA replication origin licensing in human cells by protecting Cdc6 from APC/C-dependent proteolysis. Cell 2005, 122, 915–926. [Google Scholar] [CrossRef]
- Petersen, B.O.; Lukas, J.; Sørensen, C.S.; Bartek, J.; Helin, K. Phosphorylation of mammalian CDC6 by cyclin A/CDK2 regulates its subcellular localization. EMBO J. 1999, 18, 396–410. [Google Scholar] [CrossRef]
- Jiang, W.; Wells, N.J.; Hunter, T. Multistep regulation of DNA replication by Cdk phosphorylation of HsCdc6. Proc. Natl. Acad. Sci. USA 1999, 96, 6193–6198. [Google Scholar] [CrossRef]
- Saha, P.; Chen, J.; Thome, K.C.; Lawlis, S.J.; Hou, Z.H.; Hendricks, M.; Parvin, J.D.; Dutta, A. Human CDC6/Cdc18 associates with Orc1 and cyclin-cdk and is selectively eliminated from the nucleus at the onset of S phase. Mol. Cell. Biol. 1998, 18, 2758–2767. [Google Scholar] [CrossRef]
- Walter, D.; Hoffmann, S.; Komseli, E.-S.; Rappsilber, J.; Gorgoulis, V.; Sorensen, C.S. SCFCyclin F-dependent degradation of CDC6 suppresses DNA re-replication. Nat. Commun. 2016, 7, 10530. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.; Hsiao, J.Y.; Davey, N.E.; Van Voorhis, V.A.; Foster, S.A.; Tang, C.; Morgan, D.O. Multiple mechanisms determine the order of APC/C substrate degradation in mitosis. J. Cell Biol. 2014, 207, 23–39. [Google Scholar] [CrossRef] [PubMed]
- Kamenz, J.; Mihaljev, T.; Kubis, A.; Legewie, S.; Hauf, S. Robust Ordering of Anaphase Events by Adaptive Thresholds and Competing Degradation Pathways. Mol. Cell 2015, 60, 446–459. [Google Scholar] [CrossRef] [PubMed]
- Coleman, K.E.; Grant, G.D.; Haggerty, R.A.; Brantley, K.; Shibata, E.; Workman, B.D.; Dutta, A.; Varma, D.; Purvis, J.E.; Cook, J.G. Sequential replication-coupled destruction at G1/S ensures genome stability. Genes Dev. 2015, 29, 1734–1746. [Google Scholar] [CrossRef] [PubMed]
- Mantiero, D.; Mackenzie, A.; Donaldson, A.; Zegerman, P. Limiting replication initiation factors execute the temporal programme of origin firing in budding yeast. EMBO J. 2011, 30, 4805–4814. [Google Scholar] [CrossRef] [PubMed]
- Zegerman, P.; Diffley, J.F.X. Checkpoint-dependent inhibition of DNA replication initiation by Sld3 and Dbf4 phosphorylation. Nature 2010, 467, 474–478. [Google Scholar] [CrossRef] [PubMed]
- Hiraga, S.-I.; Alvino, G.M.; Chang, F.; Lian, H.-Y.; Sridhar, A.; Kubota, T.; Brewer, B.J.; Weinreich, M.; Raghuraman, M.K.; Donaldson, A.D. Rif1 controls DNA replication by directing Protein Phosphatase 1 to reverse Cdc7-mediated phosphorylation of the MCM complex. Genes Dev. 2014, 28, 372–383. [Google Scholar] [CrossRef]
- Weinreich, M.; Stillman, B. Cdc7p-Dbf4p kinase binds to chromatin during S phase and is regulated by both the APC and the RAD53 checkpoint pathway. EMBO J. 1999, 18, 5334–5346. [Google Scholar] [CrossRef]
- Mattarocci, S.; Shyian, M.; Lemmens, L.; Damay, P.; Altintas, D.M.; Shi, T.; Bartholomew, C.R.; Thomä, N.H.; Hardy, C.F.J.; Shore, D. Rif1 Controls DNA Replication Timing in Yeast through the PP1 Phosphatase Glc7. Cell Rep. 2014, 7, 62–69. [Google Scholar] [CrossRef]
- Davé, A.; Cooley, C.; Garg, M.; Bianchi, A. Protein Phosphatase 1 Recruitment by Rif1 Regulates DNA Replication Origin Firing by Counteracting DDK Activity. Cell Rep. 2014, 7, 53–61. [Google Scholar] [CrossRef]
- Wei, L.; Zhao, X. A new MCM modification cycle regulates DNA replication initiation. Nat. Struct. Mol. Biol. 2016, 23, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Nougarède, R.; Della Seta, F.; Zarzov, P.; Schwob, E. Hierarchy of S-phase-promoting factors: Yeast Dbf4-Cdc7 kinase requires prior S-phase cyclin-dependent kinase activation. Mol. Cell. Biol. 2000, 20, 3795–3806. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reusswig, K.-U.; Pfander, B. Control of Eukaryotic DNA Replication Initiation—Mechanisms to Ensure Smooth Transitions. Genes 2019, 10, 99. https://doi.org/10.3390/genes10020099
Reusswig K-U, Pfander B. Control of Eukaryotic DNA Replication Initiation—Mechanisms to Ensure Smooth Transitions. Genes. 2019; 10(2):99. https://doi.org/10.3390/genes10020099
Chicago/Turabian StyleReusswig, Karl-Uwe, and Boris Pfander. 2019. "Control of Eukaryotic DNA Replication Initiation—Mechanisms to Ensure Smooth Transitions" Genes 10, no. 2: 99. https://doi.org/10.3390/genes10020099
APA StyleReusswig, K.-U., & Pfander, B. (2019). Control of Eukaryotic DNA Replication Initiation—Mechanisms to Ensure Smooth Transitions. Genes, 10(2), 99. https://doi.org/10.3390/genes10020099




