Abstract
The age-related decline in skeletal muscle mass, strength and function known as ‘sarcopenia’ is associated with multiple adverse health outcomes, including cardiovascular disease, stroke, functional disability and mortality. While skeletal muscle properties are known to be highly heritable, evidence regarding the specific genes underpinning this heritability is currently inconclusive. This review aimed to identify genetic variants known to be associated with muscle phenotypes relevant to sarcopenia. PubMed, Embase and Web of Science were systematically searched (from January 2004 to March 2019) using pre-defined search terms such as “aging”, “sarcopenia”, “skeletal muscle”, “muscle strength” and “genetic association”. Candidate gene association studies and genome wide association studies that examined the genetic association with muscle phenotypes in non-institutionalised adults aged ≥50 years were included. Fifty-four studies were included in the final analysis. Twenty-six genes and 88 DNA polymorphisms were analysed across the 54 studies. The ACTN3, ACE and VDR genes were the most frequently studied, although the IGF1/IGFBP3, TNFα, APOE, CNTF/R and UCP2/3 genes were also shown to be significantly associated with muscle phenotypes in two or more studies. Ten DNA polymorphisms (rs154410, rs2228570, rs1800169, rs3093059, rs1800629, rs1815739, rs1799752, rs7412, rs429358 and 192 bp allele) were significantly associated with muscle phenotypes in two or more studies. Through the identification of key gene variants, this review furthers the elucidation of genetic associations with muscle phenotypes associated with sarcopenia.
1. Introduction
Sarcopenia refers to the progressive deterioration in skeletal muscle mass, strength and physical function with advancing age [1]. The simultaneous presence of low muscle strength, muscle mass and/or physical function forms the diagnostic basis of the recommendations from the European Working Group on Sarcopenia in Older People [2]. These criteria are strong predictors of a multitude of adverse health outcomes, such as cardiovascular disease [3], functional disability [4], fall incidence [5], hospitalisation [6], stroke [7] and mortality [8]. Up to 10% of individuals aged 60–69 years are affected by sarcopenia, with this proportion rising considerably to 40% for adults over 80 years of age [9,10]. The fundamental loss of independence and susceptibility to additional diseases caused by sarcopenia also places a significant burden on public health systems worldwide. This burden is anticipated to grow considerably in coming decades, in line with increases in longevity and the consequent rise in the proportion of elderly [11]. Thus, the consequences of age-related muscle deterioration will become increasingly relevant globally.
While sarcopenia is generally more prevalent among individuals over the age of 60, strong evidence suggests that pronounced changes in muscle tissue begin from around 50 years of age [12]. From this age, muscle mass and strength begin to deteriorate at an annual rate of 1–2% and 1.5–5% respectively [12,13,14]. Developing an understanding of why and how skeletal muscle deteriorates from this age will be critical to reducing the burden of sarcopenia for patients as well as public health systems.
Currently, it is known that inter-individual variation in muscle phenotypes may be attributed to genetic factors, environmental factors and/or, gene-environment interactions [15,16]. While environmental factors such as physical activity, protein intake [17], sleep quality [18], smoking status [15] and alcohol consumption [19] have been shown to affect muscle phenotypes, heritability studies have highlighted the importance of genetic factors in determining inter-individual variability in skeletal muscle traits [20,21]. These studies have found that genetic factors account for 46–76% and 32–67% of fat-free mass (FFM) and muscle strength variability, respectively [20,21]. Additional longitudinal studies have observed heritability estimates of 64% for change in muscle strength with advancing age [22]. However, while the overall heritability of skeletal muscle phenotypes is well established, the genetic mechanisms underpinning this heritability remain unclear.
Thus, developing a deeper understanding of genetic associations underpinning skeletal muscle phenotypes is of paramount importance in the development of effective treatment interventions to manage age-related changes in muscle structure and function. Furthermore, understanding the genetic mechanisms regulating muscle accrual and loss will help facilitate early screening for susceptibility to sarcopenia, which could allow for preventative measures to be implemented prior to predicted muscle degradation.
Therefore, the purpose of this systematic review was to identify and synthesize the genetic variants associated with muscle phenotypes relevant to sarcopenia in humans.
2. Materials and Methods
Reporting followed the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement [23].
2.1. Literature Search and Eligibility Criteria
2.1.1. Inclusion and Exclusion Criteria
To be included in this review, studies had to meet the following criteria:
- Published between January 2004 and March 2019.
- Full English text available.
- Participants must be non-institutionalised human adults, aged 50 years or above.
- Subjects must have been free from any significant cardiovascular, metabolic or musculoskeletal disorders at the time of the study.
- Candidate gene association study or genome wide association study (GWAS).
2.1.2. Search Strategy
A systematic literature search of three online databases, PubMed, EMBASE and Web of Science, was conducted on 18 March 2019, for the period between January 2004 and March 2019. This time limit ensured the inclusion of the most pertinent literature. Search terms were selected based off the PEO framework and combined using Boolean operators (“AND”, “OR”). Filters were used to limit results to those using human subjects, written in the English language and published within the desired time-frame. The search strategy used was as follows: (“ageing” OR “aged” OR “elderly” OR “older persons” OR “community dwelling”) AND (“sarcopenia” OR “skeletal muscle” OR “muscle phenotype” OR “muscle mass” OR “muscle atrophy” OR “muscle strength” OR “grip strength” OR “physical performance” OR “muscle quality” OR “lean mass”) AND (“single nucleotide polymorphism” OR “genetic polymorphism” OR “allele” OR “genetic variation” OR “gene variant” OR “mutation” OR “genes” OR “chromosome” OR “genetic predisposition” OR “genetic susceptibility”) AND (“genetic association studies” OR “genome-wide association study” OR “GWAS” OR “candidate gene study” OR “genotype” OR “haplotype” OR “heritability”). The scope of the online search was further expanded by assessing bibliographic references of the eligible full text articles for relevant studies.
2.2. Study Selection and Data Extraction
Following the removal of duplicates, titles and abstracts were screened for relevance to the scope of this review. To determine inclusion in this review, the full text of every potentially relevant article was scrutinised for overall content and compliance with the eligibility criteria outlined above. The following data were extracted from each eligible article: authors, year of publication, study design, studied population (number, ethnicity, nationality, sex), gene name, polymorphism, muscle phenotype, main findings of the study.
2.3. Phenotypes
Phenotypic outcomes included in this systematic review were skeletal muscle mass, muscle strength, physical function and sarcopenia prevalence.
2.4. Quality Assessment
The quality and risk of bias of the included studies were assessed using the Quality of Genetic Association Studies (Q-Genie) tool [24]. The Q-Genie tool consists of 11 items that cover the following areas: “rationale for study”, “selection and definition of outcome of interest”, “selection and comparability of comparison groups”, “technical classification of the exposure”, “non-technical classification of the exposure”, “other source of bias”, “sample size and power”, “a priori planning of analysis”, “statistical methods and control for confounding”, “testing of assumptions and inferences for genetic analysis” and “appropriateness of inferences drawn from results”. Each area was rated using a 7-point Likert scale (“1 = poor”; “2”, “3 = good”; “4”, “5 = very good”; “6”, “7 = excellent”). The overall quality of the included articles was classified by collating the scores for each theme. Studies with control groups were classified as “poor quality” if the score was ≤ 35, “moderate quality” if the score was > 35 and ≤ 45, and “good quality” if the score was > 45. For studies without control groups, scoring ≤ 32, > 32 and ≤ 40, and > 40 reflected classifications of “poor quality”, “moderate quality” and “good quality”, respectively.
3. Results
3.1. Search Strategy
The systematic search of the online databases identified 771 papers. Following the addition of filters, removal of duplicates and screening for eligibility, 48 studies remained. Six additional articles were retrieved through the manual search of reference lists, leaving a total of 54 articles to be included in this systematic review. Figure 1 highlights the identification and selection process in accordance with the PRISMA statement.
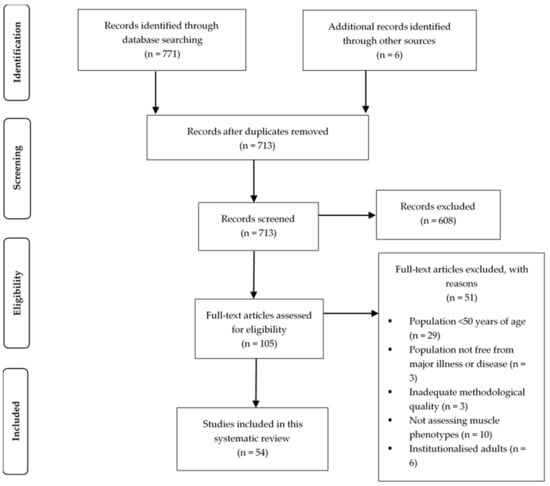
Figure 1.
PRISMA flow chart presenting the identification and selection process of articles.
3.2. Quality Assessment
A detailed quality classification for each article is displayed in Table 1. Studies scored between 33 and 50 in the Q-Genie checklist. For studies with control groups (n = 12), five were classified as “moderate quality” and seven as “good quality”. For non-control group studies (n = 42), 17 were classified as “moderate quality” and 25 as “good quality”.

Table 1.
Q-Genie quality assessment scores for the included studies.
3.3. Study and Subject Characteristics
Of the 54 studies included in this systematic review, 35 were cross sectional studies while the remaining 19 were longitudinal. A comprehensive description of the characteristics of the cross-sectional studies are presented in Table 2. Of the longitudinal studies, 11 were intervention studies while 8 were observational follow-up studies. The average intervention length was 21.3 weeks (range 10–72 weeks) while the average follow-up was 4.2 years (range 1–10 years). Table 3 presents a detailed description of the characteristics of the longitudinal studies. Out of the 54 studies, 53 were candidate gene association studies and the remaining article was a genome-wide association study.

Table 2.
Cross-sectional studies on genetic associations with muscle phenotypes.

Table 3.
Longitudinal studies on genetic association with muscle phenotypes.
A total of 38,112 subjects participated across the 54 studies. Of these, 24,890 (65.3%) were female and 13,222 (34.7%) were male. Thirty-two studies included Caucasians, 13 assessed Asian subjects and the remaining nine studies included Hispanic and African-American participants. As described in the inclusion criteria, all subjects were older than 50 years of age. Thirteen studies included subjects over 50 years of age, 22 studies recruited subjects over 60 years of age and 19 studies included individuals aged 70 years or older.
3.4. Phenotypes and Genotypes
Of the included studies, 26 reported skeletal muscle mass outcomes, 39 studies included muscle strength testing, 27 articles analysed physical function and six examined sarcopenia prevalence. A full description of the phenotypic outcomes in each study are presented in Table 2 and Table 3.
In total, 88 DNA polymorphisms in or near to 26 different genes were analysed across the 54 studies included in this review. The Alpha-actinin 3 (ACTN3), Angiotensin Converting Enzyme (ACE), and Vitamin D Receptor (VDR) genes were the most frequently researched, present in 14, 13 and nine articles, respectively. For clarity and ease of interpretation in the present review, genes are categorised into three main groups: hormone genes, growth factor and cytokine genes and structural and metabolic genes.
3.5. Synthesis of Results
3.5.1. Hormone Genes
VDR
Nine studies analysed the association between VDR polymorphisms and muscle phenotypes. The first, conducted in 2004 by Roth et al. [67], highlighted a significant association between the rs2228570 (Fok1) polymorphism and FFM. Male FF homozygotes had significantly less FFM, appendicular fat-free mass (AFFM) and skeletal muscle index (SMI) compared to f allele carriers (p = 0.002, p = 0.009, p = 0.001 respectively). Furthermore, when classified as sarcopenic, FF carriers were at a two-fold higher risk of being sarcopenic when compared to carriers of the f allele (p = 0.03). Hopkinson et al. [45] also found significant interactions between the rs2228570 polymorphism and muscle phenotypes with male FF homozygotes displaying significantly lower knee extensor (KE) strength than f allele carriers (p = 0.007). Similarly, Xia et al. [74] found subjects carrying one or more F alleles to have significantly lower handgrip (HG) strength, and FFM (p = 0.03, p = 0.04 respectively). Furthermore, these individuals had a significantly higher risk of sarcopenia than ff homozygotes (p = 0.03). In contrast, a study conducted by Gussago et al. [42] found FF homozygotes to have significantly greater HG strength than f allele carriers (p = 0.021).
Significant associations were also identified between the rs1544410 (Bsm1) polymorphism and muscle performance phenotypes although, in keeping with the above findings, results were conflicting. In a study conducted by Onder et al. [62], bb homozygotes were significantly less likely to fall than carriers of the B allele (p = 0.02). Similarly, in 2010, Barr et al. [27] found females who were homozygous for the b allele to have a significantly lower risk of falling than Bb/BB carriers. These individuals also performed significantly better in the rise from chair and power tests when compared to carriers of B allele (p = 0.03, p = 0.044 respectively). Contrarily to the above studies, Bahat et al. [26] found KE strength to be significantly higher in BB homozygotes compared to carriers of one or more b alleles (p = 0.038).
Additional VDR polymorphisms rs7136534 and rs7975232 (Apa1) were significantly associated with fall incidence and HG strength respectively (p = 0.002, p < 0.05) [28,73]. No significant associations were found for the rs731236 (Taq1) polymorphism.
Other Genes
3.5.2. Growth Factor and Cytokine Genes
IGF1 and IGFBP3
The interaction between the Insulin-like Growth Factor 1 (IGF1) gene and muscle phenotypes was particularly evident in the intervention studies (Table 3). Both Kostek et al. [50] and Hand et al. [43] demonstrated that carriers of one or more 192 alleles achieved significantly greater KE strength improvements in response to resistance training (RT), compared to non-carriers (p = 0.02, p < 0.01). However, in a cross-sectional study conducted by Mora et al. [61], no significant differences were observed in muscle strength between carriers and non-carriers of the 192 allele (p = 0.024).
Significant associations were also noted for polymorphisms rs35767 of the IGF1 gene and rs2854744 of the Insulin-like Growth Factor Binding Protein 3 (IGFBP3) gene. Kostek et al. [51] observed black females carrying the CC genotype of the rs35767 polymorphism to have significantly less total FFM and muscle cross sectional area (CSA) than TT carriers (both p < 0.05). Furthermore, male CC homozygotes performed significantly worse in the single leg chair stand test than carriers of the T allele (p < 0.05). In a study conducted by Yang et al. [75], CC carriers of the rs2854744 polymorphism had a 4.3 times higher risk of having low SMI compared to AA carriers (p < 0.05).
CNTF and CNTFR
Two studies examined the Ciliary Neurotrophic Factor (CNTF) and Ciliary Neurotrophic Factor Receptor (CNTFR) genes (Table 2). In 2006, Arking et al. [25] observed five DNA polymorphisms (rs948562, rs1800169, rs550942, rs4319530, rs1938596) of the CNTF gene to be significantly associated with HG strength (p < 0.05). Further haplotype analysis revealed the null allele (A) of rs1800169 to fully explain this relationship under a recessive model. Individuals homozygous for the A allele had 3.8 kg lower HG strength than G allele carriers (p < 0.01). Interestingly, De Mars et al. [36] found only G/A carriers of the rs1800169 polymorphism to have significantly lower KE strength than G/G or A/A carriers (p = 0.0229). Additionally, male T allele carriers of the rs3808871 polymorphism produced significantly higher KE and knee flexor (KF) isometric torque at 120° when compared to CC homozygotes (p < 0.05). Furthermore, females who carried the T allele of the rs2070802 polymorphism produced greater KF concentric torques than the A/A homozygotes (p = 0.04).
TNFα
Three studies were included in this review which investigated the Tumour Necrosis Factor Alpha (TNFα) gene, each with significant findings. In 2013, Pereira et al. [65] observed that G allele homozygotes of the rs1800629 polymorphism achieved significantly faster timed up and go (TUG) test results in response to 10 weeks of RT compared to A allele carriers (p < 0.001). Additionally, Tiainen et al. [69] found the A allele of the rs361525 polymorphism to be associated with a significantly better physical performance level compared to GG homozygotes (p < 0.001). Finally, Li et al. [53] highlighted the interaction between the A allele of the rs1799964 polymorphism with either the G allele of the Tumour Necrosis Factor Beta (TNF-β) rs909253 polymorphism or the A allele of the TNF-β rs1041981 polymorphism to result in significantly lower handgrip strength among females (p = 0.005, p = 0.006 respectively).
Other Genes
Polymorphisms rs2276541 of the activin A type IIB receptor (ACVR2B) gene, rs3807987 of Caveolin 1 (CAV1) gene and rs1805086 of the Myostatin (MSTN) gene were all significantly associated with FFM (Table 2) [41,49,56].
3.5.3. Structural and Metabolic Genes
ACTN3 (The Sprint Gene)
In this review, fourteen studies were included which examined the association between the ACTN3 rs1815739 (R577X) polymorphism and skeletal muscle phenotypes. Carrying the X allele was often associated with lower baseline muscle strength and function (Table 2). For example, in a study conducted by Kikuchi et al. [48], homozygosity for the X allele was associated with significantly poorer performance in the chair stand test compared to RR carriers (p = 0.024). Ma et al. [58] also found XX homozygotes to perform significantly worse in HG strength (p = 0.012), 5 m walk (p = 0.011) and TUG (p = 0.039) tests and to also have a significantly higher frailty index (p = 0.004). Similar results were observed by Judson et al. [46] in a group of 4163 females where RX and XX genotypes were significantly associated with fall incidence (p = 0.049, p = 0.02 respectively). In contrast, Delmonico et al. [37] found female XX homozygotes to have significantly higher absolute and relative KE peak power and peak velocity than carriers of the R allele (p < 0.05).
Individuals carrying the XX genotype were also shown to have significantly lower improvements in one repetition maximum (1RM) bench press and leg extension, vertical jump and sit-to-stand performance in response to speed and power training when compared to RR carriers (all p < 0.05) [63]. Pereira et al. [64] also demonstrated XX carriers to have significantly poorer improvements in 10 m sprint times in response to high speed and power training compared to RR homozygotes (p = 0.044). Similarly, female XX carriers were observed to have significantly lower improvements in relative KE peak power following RT compared to RR homozygotes (p = 0.02) [37]. In the male population, change in absolute KE peak power post RT approached significance when comparing RR and XX genotypes (p = 0.07) [37]. In contrast to the above studies, Delmonico et al. [38] found male XX homozygotes had a significantly greater increase in 400 m walk time when compared to RX/RR carriers (p = 0.03).
In a study conducted by Zempo et al. [77] XX homozygotes were observed to have significantly lower thigh muscle CSA compared to RR carriers (p = 0.04). Interestingly, in a secondary analysis comparing a middle age group with an old age group, XX homozygosity was only associated with low thigh muscle CSA in the old age group (p < 0.05), suggesting that the influence of ACTN3 deficiency is heightened with age [78]. Similar results were noted in 2017 by Cho et al. [32], where sarcopenia prevalence was significantly associated with the XX genotype (p = 0.038). In contrast, Lima et al. [54] found X allele carriers to have significantly more relative total FFM than RR homozygotes (p = 0.04).
Three studies found no significant differences in muscle phenotypes between ACTN3 rs1815739 genotypes [30,39,59].
ACE
The relationship between the ACE rs1799752 (insertion/deletion) polymorphism and skeletal muscle traits has been extensively investigated since the original study of Montgomery et al. in 1998 [79]. Thirteen articles are included in this review. Firstly, Charbonneau et al. [31] found that carriers of the DD genotype had significantly greater total FFM (p < 0.05) and lower limb muscle volume (p = 0.01) than II homozygotes. Similarly, in a study of 246 Brazilian females, Lima et al. [54] noted DD homozygotes to have a significantly greater SMI than I allele carriers (p = 0.044). These findings were further strengthened by Da Silva et al. [34], who demonstrated sarcopenia prevalence to be significantly higher in II genotype carriers compared to D allele carriers (p = 0.015) (Table 2). Interestingly, Lima et al. [54] showed that in response to RT, only ACE II homozygotes significantly increased AFFM (p < 0.001).
The II genotype was also associated with lower muscle strength and functional performance. For example, within a group of 431 Japanese individuals, Yoshihara et al. [76] found II homozygosity to be associated with significantly lower HG strength compared to D allele carriers (p = 0.004). Homozygosity for the I allele was also shown to associate with significantly poorer performance in the 6-min walking test and 8 ft TUG test (p = 0.008, p < 0.001 respectively) when compared to ID/DD genotypes. Furthermore, in response to RT, DD carriers achieved significantly greater improvements in 1RM bench press and sit-to-stand performance (p = 0.019, p = 0.013 respectively) [63]. Giaccaglia et al. [40] also found that DD genotype carriers achieved significantly greater improvements in concentric KE strength in response to RT compared to II homozygotes (p < 0.05). Similarly, Pereira et al. [64] observed that DD homozygotes became significantly quicker performing 10 m sprints (p = 0.012) compared to II carriers. Buford et al. [29] also reported that a 12-month exercise intervention evoked significant improvements in 400 m walking speed (p = 0.018) and short physical performance battery test (SPPB) scores (p = 0.015), but only in D allele carriers. Interestingly, II homozygosity was also significantly associated with developing mobility limitation at a 45% faster rate when compared to ID/DD carriers (p = 0.01) [52].
As with the ACTN3 rs1815739 genotypes, three studies found rs1799752 genotypes to have no significant influence on skeletal muscle traits [30,39,59].
APOE
Three studies demonstrated significant associations between the Apolipoprotein E (APOE) gene and muscle phenotypes (Table 3). A 6-year follow-up study conducted by Melzer et al. [60] found that e4 carriers displayed significantly slower gait speed and chair stand performance (p = 0.006, p = 0.015 respectively) at baseline and significantly slower chair stand performance (p = 0.034) at the end of the 6-year follow-up, compared to e3 carriers. The APOE e4 allele was also shown to be associated with a significantly larger decline in HG strength between the ages of 75 and 79 over a 4-year period, compared to non-carriers (p = 0.015) [68]. Furthermore, carriers of the e4 allele had significantly lower HG strength at age 79 compared to non-carriers (p = 0.006). Interestingly, the effect of the e4 allele on HG strength was significantly larger at age 79 than age 75 (p = 0.033), suggesting that the e4 allele becomes increasingly influential with age. In a 3-year follow-up study conducted by Verghese et al. [71], males carrying the e4 allele showed a significantly more rapid decline in gait speed and greater risk of disability than male non-carriers (p = 0.04, p = 0.007 respectively).
UCP2 and UCP3
Three studies reported significant interactions between Uncoupling Proteins 2/3 (UCP2/3) polymorphisms and skeletal muscle traits. Firstly, in a group of 432 Caucasians, Crocco et al. [33] found carriers of the CC genotype of the UCP3 rs1800849 polymorphism to exhibit significantly lower HG strength than carriers of the T allele (p = 0.010). Dato et al. [35], then showed that individuals carrying the AA genotype of UCP3 rs11235972 polymorphism have significantly lower HG strength than GG homozygotes (p < 0.001). In 2015, Keogh et al. [47] demonstrated that GG carriers of UCP2 rs659366 polymorphism perform significantly worse in the 8 ft TUG test compared with AA/GA genotypes (p = 0.045). However, post RT intervention, GG homozygotes of UCP2 rs659366 had the greatest improvements in 8 ft TUG performance (p = 0.023).
Genome-wide Studies
Other genes that demonstrated significant associations with muscle phenotypes included the PR domain containing 16 (PRDM16) gene, Zinc finger protein 295 (ZNF295) gene and C2 calcium dependent domain containing 2 (C2CD2) gene (Table 2 and Table 3) [44,70].
Moreover, a recent GWAS by Hernandez-Cordero et al. [80] evaluated genetic contribution to ALM in the UK Biobank dataset, comparing middle-aged (38–49 years) and elderly (60–74 years) individuals. A total of 182 genome-wide significant regions, many with multiple variants within them, were associated with ALM in middle-aged individuals. Of these, 78% were also associated with ALM in elderly individuals. Variants at three genes, VCAM, ADAMTSL3 and FTO, had previously been associated with lean body mass in the UK Biobank [81]. Hernandez Cortez et al. also confirmed, in vitro, a functional role for CPNE1 and STC2 in myogenesis. In addition, the study highlighted five genomic regions, containing multiple genes, that are associated with muscle mass in both mice and humans.
4. Discussion
To the best of the authors’ knowledge, this is the first systematic review to collate literature on genetic associations with muscle phenotypes relevant to sarcopenia. To date, most research targeting genetic associations with muscle phenotypes has not focused on elderly subjects, and thus, the genetic mechanisms underpinning the age-related changes in skeletal muscle traits are largely uncharted.
Given that the deterioration of skeletal muscle with advancing age can have profound consequences for patients and public health systems, improving our understanding of how genes influence this process is of paramount importance. This review has enhanced our knowledge surrounding the key genes and gene variants that may prove crucial in further developing our understanding of the pathogenesis of sarcopenia and improving prognosis and treatment interventions alike.
4.1. Summary of Findings
The systematic literature search identified 24 genes and 46 DNA polymorphisms whose expression was significantly associated with muscle phenotypes in older adults. Ten of these DNA polymorphisms (rs154410, rs2228570, rs1800169, rs3093059, rs1800629, rs1815739, rs1799752, rs7412, rs429358 and 192 bp allele) were significantly associated with muscle phenotypes in two or more studies. The complex and multifactorial mechanisms underpinning muscle regulation suggest that the accrual and loss of muscle mass and muscle strength is not reducible to one single gene or gene variant. The dynamic interactions between inhibitory and promotory pathways within the human body further highlight the importance of a holistic approach when considering genetic associations with skeletal muscle traits.
Nevertheless, the findings of this systematic review demonstrate that the most compelling current evidence in the field exists for the ACTN3, ACE and VDR genotypes.
4.1.1. ACTN3 (The Sprint Gene)
The ACTN3 gene is among the most extensively researched genes in relation to muscle phenotypes, and appeared most frequently within this review. The ACTN3 protein encoded by the ACTN3 gene forms an integral part of the sarcomere Z-line in fast twitch muscle fibres and further aids in coordinating myofiber contractions [82,83]. Up to 20% of humans are deficient in this protein, due to homozygosity for the premature stop codon at the rs1815739 polymorphism [84]. This significant proportion of ACTN3 deficiency among the population suggests that X allele status is a key factor in variability in muscle phenotypes. In this regard, much of the research surrounding the ACTN3 genotype has focused on athletic performance [85]. Association studies have repeatedly found reduced X allele frequency among elite sprint/power athletes [85,86,87]. This suggests that the presence of ACTN3 is crucial for the optimal generation of force. Considering that fast twitch muscle fibres are particularly susceptible to age-related atrophy [88], it is plausible that regulation of this protein may also be an important factor in understanding age-related changes in muscle phenotypes. To date, however, limited research has been conducted within elderly populations, with the result that the true impact of the ACTN3 gene on age-related changes in muscle phenotypes remains inconclusive. Despite this, fourteen of the studies included in this review examining the ACTN3 genotype reported promising findings. Carriers of the X allele were often found to display lower skeletal muscle mass, strength and functional abilities. This was particularly evident among the Asian population. All five cross-sectional studies that examined Asian participants found significant associations between X allele status and muscle phenotypes [32,48,58,77]. No such association was found in the other three cross-sectional studies that targeted Caucasian individuals [30,39,59], therefore suggesting ethnicity may determine the degree to which ACTN3 genotypes effect aging muscle. This coincides with existing research whereby X allele frequency and fast twitch fibre composition have been shown to vary across different ethnic groups [89,90,91,92]. The Asian population have the highest frequency of the X allele [89], while having the lowest percentage of fast twitch muscle fibres [90,91,92], two likely contributing factors in the ethnic group having the highest sarcopenia prevalence globally [93]. Unlike above, X allele status was significantly associated with training adaptation within Caucasian, North-American and South-American individuals. Thus, the inconsistencies within this review highlight the need for future research to provide clarification on how ethnicity, ACTN3 genotypes and muscle phenotypes are associated within the elderly.
4.1.2. ACE
Like the ACTN3 gene, the ACE gene has been widely researched within athletic populations, and knowledge within older populations is limited. There are, however, compelling molecular pathways controlled by the ACE gene that suggest its importance in age-related changes in muscle phenotypes. The ACE is expressed by skeletal muscle endothelial cells, and catalyses the production of angiotensin II, known to enhance skeletal muscle hypertrophy [94,95]. To date, research in relation to muscle phenotypes has centred around the ACE rs1799752 polymorphism. The D and I alleles have been associated with higher and lower ACE activity respectively [96,97,98]. The D allele is suggested, therefore, to associate with greater muscle performance. To support this hypothesis, recent studies have focused on the rs1799752 polymorphism in elite athletes, with interesting findings. The I allele has been repeatedly associated with endurance performance, while the D allele associates with strength/power capabilities [99,100]. Findings from this systematic review further strengthen these observations. The D allele was consistently associated with higher baseline muscle strength and functional performance, as well as greater improvements in muscle strength and function in response to RT. Evidence of the association between the ACE rs1799752 polymorphism and muscle mass is less definitive. While the D allele was often associated with greater amounts of FFM, contradictory findings were also in evidence, and thus, further research is needed in this area to reach a consensus. Like with ACTN3 genotypes, frequency of the I and D allele of the ACE gene are highly determined by ethnic background. Asians have been shown to have the highest frequency for the undesirable I allele [101], while African-American have the lowest [101], aligning with global sarcopenia prevalence estimates where Asians and African-Americans have the highest and lowest risk respectively [93]. While evidence in this review is insufficient in highlighting a true ethnic impact on the association between ACE genotypes and aging muscle phenotypes, the disparity in allele frequency among different ethnicities is promising.
4.1.3. VDR
The true significance of the association between the VDR gene and muscle phenotypes is currently unknown. While the VDR gene has been extensively researched, findings are often contradictory. Furthermore, due to its crucial role in regulating calcium absorption, much of the existing research has focused on the association between VDR genotypes and bone health [102]. However, the VDR gene is also known to stimulate changes in muscle protein synthesis through its key regulatory role in the transcription of messenger RNA [103], and thus, the potential of the VDR gene as a candidate gene for muscle phenotype associations has been suggested. More specifically, the rs2228570 polymorphism is the only known VDR polymorphism where variation results in structural changes within the VDR protein due to differences in translational initiation sites [104]. The VDR f allele results in a full length VDR protein of 427 amino acids [105], while a VDR F allele results in a truncated VDR protein with three amino acids less [106]. Interestingly, three of four studies that examined the rs2228570 polymorphism in this review found F allele carriers to perform significantly worse across a range of muscle phenotypes [45,67,74], suggesting the potential importance of the rs2228570 polymorphism.
While compelling evidence exists supporting the importance of the VDR gene for muscle phenotypes, many studies have failed to replicate earlier results, and thus, the strength of this association remains to be established [107,108]. Unlike for ACTN3 and ACE polymorphisms, evidence of an ethnic influence on VDR polymorphism frequency is conflicting [109,110]. As with most genetic association studies, much of the research surrounding VDR polymorphisms and muscle phenotypes has been conducted using Caucasian subjects. Only nine articles examining VDR genotypes were included in this review, seven of which focused on Caucasian individuals [26,27,28,42,45,62,67]. Furthermore, as with the ACTN3 and ACE genes, limited research has been conducted within an elderly population, further limiting the transferability of findings for older adults.
4.1.4. Other Genes of Interest
Other genes with convincing molecular pathways and findings, that warrant future investigation include the IGF1/IGFBP3, TNFα, APOE, CNTF/R and UCP2/3 genes.
4.1.5. IGF1 and IGFBP3
The IGF family of genes encode peptides that are crucial in regulating cell proliferation, apoptosis and differentiation [111]. The mitogenic effect of IGF1 is integral to the facilitation of growth in multiple tissues, including skeletal muscle [112]. Considering that advancing age is associated with a decline in circulating IGF1 levels, the IGF1 gene is a likely candidate to effect muscle phenotypes among the elderly [113]. The current review found significant associations between IGF1 variants and skeletal muscle mass and strength. Associations were particularly convincing in longitudinal studies, suggesting that the IGF1 192 polymorphism may be particularly influential in the strength-training response of skeletal muscle phenotypes as opposed to baseline measurements.
The function of IGF1 is mediated through interactions with binding proteins, mainly, IGFBP3. Research has demonstrated that IGFBP3 is the most prolific potentiator of IGF1, therefore suggesting its importance in explaining inter-individual variation in muscle phenotypes [114]. While only Yang et al. [75] have investigated the impact of the IGFBP3 gene in an elderly population, the significant findings of that study combined with the relevant gene mechanisms warrants future research.
4.1.6. TNFα
Like the IGF family, the TNFα gene aids in the regulation of a multitude of biological processes such as cell proliferation, differentiation and apoptosis, and is thus an important candidate gene for aging skeletal muscle [115]. TNFα is also known to be an integral mediator of the inflammatory response to muscle damage [116]. Considering that inflammation is a vital response to RT in facilitating muscle regeneration, the TNFα gene is likely to affect the response of skeletal muscle tissue to RT [117]. This is supported by the findings of Pereira et al. [65] who observed that TNFα genotypes associate significantly with TUG performance adaptation. While Tiainen et al. [69] also highlighted significant cross-sectional associations, these were based on self-reported measures and should be interpreted with caution. Thus, longitudinal studies focusing on RT response of skeletal muscle may prove most beneficial in understanding the effect of TNFα genotypes on the aging muscle.
4.1.7. APOE
APOE protein encoded by the APOE gene, is involved in lipid metabolism and is a well-established risk factor for Alzheimer’s disease and various other aging disorders such as cardiovascular disease, atherosclerosis, stroke and impaired cognitive function [118]. Considering the associations between muscle phenotypes such as HG strength and these disorders, research has begun to investigate the relationship between the APOE gene and skeletal muscle traits. The gene has three common alleles (e2, e3 and e4), with e2 and e4 carriers having the lowest and highest risk of developing such aging disorders respectively [119]. As a result, much of the research in relation to skeletal muscle has centred around the e4 allele. The e4 allele was consistently associated with unfavourable skeletal muscle traits within this review, and therefore, supports the possibility of APOE as a candidate gene for explaining variation in muscle phenotypes with advancing age. Interestingly, like for ACTN3 and ACE genotypes, prevalence of the e4 allele is known to be highly varied among different populations [120]. With only three studies were included in this review, the effect of ethnicity on e4 allele frequency and the resulting association with muscle phenotypes is yet to be confirmed.
4.1.8. CNTF and CNTFR
The CNTF and CNTFR genes are both mediated through a common signal-transducing component, and thus are often examined in parallel [121]. CNTF, located in glial cells, aids in the promotion of motor neuron survival, and is therefore suggested to limit age-related atrophy of skeletal muscle caused by denervation [122]. The CNTFR is largely expressed in skeletal muscle, promoting research to examine the role of the CNTF and CNTFR genes in the regulation of muscle phenotypes [123]. To date, however, much of this research has been conducted using rats, with limited research being conducted with human populations. Thus, while the current review has highlighted some significant associations with muscle phenotypes, additional research is required to further understand the mechanisms underpinning this association in humans.
4.1.9. UCP2 and UCP3
Uncoupling proteins (UCPs) are mitochondrial transporters, best known for their involvement in thermogenesis and energy utilisation. As a result, UCPs are most commonly researched in relation to obesity-related phenotypes [124,125]. There is, however, evidence that suggests their importance in regulating muscle phenotypes. UCP2 and UCP3 have both been shown to effect skeletal muscle performance through the inhibition of mitochondrial ATP synthesis [126]. Additionally, UCP2 and UCP3 genes serve a key purpose in the protection of cells by attenuating mitochondrial reactive oxygen species (ROS) production, known to exert damaging effects on cells [127]. While loss of skeletal muscle mitochondrial content is known to occur with advancing age [128], evidence suggests UCPs are particularly active in the latter stages of life due to an increase in ROS and the associated rise in mitochondrial superoxide [129]. Therefore, UCP2 and UCP3 genes may affect how metabolic function of skeletal muscle is retained during the aging process. While the three studies included in this review found significant associations between UCP2 and UCP3 variants and muscle phenotypes, other data from human studies are scarce and as a result, the strength of this association remains to be elucidated.
4.2. Strengths and Limitations
This is the first systematic literature review to explore the genetic association with muscle phenotypes among the elderly. Only healthy subjects were included in the review, allowing for any association to be solely attributed to genotype-phenotype interactions rather than disease. All subjects were over the age of 50 years, ensuring relevance towards developing the understanding of the pathogenesis of sarcopenia. While some methodological weaknesses exist, most studies were well designed and conducted.
Findings within this review were at times conflicting. This incongruity may be partly explained by between-study disparities in methodological aspects such as sample size, subject characteristics and false-positive reporting. Furthermore, not all studies utilised the same measure for each muscle phenotype. For example, muscle strength measured through handgrip or leg extension may lead to different results. Evidently, there is a need for genetic association studies to implement more comprehensive and stringent methodology to maximise the potential of identifying genetic variants relevant to aging muscle phenotypes.
Finally, while not necessarily a limitation of this review itself, the overall lack of research currently available regarding the association between genetic variants and muscle phenotypes within the elderly prevents more definitive inferences to be made. As evidenced in this review, most research to date has focused on European populations, thus limiting the transferability of findings to other ethnic groups. Considering the promising ethnic differences in polymorphism frequency previously highlighted, future genetic studies may benefit from including individuals from a variety of ethnic backgrounds. The distinct lack of GWAS targeting aging muscle phenotypes is also contributive towards the uncertainty surrounding this area. A large body of research has utilised a candidate gene approach. Historically, many candidate gene studies have been statistically underpowered, the replication of findings has been problematic and there has been a suspected bias against publication of negative results, which may lead to conflicting findings [130]. Many of these issues have been overcome by GWAS in large, well characterised cohorts [80,131,132,133]. Therefore, future GWAS may help to further illuminate the genetic basis of aging muscle phenotypes.
5. Conclusions
The ability to maintain skeletal muscle mass, strength and function with advancing age is essential in preventing sarcopenia. Thus, the elucidation of the genetic variants associated with these phenotypes is of paramount importance. Evidently, skeletal muscle mass, strength and function are multifaceted characteristics that vary widely among the elderly. While heritability studies have highlighted that significant proportions of this inter-individual variability are determined by genetic factors, the specific genes involved remain mostly unknown.
The genetic association with muscle phenotypes is relatively under-researched, with only a limited number of candidate genes being explored to date. This review identified and systematically compiled the key genes shown to be significantly associated with muscle phenotypes within an elderly population. While relatively few genes have been identified which significantly contribute towards variation in muscle phenotypes, promising findings pointing to more extensive associations exist. Evidence is particularly supportive of the ACTN3, ACE and VDR genes, while the IGF1/IGFBP3, TNFα, APOE, CNTF/R and UCP2/3 genes have also been shown to be significantly associated with skeletal muscle phenotypes in two or more studies.
To conclude, the findings from this review helped to further illuminate the genetic basis of sarcopenia. While the molecular genetic pathways are often compelling, the limited volume of research within this field is as yet insufficient to demonstrate a clear genetic basis for sarcopenia. Future GWAS could facilitate the identification of novel genetic variants that may have key regulatory roles in aging muscle phenotypes. Further still, a more extensive exploration of the candidate genes highlighted in this review should provide further insight into the pathogenesis of sarcopenia and further aid in the development of effective prognosis, preventive and treatment protocols to combat the profound consequences of sarcopenia for patients and health systems worldwide.
Author Contributions
Conceptualization, J.P.; Literature search and validation, J.P., G.D.V. and C.B.; Analysis, J.P.; Writing—original draft preparation, J.P.; Writing—review and editing, J.P., G.D.V., C.B., S.E. and A.W.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Rosenburg, I.H. Sarcopenia: Origins and clinical relevance. J. Nutr. 1997, 127, 990S–S991S. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Jentoft, A.J.; Bahat, G.; Bauer, J.; Boirie, Y.; Bruyère, O.; Cederholm, T.; Cooper, C.; Landi, F.; Rolland, Y.; Sayer, A.A.; et al. Sarcopenia: Revised European consensus on definition and diagnosis. Age Ageing 2019, 48, 16–31. [Google Scholar] [CrossRef] [PubMed]
- Chin, S.O.; Rhee, S.Y.; Chon, S.; Hwang, Y.C.; Jeong, I.K.; Oh, S.; Ahn, K.J.; Chung, H.Y.; Woo, J.T.; Kim, S.W.; et al. Sarcopenia Is independently associated with cardiovascular disease in older Korean adults: The Korea national health and nutrition examination survey (KNHANES) from 2009. PLoS ONE 2013, 8, e60119. [Google Scholar] [CrossRef] [PubMed]
- Janssen, I.; Heymsfield, S.B.; Ross, R. Low relative skeletal muscle mass (sarcopenia) in older persons is associated with functional impairment and physical disability. J. Am. Geriatr. Soc. 2002, 50, 889–896. [Google Scholar] [CrossRef] [PubMed]
- Landi, F.; Liperoti, R.; Russo, A.; Giovannini, S.; Tosato, M.; Capoluongo, E. Sarcopenia as a risk factor for falls in elderly individuals: Results from the ilSIRENTE study. Clin. Nutr. 2012, 31, 652–658. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, W.; Wang, C.; Tao, W.; Dou, Q.; Yang, Y. Sarcopenia as a predictor of hospitalization among older people: A systematic review and meta-analysis. BMC Geriatr. 2018, 18, 188. [Google Scholar] [CrossRef]
- Kim, T.N.; Choi, K.M. The implications of sarcopenia and sarcopenic obesity on cardiometabolic disease. J. Cell Biochem. 2015, 116, 1171–1178. [Google Scholar] [CrossRef]
- Brown, J.C.; Harhay, M.O.; Harhay, M.N. Sarcopenia and mortality among a population based sample of community-dwelling older adults. J. Cachexia Sarcopenia Muscle 2016, 7, 290–298. [Google Scholar] [CrossRef]
- Shafiee, G.; Keshtkar, A.; Soltani, A.; Ahadi, Z.; Larijani, B.; Heshmat, R. Prevalence of sarcopenia in the world: A systematic review and meta- analysis of general population studies. J. Diabetes Metab. Disord. 2017, 16, 16–21. [Google Scholar] [CrossRef]
- Melton, L.J.; Khosla, S.; Crowson, C.S.; O’Connor, M.K.; O’Fallon, W.M.; Riggs, B.L. Epidemiology of sarcopenia. J. Am. Geriatr. Soc. 2000, 48, 625–630. [Google Scholar] [CrossRef]
- Ethgen, O.; Beaudart, C.; Buckinx, F.; Bruyère, O.; Reginster, J.Y. The future prevalence of sarcopenia in Europe: A claim for public health action. Calcif. Tissue Int. 2017, 100, 229–234. [Google Scholar] [CrossRef] [PubMed]
- Keller, K.; Engelhardt, M. Strength and muscle mass loss with aging process. Age and strength loss. Muscles Ligaments Tendons J. 2014, 3, 346–350. [Google Scholar] [CrossRef] [PubMed]
- Deschenes, M.R. Effects of aging on muscle fibre type and size. Sports Med. 2004, 34, 809–824. [Google Scholar] [CrossRef] [PubMed]
- Doherty, T.J. The influence of aging and sex on skeletal muscle mass and strength. Curr. Opin. Clin. Nutr. Metab. Care 2001, 4, 503–508. [Google Scholar] [CrossRef]
- Prior, S.J.; Roth, S.M.; Wang, X.; Kammerer, C.; Miljkovic-Gacic, I.; Bunker, C.H.; Wheeler, V.W.; Patrick, A.L.; Zmuda, J.M. Genetic and environmental influences on skeletal muscle phenotypes as a function of age and sex in large, multigenerational families of African heritage. J. Appl. Physiol. 2007, 103, 1121–1127. [Google Scholar] [CrossRef]
- Kemp, G.J.; Birrell, F.; Clegg, P.D.; Cuthbertson, D.J.; De Vito, G.; Van Dieën, J.H.; Del Din, S.; Eastell, R.; Garnero, P.; Goljanek–Whysall, K.; et al. Developing a toolkit for the assessment and monitoring of musculoskeletal ageing. Age Ageing 2018, 47, 1–19. [Google Scholar] [CrossRef]
- Franzke, B.; Neubauer, O.; Cameron-Smith, D.; Wagner, K.H. Dietary protein, muscle and physical function in the very old. Nutrients 2018, 10, 935. [Google Scholar] [CrossRef]
- Buchmann, N.; Spira, D.; Norman, K.; Demuth, I.; Eckardt, R.; Steinhagen-Thiessen, E. Sleep, muscle mass and muscle function in older people. Dtsch. Arzteblatt Int. 2016, 113, 253–260. [Google Scholar] [CrossRef]
- Yoo, J.I.; Ha, Y.C.; Lee, Y.K.; Hana-Choi; Yoo, M.J.; Koo, K.H. High prevalence of sarcopenia among binge drinking elderly women: A nationwide population-based study. BMC Geriatr. 2017, 17, 114. [Google Scholar] [CrossRef]
- Abney, M.; McPeek, M.S.; Ober, C. Broad and narrow heritabilities of quantitative traits in a founder population. Am. J. Hum. Genet. 2001, 68, 1302–1307. [Google Scholar] [CrossRef]
- Zempo, H.; Miyamoto-Mikami, E.; Kikuchi, N.; Fuku, N.; Miyachi, M.; Murakami, H. Heritability estimates of muscle strength-related phenotypes: A systematic review and meta-analysis. Scand. J. Med. Sci. Sports 2017, 27, 1537–1546. [Google Scholar] [CrossRef] [PubMed]
- Zhai, G.; Ding, C.; Stankovich, J.; Cicuttini, F.; Jones, G. The genetic contribution to longitudinal changes in knee structure and muscle strength: A sibpair study. Arthritis Rheum. 2005, 52, 2830–2834. [Google Scholar] [CrossRef] [PubMed]
- Liberati, A.; Altman, D.; Tetzlaff, J.; Mulrow, C.; Gøtzsche, P.; Ioannidis, J.; Clarke, M.; Devereaux, P.; Kleijnen, J.; Moher, D. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: Explanation and elaboration. BMJ 2009, 339, b2700. [Google Scholar] [CrossRef] [PubMed]
- Sohani, Z.N.; Meyre, D.; de Souza, R.J.; Joseph, P.G.; Gandhi, M.; Dennis, B.B.; Norman, G.; Anand, S.S. Assessing the quality of published genetic association studies in meta-analyses: The quality of genetic studies (Q-Genie) tool. BMC Genet. 2015, 16, 50. [Google Scholar] [CrossRef] [PubMed]
- Arking, D.E.; Fallin, D.M.; Fried, L.P.; Li, T.; Beamer, B.A.; Xue, Q.L.; Chakravarti, A.; Walston, J. Variation in the ciliary neurotrophic factor gene and muscle strength in older Caucasian women. J. Am. Geriatr. Soc. 2006, 54, 823–826. [Google Scholar] [CrossRef]
- Bahat, G.; Saka, B.; Erten, N.; Ozbek, U.; Coskunpinar, E.; Yildiz, S.; Sahinkaya, T.; Karan, M.A. BsmI polymorphism in the vitamin D receptor gene is associated with leg extensor muscle strength in elderly men. Aging Clin. Exp. Res. 2010, 22, 198–205. [Google Scholar] [CrossRef]
- Barr, R.; Macdonald, H.; Stewart, A.; McGuigan, F.; Rogers, A.; Eastell, R.; Felsenberg, D.; Glüer, C.; Roux, C.; Reid, D.M. Association between vitamin D receptor gene polymorphisms, falls, balance and muscle power: Results from two independent studies (APOSS and OPUS). Osteoporos. Int. 2010, 21, 457–466. [Google Scholar] [CrossRef]
- Björk, A.; Ribom, E.; Johansson, G.; Scragg, R.; Mellström, D.; Grundberg, E.; Ohlsson, C.; Karlsson, M.; Ljunggren, Ö.; Kindmark, A. Variations in the vitamin D receptor gene are not associated with measures of muscle strength, physical performance, or falls in elderly men. Data from MrOS Sweden. J. Steroid Biochem. Mol. Biol. 2019, 187, 160–165. [Google Scholar] [CrossRef]
- Buford, T.W.; Hsu, F.C.; Brinkley, T.E.; Carter, C.S.; Church, T.S.; Dodson, J.A.; Goodpaster, B.H.; McDermott, M.M.; Nicklas, B.J.; Yank, V.; et al. Genetic influence on exercise-induced changes in physical function among mobility-limited older adults. Physiol. Genomics 2014, 46, 149–158. [Google Scholar] [CrossRef]
- Bustamante-Ara, N.; Santiago, C.; Verde, Z.; Yvert, T.; Gómez-Gallego, F.; Rodríguez-Romo, G.; González-Gil, P.; Serra-Rexach, J.A.; Ruiz, J.R.; Lucia, A. ACE and ACTN3 genes and muscle phenotypes in nonagenarians. Int. J. Sports Med. 2010, 31, 221–224. [Google Scholar] [CrossRef]
- Charbonneau, D.E.; Hanson, E.D.; Ludlow, A.T.; Delmonico, M.J.; Hurley, B.F.; Roth, S.M. ACE genotype and the muscle hypertrophic and strength responses to strength training. Med. Sci. Sports Exerc. 2008, 40, 677–683. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.; Lee, I.; Kang, H. ACTN3 Gene and susceptibility to sarcopenia and osteoporotic status in older Korean adults. Biomed. Res. Int. 2017, 2017, 4239648. [Google Scholar] [CrossRef] [PubMed]
- Crocco, P.; Montesanto, A.; Passarino, G.; Rose, G. A common polymorphism in the UCP3 promoter influences hand grip strength in elderly people. Biogerontology 2011, 12, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, J.R.D.; Freire, I.V.; Ribeiro, Í.J.; dos Santos, C.S.; Casotti, C.A.; dos Santos, D.B.; Barbosa, A.A.L.; Pereira, R. Improving the comprehension of sarcopenic state determinants: An multivariate approach involving hormonal, nutritional, lifestyle and genetic variables. Mech. Ageing Dev. 2018, 173, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Dato, S.; Soerensen, M.; Montesanto, A.; Lagani, V.; Passarino, G.; Christensen, K.; Christiansen, L. UCP3 polymorphisms, hand grip performance and survival at old age: Association analysis in two Danish middle aged and elderly cohorts. Mech. Ageing Dev. 2012, 133, 530–537. [Google Scholar] [CrossRef]
- De Mars, G.; Windelinckx, A.; Beunen, G.; Delecluse, C.; Lefevre, J.; Thomis, M.A. Polymorphisms in the CNTF and CNTF receptor genes are associated with muscle strength in men and women. J. Appl. Physiol. 2007, 102, 1824–1831. [Google Scholar] [CrossRef]
- Delmonico, M.J.; Kostek, M.C.; Doldo, N.A.; Hand, B.D.; Walsh, S.; Conway, J.M.; Carignan, C.R.; Roth, S.M.; Hurley, B.F. Alpha-actinin-3 (ACTN3) R577X polymorphism influences knee extensor peak power response to strength training in older men and women. J. Gerontol. A Biol. Sci. Med. Sci. 2007, 62, 206–212. [Google Scholar] [CrossRef]
- Delmonico, M.J.; Zmuda, J.M.; Taylor, B.C.; Cauley, J.A.; Harris, T.B.; Manini, T.M.; Schwartz, A.; Li, R.; Roth, S.M.; Hurley, B.F.; et al. Association of the ACTN3 genotype and physical functioning with age in older adults. J. Gerontol. A Biol. Sci. Med. Sci. 2008, 63, 1227–1234. [Google Scholar] [CrossRef]
- Garatachea, N.; Fiuza-Luces, C.; Torres-Luque, G.; Yvert, T.; Santiago, C.; Gómez-Gallego, F.; Ruiz, J.R.; Lucia, A. Single and combined influence of ACE and ACTN3 genotypes on muscle phenotypes in octogenarians. Eur. J. Appl. Physiol. 2012, 112, 2409–2420. [Google Scholar] [CrossRef]
- Giaccaglia, V.; Nicklas, B.; Kritchevsky, S.; Mychalecky, J.; Messier, S.; Bleecker, E.; Pahor, M. Interaction between angiotensin converting enzyme insertion/deletion genotype and exercise training on knee extensor strength in older individuals. Int. J. Sports Med. 2008, 29, 40–44. [Google Scholar] [CrossRef]
- González-Freire, M.; Rodríguez-Romo, G.; Santiago, C.; Bustamante-Ara, N.; Yvert, T.; Gómez-Gallego, F.; Rexach, J.A.S.; Ruiz, J.R.; Lucia, A. The K153R variant in the myostatin gene and sarcopenia at the end of the human lifespan. Age 2010, 32, 405–409. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gussago, C.; Arosio, B.; Guerini, F.R.; Ferri, E.; Costa, A.S.; Casati, M.; Bollini, E.M.; Ronchetti, F.; Colombo, E.; Bernardelli, G.; et al. Impact of vitamin D receptor polymorphisms in centenarians. Endocrine 2016, 53, 558–564. [Google Scholar] [CrossRef] [PubMed]
- Hand, B.D.; Kostek, M.C.; Ferrell, R.E.; Delmonico, M.J.; Douglass, L.W.; Roth, S.M.; Hagberg, J.M.; Hurley, B.F. Influence of promoter region variants of insulin-like growth factor pathway genes on the strength-training response of muscle phenotypes in older adults. J. Appl. Physiol. 2007, 103, 1678–1687. [Google Scholar] [CrossRef] [PubMed]
- Heckerman, D.; Traynor, B.J.; Picca, A.; Calvani, R.; Marzetti, E.; Hernandez, D.; Nalls, M.; Arepali, S.; Ferrucci, L.; Landi, F. Genetic variants associated with physical performance and anthropometry in old age: A genome-wide association study in the ilSIRENTE cohort. Sci. Rep. 2017, 7, 15879. [Google Scholar] [CrossRef] [PubMed]
- Hopkinson, N.S.; Li, K.W.; Kehoe, A.; Humphries, S.E.; Roughton, M.; Moxham, J.; Montgomery, H.; Polkey, M.I. Vitamin D receptor genotypes influence quadriceps strength in chronic obstructive pulmonary disease. Am. J. Clin. Nutr. 2008, 87, 385–390. [Google Scholar] [CrossRef]
- Judson, R.N.; Wackerhage, H.; Hughes, A.; Mavroeidi, A.; Barr, R.J.; Macdonald, H.M.; Ratkevicius, A.; Reid, D.M.; Hocking, L.J. The functional ACTN3 577X variant increases the risk of falling in older females: Results from two large independent cohort studies. J. Gerontol. A Biol. Sci. Med. Sci. 2011, 66, 130–135. [Google Scholar] [CrossRef]
- Keogh, J.W.L.; Palmer, B.R.; Taylor, D.; Kilding, A.E. ACE and UCP2 gene polymorphisms and their association with baseline and exercise-related changes in the functional performance of older adults. PeerJ 2015, 3, e980. [Google Scholar] [CrossRef]
- Kikuchi, N.; Yoshida, S.; Min, S.K.; Lee, K.; Sakamaki-Sunaga, M.; Okamoto, T.; Nakazato, K. The ACTN3 R577X genotype is associated with muscle function in a Japanese population. Appl. Physiol. Nutr. Metab. 2015, 40, 316–322. [Google Scholar] [CrossRef]
- Klimentidis, Y.C.; Bea, J.W.; Thompson, P.; Klimecki, W.T.; Hu, C.; Wu, G.; Nicholas, S.; Ryckman, K.K.; Chen, Z. Genetic variant in ACVR2B is associated with lean mass. Med. Sci. Sports Exerc. 2016, 48, 1270–1275. [Google Scholar] [CrossRef][Green Version]
- Kostek, M.C.; Delmonico, M.J.; Reichel, J.B.; Roth, S.M.; Douglass, L.; Ferrell, R.E.; Hurley, B.F. Muscle strength response to strength training is influenced by insulin-like growth factor 1 genotype in older adults. J. Appl. Physiol. 2005, 98, 2147–2154. [Google Scholar] [CrossRef]
- Kostek, M.C.; Devaney, J.M.; Gordish-Dressman, H.; Harris, T.B.; Thompson, P.D.; Clarkson, P.M.; Angelopoulos, T.J.; Gordon, P.M.; Moyna, N.M.; Pescatello, L.S.; et al. A polymorphism near IGF1 is associated with body composition and muscle function in women from the Health, Aging, and Body Composition Study. Eur. J. Appl. Physiol. 2010, 110, 315–324. [Google Scholar] [CrossRef] [PubMed]
- Kritchevsky, S.B.; Nicklas, B.J.; Visser, M.; Simonsick, E.M.; Newman, A.B.; Harris, T.B.; Lange, E.M.; Penninx, B.W.; Goodpaster, B.H.; Satterfield, S.; et al. Angiotensin-converting enzyme insertion/deletion genotype, exercise, and physical decline. Jama 2005, 294, 691–698. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, C.I.; Li, T.C.; Liao, L.N.; Liu, C.S.; Yang, C.W.; Lin, C.H.; Hsiao, J.H.; Meng, N.H.; Lin, W.Y.; Wu, F.Y.; et al. Joint effect of gene-physical activity and the interactions among CRP, TNF-alpha, and LTA polymorphisms on serum CRP, TNF-alpha levels, and handgrip strength in community-dwelling elders in Taiwan-TCHS-E. Age 2016, 38, 46. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lima, R.M.; Leite, T.K.; Pereira, R.W.; Rabelo, H.T.; Roth, S.M.; Oliveira, R.J. ACE and ACTN3 genotypes in older women: Muscular phenotypes. Int. J. Sports Med. 2011, 32, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.C.; Wu, F.Y.; Liao, L.N.; Li, C.I.; Lin, C.H.; Yang, C.W.; Meng, N.H.; Chang, C.K.; Lin, W.Y.; Liu, C.S.; et al. Association of CRP gene polymorphisms with serum CRP level and handgrip strength in community-dwelling elders in Taiwan: Taichung Community Health Study for Elders (TCHS-E). Exp. Gerontol. 2014, 57, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.H.; Lin, C.C.; Tsai, C.W.; Chang, W.S.; Yang, M.D.; Bau, D.T. A novel caveolin-1 biomarker for clinical outcome of sarcopenia. In Vivo 2014, 28, 383–389. [Google Scholar]
- Lunardi, C.C.; Lima, R.M.; Pereira, R.W.; Leite, T.K.; Siqueira, A.B.; Oliveira, R.J. Association between polymorphisms in the TRHR gene, fat-free mass, and muscle strength in older women. Age 2013, 35, 2477–2483. [Google Scholar] [CrossRef]
- Ma, T.; Lu, D.; Zhu, Y.S.; Chu, X.F.; Wang, Y.; Shi, G.P.; Wang, Z.D.; Yu, L.; Jiang, X.Y.; Wang, X.F. ACTN3 genotype and physical function and frailty in an elderly Chinese population: The rugao longevity and ageing study. Age Ageing 2018, 47, 416–422. [Google Scholar] [CrossRef]
- McCauley, T.; Mastana, S.S.; Folland, J.P. ACE I/D and ACTN3 R/X polymorphisms and muscle function and muscularity of older Caucasian men. Eur. J. Appl. Physiol. 2010, 109, 269–277. [Google Scholar] [CrossRef]
- Melzer, D.; Dik, M.G.; van Kamp, G.J.; Jonker, C.; Deeg, D.J. The apolipoprotein E e4 polymorphism is strongly associated with poor mobility performance test results but not self-reported limitation in older people. J. Gerontol. A Biol. Sci. Med. Sci. 2005, 60, 1319–1323. [Google Scholar] [CrossRef]
- Mora, M.; Perales, M.J.; Serra-Prat, M.; Palomera, E.; Buquet, X.; Oriola, J.; Puig-Domingo, M.; Mataró Ageing Study Group. Aging phenotype and its relationship with IGF-I gene promoter polymorphisms in elderly people living in Catalonia. Growth Horm. IGF Res. 2011, 21, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Onder, G.; Capoluongo, E.; Danese, P.; Settanni, S.; Russo, A.; Concolino, P.; Bernabei, R.; Landi, F. Vitamin D receptor polymorphisms and falls among older adults living in the community: Results from the ilSIRENTE study. J. Bone Miner. Res. 2008, 23, 1031–1036. [Google Scholar] [CrossRef] [PubMed]
- Pereira, A.; Costa, A.M.; Izquierdo, M.; Silva, A.J.; Bastos, E.; Marques, M.C. ACE I/D and ACTN3 R/X polymorphisms as potential factors in modulating exercise-related phenotypes in older women in response to a muscle power training stimuli. Age 2013, 35, 1949–1959. [Google Scholar] [CrossRef] [PubMed]
- Pereira, A.; Costa, A.M.; Leitão, J.C.; Monteiro, A.M.; Izquierdo, M.; Silva, A.J.; Bastos, E.; Marques, M.C. The influence of ACE ID and ACTN3 R577X polymorphisms on lower-extremity function in older women in response to high-speed power training. BMC Geriatr. 2013, 13, 131. [Google Scholar] [CrossRef]
- Pereira, D.S.; Mateo, E.C.C.; de Queiroz, B.Z.; Assumpção, A.M.; Miranda, A.S.; Felício, D.C.; Rocha, N.P.; dos Anjos, D.M.D.C.; Pereira, D.A.G.; Teixeira, A.L.; et al. TNF-alpha, IL6, and IL10 polymorphisms and the effect of physical exercise on inflammatory parameters and physical performance in elderly women. Age 2013, 35, 2455–2463. [Google Scholar] [CrossRef]
- Prakash, J.; Herlin, M.; Kumar, J.; Garg, G.; Akesson, K.E.; Grabowski, P.S.; Skerry, T.M.; Richards, G.O.; McGuigan, F.E. Analysis of RAMP3 gene polymorphism with body composition and bone density in young and elderly women. Gene X 2019, 2, 100009. [Google Scholar] [CrossRef]
- Roth, S.M.; Zmuda, J.M.; Cauley, J.A.; Shea, P.R.; Ferrell, R.E. Vitamin D receptor genotype is associated with fat-free mass and sarcopenia in elderly men. J. Gerontol. A Biol. Sci. Med. Sci. 2004, 59, 10–15. [Google Scholar] [CrossRef]
- Skoog, I.; Hörder, H.; Frändin, K.; Johansson, L.; Östling, S.; Blennow, K.; Zetterberg, H.; Zettergren, A. Association between APOE genotype and change in physical function in a population-based swedish cohort of older individuals followed over four years. Front. Aging Neurosci. 2016, 8, 225. [Google Scholar] [CrossRef]
- Tiainen, K.; Thinggaard, M.; Jylha, M.; Bladbjerg, E.; Christensen, K.; Christiansen, L. Associations between inflammatory markers, candidate polymorphisms and physical performance in older Danish twins. Exp. Gerontol. 2012, 47, 109–115. [Google Scholar] [CrossRef]
- Urano, T.; Shiraki, M.; Sasaki, N.; Ouchi, Y.; Inoue, S. Large-scale analysis reveals a functional single-nucleotide polymorphism in the 5’-flanking region of PRDM16 gene associated with lean body mass. Aging Cell 2014, 13, 739–743. [Google Scholar] [CrossRef]
- Verghese, J.; Holtzer, R.; Wang, C.; Katz, M.J.; Barzilai, N.; Lipton, R.B. Role of APOE genotype in gait decline and disability in aging. J. Gerontol. A Biol. Sci. Med. Sci. 2013, 68, 1395–1401. [Google Scholar] [CrossRef] [PubMed]
- Walsh, S.; Zmuda, J.M.; Cauley, J.A.; Shea, P.R.; Metter, E.J.; Hurley, B.F.; Ferrell, R.E.; Roth, S.M. Androgen receptor CAG repeat polymorphism is associated with fat-free mass in men. J. Appl. Physiol. 2005, 98, 132–137. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.Y.; Liu, C.S.; Liao, L.N.; Li, C.I.; Lin, C.H.; Yang, C.W.; Meng, N.H.; Lin, W.Y.; Chang, C.K.; Hsiao, J.H.; et al. Vitamin D receptor variability and physical activity are jointly associated with low handgrip strength and osteoporosis in community-dwelling elderly people in Taiwan: The Taichung Community Health Study for Elders (TCHS-E). Osteoporos. Int. 2014, 25, 1917–1929. [Google Scholar] [CrossRef] [PubMed]
- Xia, Z.; Man, Q.; Li, L.; Song, P.; Jia, S.; Song, S.; Meng, L.; Zhang, J. Vitamin D receptor gene polymorphisms modify the association of serum 25-hydroxyvitamin D levels with handgrip strength in the elderly in Northern China. Nutrition 2019, 57, 202–207. [Google Scholar] [CrossRef]
- Yang, C.W.; Li, T.C.; Li, C.I.; Liu, C.S.; Lin, C.H.; Lin, W.Y.; Lin, C.C. Insulin like growth factor-1 and its binding protein-3 polymorphisms predict circulating IGF-1 level and appendicular skeletal muscle mass in Chinese elderly. J. Am. Med. Dir. Assoc. 2015, 16, 365–370. [Google Scholar] [CrossRef]
- Yoshihara, A.; Tobina, T.; Yamaga, T.; Ayabe, M.; Yoshitake, Y.; Kimura, Y.; Shimada, M.; Nishimuta, M.; Nakagawa, N.; Ohashi, M.; et al. Physical function is weakly associated with angiotensin-converting enzyme gene I/D polymorphism in elderly Japanese subjects. Gerontology 2009, 55, 387–392. [Google Scholar] [CrossRef]
- Zempo, H.; Tanabe, K.; Murakami, H.; Iemitsu, M.; Maeda, S.; Kuno, S. ACTN3 polymorphism affects thigh muscle area. Int. J. Sports Med. 2010, 31, 138–142. [Google Scholar] [CrossRef]
- Zempo, H.; Tanabe, K.; Murakami, H.; Iemitsu, M.; Maeda, S.; Kuno, S. Age differences in the relation between ACTN3 R577X polymorphism and thigh-muscle cross-sectional area in women. Genet. Test. Mol. Biomark. 2011, 15, 639–643. [Google Scholar] [CrossRef]
- Montgomery, H.E.; Marshall, R.; Hemingway, H.; Myerson, S.; Clarkson, P.; Dollery, C.; Hayward, M.; Holliman, D.E.; Jubb, M.; World, M.; et al. Human gene for physical performance. Nature 1998, 393, 221–222. [Google Scholar] [CrossRef]
- Cordero, A.I.H.; Gonzales, N.M.; Parker, C.C.; Sokoloff, G.; Vandenbergh, D.J.; Cheng, R.; Abney, M.; Skol, A.; Douglas, A.; Palmer, A.A.; et al. Genome-wide associations reveal human-mouse genetic convergence and modifiers of myogenesis, CPNE1 and STC2. Am. J. Hum. Genet. 2019, 105, 1222–1236. [Google Scholar] [CrossRef]
- Zillikens, M.C.; Demissie, S.; Hsu, Y.H.; Yerges-Armstrong, L.M.; Chou, W.C.; Stolk, L.; Livshits, G.; Broer, L.; Johnson, T.; Koller, D.L.; et al. Large meta-analysis of genome-wide association studies identifies five loci for lean body mass. Nat. Commun. 2017, 8, 80. [Google Scholar] [CrossRef] [PubMed]
- Houweling, P.J.; North, K.N. Sarcomeric α-actinins and their role in human muscle disease. Future Neurol. 2009, 4, 731–741. [Google Scholar] [CrossRef]
- Blanchard, A.; Ohanian, V.; Critchley, D. The structure and function of α-actinin. J. Muscle Res. Cell Motil. 1989, 10, 280–289. [Google Scholar] [CrossRef] [PubMed]
- North, K.N.; Yang, N.; Wattanasirichaigoon, D.; Mills, M.; Easteal, S.; Beggs, A.H. A common nonsense mutation results in alpha-actinin-3 deficiency in the general population. Nat. Genet. 1999, 21, 353–354. [Google Scholar] [CrossRef]
- Yang, N.; MacArthur, D.G.; Gulbin, J.P.; Hahn, A.G.; Beggs, A.H.; Easteal, S.; North, K. ACTN3 genotype is associated with human elite athletic performance. Am. J. Hum. Genet. 2003, 73, 627–631. [Google Scholar] [CrossRef]
- Druzhevskaya, A.M.; Ahmetov, I.I.; Astratenkova, I.V.; Rogozkin, V.A. Association of the ACTN3 R577X polymorphism with power athlete status in Russians. Eur. J. Appl. Physiol. 2008, 103, 631–634. [Google Scholar] [CrossRef]
- Papadimitriou, I.D.; Lucia, A.; Pitsiladis, Y.P.; Pushkarev, V.P.; Dyatlov, D.A.; Orekhov, E.F.; Artioli, G.G.; Guilherme, J.P.L.; Lancha, A.H.; Ginevičienė, V.; et al. ACTN3 R577X and ACE I/D gene variants influence performance in elite sprinters: A multi-cohort study. BMC Genomics 2016, 17, 285. [Google Scholar] [CrossRef]
- Tieland, M.; Trouwborst, I.; Clark, B.C. Skeletal muscle performance and ageing. J. Cachexia Sarcopenia Muscle 2018, 9, 3–19. [Google Scholar] [CrossRef]
- Pickering, C.; Kiely, J. ACTN3: More than just a gene for speed. Front. Physiol. 2017, 8, 1080. [Google Scholar] [CrossRef]
- Kumagai, H.; Tobina, T.; Ichinoseki-Sekine, N.; Kakigi, R.; Tsuzuki, T.; Zempo, H.; Shiose, K.; Yoshimura, E.; Kumahara, H.; Ayabe, M.; et al. Role of selected polymorphisms in determining muscle fiber composition in Japanese men and women. J. Appl. Physiol. 2018, 124, 1377–1384. [Google Scholar] [CrossRef]
- Nielsen, J.; Christensen, D.L. Glucose intolerance in the West African Diaspora: A skeletal muscle fibre type distribution hypothesis. Acta Physiol. 2011, 202, 605–616. [Google Scholar] [CrossRef] [PubMed]
- Ama, P.F.; Simoneau, J.A.; Boulay, M.R.; Serresse, O.; Thériault, G.; Bouchard, C. Skeletal muscle characteristics in sedentary black and Caucasian males. J. Appl. Physiol. 1986, 61, 1758–1761. [Google Scholar] [CrossRef] [PubMed]
- Jeng, C.; Zhao, L.; Wu, K.; Zhou, Y.; Chen, T.; Deng, H.W. Race and socioeconomic effect on sarcopenia and sarcopenic obesity in the Louisiana Osteoporosis Study (LOS). JCSM Clin. Rep. 2018, 3, e00027. [Google Scholar] [CrossRef] [PubMed]
- Rigat, B.; Hubert, C.; Alhenc-Gelas, F.; Cambien, F.; Corvol, P.; Soubrier, F. An insertion/deletion polymorphism in the angiotensin I-converting enzyme gene accounting for half the variance of serum enzyme levels. J. Clin. Invest. 1990, 86, 1343–1346. [Google Scholar] [CrossRef]
- Gordon, S.E.; Davis, B.S.; Carlson, C.J.; Booth, F.W. ANG II is required for optimal overload-induced skeletal muscle hypertrophy. Am. J. Physiol. Endocrinol. Metab. 2001, 280, E150–E159. [Google Scholar] [CrossRef]
- Danser, A.J.; Schalekamp, M.A.; Bax, W.A.; van den Brink, A.M.; Saxena, P.R.; Riegger, G.A.; Schunkert, H. Angiotensin-converting enzyme in the human heart. Effect of the deletion/insertion polymorphism. Circulation 1995, 92, 1387–1388. [Google Scholar] [CrossRef]
- Tiret, L.; Rigat, B.; Visvikis, S.; Breda, C.; Corvol, P.; Cambien, F.; Soubrier, F. Evidence, from combined segregation and linkage analysis, that a variant of the angiotensin I-converting enzyme (ACE) gene controls plasma ACE levels. Am. J. Hum. Genet. 1992, 51, 197–205. [Google Scholar]
- Williams, A.G.; Day, S.H.; Folland, J.P.; Gohlke, P.; Dhamrait, S.; Montgomery, H.E. Circulating angiotensin converting enzyme activity is correlated with muscle strength. Med. Sci. Sports Exerc. 2005, 37, 944–948. [Google Scholar] [CrossRef]
- Myerson, S.; Hemingway, H.; Budget, R.; Martin, J.; Humphries, S.; Montgomery, H. Human angiotensin I-converting enzyme gene and endurance performance. J. Appl. Physiol. 1999, 87, 1313–1316. [Google Scholar] [CrossRef]
- Puthucheary, Z.; Skipworth, J.R.; Rawal, J.; Loosemore, M.; Van Someren, K.; Montgomery, H.E. The ACE gene and human performance: 12 Years on. Sports Med. 2011, 41, 433–448. [Google Scholar] [CrossRef]
- Al-Hinai, A.T.; Hassan, M.O.; Simsek, M.; Al-Barwani, H.; Bayoumi, R. Genotypes and allele frequencies of angiotensin converting enzyme (ACE) insertion/deletion polymorphism among Omanis. J. Sci. Res. Med. Sci. 2002, 4, 25–27. [Google Scholar] [PubMed]
- Wishart, J.M.; Horowitz, M.; Need, A.G.; Scopacasa, F.; Morris, H.A.; Clifton, P.M. Relations between calcium intake, calcitriol, polymorphisms of the vitamin D receptor gene, and calcium absorption in premenopausal women. Am. J. Clin. Nutr. 1997, 65, 798–802. [Google Scholar] [CrossRef] [PubMed]
- Pfeifer, M.; Begerow, B.; Minne, H.W. Vitamin D and muscle function. Osteoporos. Int. 2002, 13, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Whitfield, G.K.; Remus, L.S.; Jurutka, P.W.; Zitzer, H.; Oza, A.K.; Dang, H.T.; Haussler, C.A.; Galligan, M.A.; Thatcher, M.L.; Dominguez, C.E.; et al. Functionally relevant polymorphisms in the human nuclear vitamin D receptor gene. Mol. Cell Endocrinol. 2001, 177, 145–159. [Google Scholar] [CrossRef]
- Baker, A.R.; McDonnell, D.P.; Hughes, M.; Crisp, T.M.; Mangelsdorf, D.J.; Haussler, M.R.; Pike, J.W.; Shine, J.; O’Malley, B.W. Cloning and expression of full-length cDNA encoding human vitamin D receptor. Proc. Natl. Acad. Sci. USA 1988, 85, 3294–3298. [Google Scholar] [CrossRef]
- Arai, H.; Miyamoto, K.I.; Taketani, Y.; Yamamoto, H.; Iemori, Y.; Morita, K.; Tonai, T.; Nishisho, T.; Mori, S.; Takeda, E. A vitamin D receptor gene polymorphism in the translation initiation codon: Effect on protein activity and relation to bone mineral density in Japanese women. J. Bone Miner. Res. 1997, 12, 915–921. [Google Scholar] [CrossRef]
- Moreno Lima, R.; De Abreu, B.S.; Gentil, P.; de Lima Lins, T.C.; Grattapaglia, D.; Pereira, R.W.; De Oliveira, R.J. Lack of association between vitamin D receptor genotypes and haplotypes with fat-free mass in postmenopausal Brazilian women. J. Gerontol. A Biol. Sci. Med. Sci. 2007, 62, 966–972. [Google Scholar] [CrossRef]
- Iki, M.; Saito, Y.; Dohi, Y.; Kajita, E.; Nishino, H.; Yonemasu, K.; Kusaka, Y. Greater trunk muscle torque reduces postmenopausal bone loss at the spine independently of age, body size, and vitamin D receptor genotype in Japanese women. Calcif. Tissue Int. 2002, 71, 300–307. [Google Scholar] [CrossRef]
- Nelson, D.A.; Vande-Vord, P.J.; Wooley, P.H. Polymorphism in the vitamin D receptor gene and bone mass in African-American and white mothers and children: A preliminary report. Ann. Rheum. Dis. 2000, 59, 626–630. [Google Scholar] [CrossRef]
- Fleet, J.C.; Harris, S.S.; Wood, R.J.; Dawson-Hughes, B. The BsmI vitamin D receptor restriction fragment length polymorphism (BB) predicts low bone density in premenopausal black and white women. J. Bone Miner. Res. 1995, 10, 985–990. [Google Scholar] [CrossRef]
- O’Dell, S.D.; Day, I.N. Insulin-like growth factor II (IGF-II). Int. J. Biochem. Cell Biol. 1998, 30, 767–771. [Google Scholar] [CrossRef]
- Stewart, C.E.; Rotwein, P. Growth, differentiation, and survival: Multiple physiological functions for insulin-like growth factors. Physiol. Rev. 1996, 76, 1005–1026. [Google Scholar] [CrossRef] [PubMed]
- Junnila, R.K.; List, E.O.; Berryman, D.E.; Murrey, J.W.; Kopchick, J.J. The GH/IGF-1 axis in ageing and longevity. Nat. Rev. Endocrinol. 2013, 9, 366–376. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.I.; Clemmons, D.R. Insulin-like growth factors and their binding proteins: Biological actions. Endocr. Rev. 1995, 16, 3–34. [Google Scholar] [CrossRef]
- Baxter, G.T.; Kuo, R.C.; Jupp, O.J.; Vandenabeele, P.; MacEwan, D.J. Tumor necrosis factor-alpha mediates both apoptotic cell death and cell proliferation in a human hematopoietic cell line dependent on mitotic activity and receptor subtype expression. J. Biol. Chem. 1999, 274, 9539–9547. [Google Scholar] [CrossRef]
- Bradley, J.R. TNF-mediated inflammatory disease. J. Pathol. 2008, 214, 149–160. [Google Scholar] [CrossRef]
- Costamagna, D.; Costelli, P.; Sampaolesi, M.; Penna, F. Role of inflammation in muscle homeostasis and myogenesis. Mediat. Inflamm. 2015, 2015, 805172. [Google Scholar] [CrossRef]
- Smith, J.D. Apolipoproteins and aging: Emerging mechanisms. Ageing Res. Rev. 2002, 1, 345–365. [Google Scholar] [CrossRef]
- Bertram, L.; McQueen, M.B.; Mullin, K.; Blacker, D.; Tanzi, R.E. Systematic meta-analyses of Alzheimer disease genetic association studies: The AlzGene database. Nat. Genet. 2007, 39, 17–23. [Google Scholar] [CrossRef]
- Fullerton, S.M.; Clark, A.G.; Weiss, K.M.; Nickerson, D.A.; Taylor, S.L.; Stengård, J.H.; Salomaa, V.; Vartiainen, E.; Perola, M.; Boerwinkle, E.; et al. Apolipoprotein E variation at the sequence haplotype level: Implications for the origin and maintenance of a major human polymorphism. Am. J. Hum. Genet. 2000, 67, 881–900. [Google Scholar] [CrossRef]
- Kami, K.; Morikawa, Y.; Sekimoto, M.; Senba, E. Gene expression of receptors for IL-6, LIF, and CNTF in regenerating skeletal muscles. J. Histochem. Cytochem. 2000, 48, 1203–1213. [Google Scholar] [CrossRef] [PubMed]
- Sendtner, M.; Kreutzberg, G.W.; Thoenen, H. Ciliary neurotrophic factor prevents the degeneration of motor neurons after axotomy. Nature 1990, 345, 440–441. [Google Scholar] [CrossRef] [PubMed]
- Sleeman, M.W.; Anderson, K.D.; Lambert, P.D.; Yancopoulos, G.D.; Wiegand, S.J. The ciliary neurotrophic factor and its receptor, CNTFR alpha. Pharm. Acta Helv. 2000, 74, 265–272. [Google Scholar] [CrossRef]
- Schrauwen, P.; Hesselink, M. UCP2 and UCP3 in muscle controlling body metabolism. J. Exp. Biol. 2002, 205, 2275–2285. [Google Scholar]
- Schrauwen, P.; Hoeks, J.; Hesselink, M.K. Putative function and physiological relevance of the mitochondrial uncoupling protein-3: Involvement in fatty acid metabolism? Prog. Lipid Res. 2006, 45, 17–41. [Google Scholar] [CrossRef]
- Dhamrait, S.S.; Williams, A.G.; Day, S.H.; Skipworth, J.; Payne, J.R.; World, M.; Humphries, S.E.; Montgomery, H.E. Variation in the uncoupling protein 2 and 3 genes and human performance. J. Appl. Physiol. 2012, 112, 1122–1127. [Google Scholar] [CrossRef]
- Brand, M.D.; Pamplona, R.; Portero-Otín, M.; Requena, J.R.; Roebuck, S.J.; Buckingham, J.A.; Clapham, J.C.; Cadenas, S. Oxidative damage and phospholipid fatty acyl composition in skeletal muscle mitochondria from mice underexpressing or overexpressing uncoupling protein 3. Biochem. J. 2002, 368, 597–603. [Google Scholar] [CrossRef]
- Seo, D.Y.; Lee, S.R.; Kim, N.; Ko, K.S.; Rhee, B.D.; Han, J. Age-related changes in skeletal muscle mitochondria: The role of exercise. Integr. Med. Res. 2016, 5, 182–186. [Google Scholar] [CrossRef]
- Brand, M.D.; Affourtit, C.; Esteves, T.C.; Green, K.; Lambert, A.J.; Miwa, S.; Pakay, J.L.; Parker, N. Mitochondrial superoxide: Production, biological effects, and activation of uncoupling proteins. Free Radic. Biol. Med. 2004, 37, 755–767. [Google Scholar] [CrossRef]
- Colhoun, H.M.; McKeigue, P.M.; Smith, D.G. Problems of reporting genetic associations with complex outcomes. Lancet 2003, 361, 865–872. [Google Scholar] [CrossRef]
- Duncan, L.E.; Ostacher, M.; Ballon, J. How genome-wide association studies (GWAS) made traditional candidate gene studies obsolete. Neuropsychopharmacology 2019, 44, 1518–1523. [Google Scholar] [CrossRef] [PubMed]
- Visscher, P.M.; Wray, N.R.; Zhang, Q.; Sklar, P.; McCarthy, M.I.; Brown, M.A.; Yang, J. 10 Years of GWAS discovery: Biology, function, and translation. Am. J. Hum. Genet. 2017, 101, 5–22. [Google Scholar] [CrossRef] [PubMed]
- Sawcer, S. The complex genetics of multiple sclerosis: Pitfalls and prospects. Brain 2008, 131, 3118–3131. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).