Abstract
Diverse signals and stress factors regulate the activity and homeostasis of ribosomes in all cells. The Saccharomyces cerevisiae protein Asc1/yRACK1 occupies an exposed site at the head region of the 40S ribosomal subunit (hr40S) and represents a central hub for signaling pathways. Asc1 strongly affects protein phosphorylation and is involved in quality control pathways induced by translation elongation arrest. Therefore, it is important to understand the dynamics of protein formations in the Asc1 microenvironment at the hr40S. We made use of the in vivo protein-proximity labeling technique Biotin IDentification (BioID). Unbiased proxiOMICs from two adjacent perspectives identified nucleocytoplasmic shuttling mRNA-binding proteins, the deubiquitinase complex Ubp3-Bre5, as well as the ubiquitin E3 ligase Hel2 as neighbors of Asc1. We observed Asc1-dependency of hr40S localization of mRNA-binding proteins and the Ubp3 co-factor Bre5. Hel2 and Ubp3-Bre5 are described to balance the mono-ubiquitination of Rps3 (uS3) during ribosome quality control. Here, we show that the absence of Asc1 resulted in massive exposure and accessibility of the C-terminal tail of its ribosomal neighbor Rps3 (uS3). Asc1 and some of its direct neighbors together might form a ribosomal decision tree that is tightly connected to close-by signaling modules.
1. Introduction
Connectivity of membrane-separated cellular processes in eukaryotes depends on signal transduction to bridge intra-cellular distances and organelle barriers. Ribosomes localize for site-specific and process-bound protein biosynthesis. They are connected to local signaling entities, for example, membrane receptors, kinase modules, and/or sources of secondary messengers, to facilitate timely reactions to stimuli. Conversely, they have to communicate their own status, like translational arrest or ribosome stalling, to initiate processes such as ribosome clearance, ribosome preservation, or ribophagy. Moreover, they must forward ribosome status messages, for example, into the nucleus for the adaptation of genome activities.
The Asc1 protein of Saccharomyces cerevisiae is a highly conserved eukaryotic seven-bladed β-propeller of the WD40-repeat protein family and named RACK1 in plants and mammals. It is a canonical ribosomal protein and gets incorporated late into the 40S ribosome during cytosolic maturation [1,2]. Cellular depletion of this protein in S. cerevisiae (asc1− cells) causes massive proteome and transcriptome changes [3,4]. RACK1 gene knock-down or mRNA/protein depletion in higher eukaryotes is lethal during early embryogenesis, and medical studies have provided ample evidence that aberrant RACK1 expression in mammals coincides with different types of cancer [5,6,7,8,9]. Yeast asc1− cells proliferate well in culture, although they display a tremendously altered phosphoproteome [4,10]. Furthermore, they show several phenotypical traits, for example, loss of dimorphism (haploid adhesive and diploid pseudohyphal growth), increased sensitivity to osmotic constraint and heat, slow-growth on non-fermentable carbon sources, increased sensitivity to translation inhibitors and genotoxic compounds, and cell cycle delay [3,4,11,12,13,14].
Asc1/RACK1 resides at the head region of the 40S ribosomal subunit (hr40S), close to the mRNA entry and exit tunnels of initiating 40S and translating/stalled 80S ribosomes [15,16]. Remodeling of protein composition and arrangement within this microenvironment is expected to account for ribosome-activity adaptations to meet cellular needs. Conversely, even the ribosomal translation status itself is expected to influence protein assembly at the hr40S. The analysis of the remodeling of cellular microenvironments is frequently done with co-immunoprecipitation and tag-affinity capture. These techniques rely on high binding affinities of proteins within complexes that have to endure cell lysis and the subsequent affinity purification procedure. They sometimes suffer from artificial protein–protein binding within cell extracts that have lost the cellular order. New approaches employ the covalent labeling of proteins of a microenvironment within living cells. This enables the subsequent capture and identification of adjacent proteins irrespectively from their behavior upon and after cell lysis. Our previous microenvironment analysis using a Biotin IDentification approach (BioID) [17] for S. cerevisiae combined with quantitative LC-MS analysis provided a first Asc1-proxiOME: beyond expected ribosomal Asc1-neighboring proteins, we identified, for example, the ubiquitin E3 ligase Hel2, the deubiquitinating complex Ubp3-Bre5, and various mRNA-binding proteins, amongst them, the mRNA-exonuclease Xrn1 [18]. Also, Def1, a protein known to act in concert with the Elongin-Cullin E3 ligase complex to trigger proteasomal RNA polymerase II subunit degradation in the nucleus, was found to be in the proximity of Asc1 [18,19].
RACK1/Asc1, and its neighbor Hel2, function in the ribosome-associated quality control (RQC) and no-go-decay (NGD) systems that are required for the degradation of aberrant polypeptides and mRNA clearance upon abnormal translation arrest. Ribosome stalling during translation elongation is caused by various translation compromising factors, such as rare codons within the translated mRNA (reviewed in [20]). Increasing data imply a RACK1/Asc1 function within early RQC to initiate the degradation of nascent aberrant peptide chains of stalled 80S ribosomes [21]. Also, the endonucleolytic cleavage of stalled non-stop mRNAs (non-stop decay, NSD) depends on RACK1/Asc1 [22]. Splitting apart the 40S and 60S-nascent-peptide-chain subunits of stalled ribosomes depends on the ubiquitination of 40S proteins through Hel2 (ortholog of human ZNF598) and is followed by Rqc2-and Hel2-dependent CATylation of split 60S-nascent-peptide-chains [21,23,24]. The C-terminal tail of the 40S ribosomal protein S3 (Rps3/uS3) binds Asc1, thereby linking it to the mRNA guiding tunnel and the mRNA decoding area in 80S ribosomes [15]. Physical interaction between mRNA and Rps3 at the entry tunnel of the small ribosomal subunit promotes NGD of aberrant mRNAs [25,26]. Mono-ubiquitination within the C-terminal tail of Rps3 is critical for RQC and correlates with translational stalling. This, in turn, depends on the balanced activities of the E3 ubiquitin ligase Hel2 and of the deubiquitinase complex Ubp3-Bre5 (ortholog of human USP10-G3BP1) [27,28]. Ubp3-Bre5 also controls ribophagy, the ultimate route towards bulk degradation of dispensable ribosomes [29]. We showed previously that phosphorylation of Bre5 at serine 282 is Asc1-dependent [10].
Here, we intended to broaden the current knowledge of the hr40S microenvironment through affinity capture from the Asc1-adjacent perspective of Rps2 (uS5). Our study focuses on the analysis of the Asc1-dependency of the hr40S composition. We went for a comparative BioID experiment expressing the Rps2-BirA* fusion protein in both wild-type ASC1 and asc1− cells. The localization of Bre5 at hr40S turned out to depend on Asc1. By contrast, Vps30 is captured upon Rps2-BirA* biotinylation only in the absence of Asc1. Vps30 (ortholog of human Beclin-1) is a subunit of the phosphatidylinositol (PtdIns) 3-kinase complexes I and II, which are involved in autophagosome formation and vacuolar protein-sorting [30]. Furthermore, the Rps2-BioID experiments confirmed the localization of Def1 to the hr40S. A picture emerges with Asc1/RACK1 acting as a context-bound signal-coupling factor that coordinates ribosome activity and homeostasis. During this process, it might couple ribosomal status changes with nuclear genome activities via ubiquitin- and phosphorylation-mediated protein translocation. The hr40S might serve as a wharf for protein arrival and departure. Twenty-five years after launching mass spectrometry-based proteomics, proxiOME approaches, like BioID and the recently developed context-specific variant of split-BioID [31], are new and promising proteomics techniques to study meaningful cellular sub-proteomes. They might have the power to illuminate otherwise invisible molecular ships passing in the night (expression with reference to Toby J. Gibsons article RACK1 research—ships passing in the night? [32]).
2. Materials and Methods
2.1. Plasmid Construction
All plasmids used in this study are listed in Table 1. The RPS2-birA* plasmid pME4799 is equivalent to the ASC1-birA* bearing plasmid pME4478 and was generated using the GeneArt® Seamless Cloning and Assembly Enzyme Mix Kit (#A14606, Thermo Fisher Scientific).

Table 1.
Plasmids used in this study.
2.2. Yeast Strains and Growth Conditions
Yeast strains used in this study are either of the Σ1278b or S288c background and are listed in Table 2. Yeasts cells were grown in liquid yeast nitrogen base medium (YNB; 0.15% YNB, 0.5% ammonium sulfate, 2% glucose) with additional supplements as required: L-arginine (20 mg/l), L-lysine HCl (30 mg/L), L-tryptophan (20 mg/L), uracil (20 mg/L), L-leucine 30 mg/L), L-valine 150 mg/L), L-isoleucine (30 mg/L). To obtain the methionine prototrophic strain RH3793, the wild-type MET15 gene was reintroduced into the genome of the Bre5-GFP expressing strain from Invitrogen. This was done to avoid suppression of the MET15 promoter that regulates the expression of RPS2-birA* and birA*. The ASC1 gene was deleted in strain RH3793 by replacing it with LEU2 (derived from plasmid pUG73) resulting in strain RH3794. Deletion cassettes for the replacement of BRE5 and UBP3 with the kanMX4 resistance marker were amplified from genomic DNA isolated from the respective Euroscarf deletion strains. The deletion cassettes were transformed into the ASC1 wild-type strain (RH2817) and the asc1− strain (RH3510), resulting in strains RH3789-3792.

Table 2.
Saccharomyces cerevisiae strains used in this study.
2.3. Western Blot Analysis
Cell lysis was performed as described by Kushnirov (2000) [36]. For each sample, an equal number of cells (OD600 of 2.5, calculated according to the OD600 of the corresponding culture) were harvested by centrifugation, washed with water, and resuspended in 200 µL 0.1 M NaOH. Samples were incubated for 5 min at room temperature and centrifuged at 4000 rpm for 50 s. The supernatant was discarded, and 50 µL of 1:4 diluted loading dye (see below for composition) was added. Samples were heated for 3 min at 95 °C, centrifuged at 16,200× g for 5 min, and 6 µL of the supernatant were subjected to SDS-PAGE followed by the transfer onto nitrocellulose membrane. Proteins on the membrane were visualized by staining with Ponceau Red (0.2% PonceauS, 3% trichloroacetic acid). After blocking, the membranes were incubated either with polyclonal rabbit anti-Asc1p [10], or polyclonal rabbit anti-Rps3 (provided by Heike Krebber), or monoclonal mouse anti-GFP (B-2, sc-9996, Santa Cruz Biotechnology) followed by incubation with the respective peroxidase-coupled secondary antibody. For the detection of biotinylated proteins, membranes were blocked overnight with 1% BSA in phosphate buffered saline (PBS, 8 mM Na2HPO4, 2 mM NaH2PO4, 150 mM NaCl). PierceTM High Sensitivity Streptavidin-HRP (#21130, Thermo Fisher Scientific, diluted 1:2000 in the blocking buffer with 0.1% Tween 20) was added for 1 h to the membranes. Chemiluminescent signals were detected using the FUSION-SL-4 (Peqlab).
2.4. BioID Experiments
Yeast cells were cultivated in 200 mL YNB medium to mid-log phase. The media contained 10μM D-biotin and differentially labeled lysine and arginine for SILAC. The following amino acids were used for differential labeling: 13C6-L-arginine HCl, 13C615N4-L-arginine HCl, 4,4,5,5-D4-L-lysine HCl, 13C6-L-lysine HCl. Cell pellets were washed twice with 10 mL of washing buffer 1 (10 mM HEPES, pH 7.9, 10 mM KCl, 1.5 mM MgCl2) and mixed in a 1:1:1 ratio of cells according to the OD600 of the cultures. Cells were lysed in breaking buffer (wash buffer with 1 cOmpleteTM protease inhibitor cocktail tablet (Merck) per 50 mL, 0.5 mM DTT, and 0.5 mM PMSF) using glass beads. SDS was added to a final w/v of 4% for the denaturation of proteins. Enrichment of biotinylated proteins was performed according to the protocol described in Opitz et al. (2017) [18]. Strep-Tactin® Sepharose® gravity flow columns with 0.2 mL instead of 1 mL bed volume were used (#2-1202-550, IBA GmbH). The cell extract was applied to the column, and the column was washed with 10 mL 1x washing buffer 2 (#2-1003-100, IBA GmbH) containing 0.4% SDS. The biotinylated proteins were eluted with 600 µL 1x washing buffer containing 10 mM D-biotin. Proteins were precipitated according to Wessel and Flügge (1984) [37] by chloroform-methanol extraction. The cell pellet was dissolved in 20 µL 8 M urea/2 M thiourea and subjected to SDS-PAGE. Gel lanes were cut into 8 pieces each, and proteins were digested in-gel with trypsin [38]. Peptides were desalted with C18 (3M) stop and go extraction (stage) tips [39,40]. Peptides were dried and then dissolved in a 20 µL sample buffer (2% acetonitrile, 0.1% formic acid) for the subsequent LC-MS analysis. Strep-Tactin®XT spin columns (#2-4150-025, IBA GmbH) were used to enrich biotinylated peptides according to the manufacturer’s instructions. Peptides were eluted using three times the 150 µL elution buffer with 50 mM biotin. Peptides were purified again using stage tips and subjected to LC-MS analysis.
For the Western blot analysis of input samples and column flow-through, 3x loading dye (0.25 M Tris-HCl pH 6.8, 30% glycerol, 15% β-mercaptoethanol, 7% SDS, 0.3% bromphenol blue) was added to the respective fractions, and the samples were heated at 65 °C for 10 min. Equal volumes of the input and flow-through samples were subjected to SDS-PAGE.
For BioID experiments with subsequent Western blot analysis, cells were cultivated in 150 mL YNB medium to mid-log phase in the presence of 10 μM D-biotin. The preparation of cell extracts and enrichment of biotinylated proteins were performed as already described, except that 100 µL Strep-Tactin® Sepharose® Slurry (50 µL bed volume, #2-1201-002, IBA GmbH) was used instead of gravity flow columns. Cell lysates were incubated with the equilibrated beads on a rotating wheel for 30 min. Beads were washed three times with 800 µL washing buffer containing 0.4% SDS. To elute proteins from the beads, 75 µL 2x loading dye were added, and samples were heated for 3 min at 95 °C.
2.5. Liquid Chromatography-Mass Spectrometry (LC-MS) Analysis
Liquid chromatography systems (UltiMate 3000 RSLCnano system) coupled to either an Orbitrap Velos Pro hybrid ion trap-Orbitrap or a Q Exactive HF mass spectrometer (all Thermo Fisher Scientific) were used for peptide analysis. Peptides in 4 µL sample solution were trapped and washed on an Acclaim PepMap 100 precolumn (C18; 100 µm, 2 cm, 3 µm, 100 Å; Thermo Fisher Scientific) at a flow rate of 25 µL/min for 6 min in 100% solvent A (98% water, 2% acetonitrile, 0.07% trifluoroacetic acid). Analytical peptide separation by reverse-phase chromatography was performed on an Acclaim PepMap RSLC column (C18; 75 µm, 50 cm, 3 µm, 100 Å; Thermo Fisher Scientific), running a gradient from 98% solvent A (0.1% formic acid) and 2% solvent B (80% acetonitrile, 0.1% formic acid) to 7% solvent B within 3 min, to 32% solvent B within 92 min, and to 65% solvent B within the next 26 min, at a flow rate of 300 nL/min (solvents and acids were from Fisher Chemical). For the chromatography of the Q Exactive HF coupled system, a similar gradient was used with minor changes. Eluting peptides were ionized online by nanoelectrospray ionization (nESI) using a Nanospray Flex ion source (Thermo Fisher Scientific) at 1.5 kV (Orbitrap Velos Pro) and 1.6 kV (Q Exactive HF) and were continuously transferred into the mass spectrometer. Full scans within the mass range of 300 to 1850 m/z were taken within the Orbitrap-FT analyzer at a resolution of 60,000 with parallel data-dependent top 15 MS2 collision induced dissociation (CID) fragmentation with the LTQ Velos Pro linear ion trap. For the Q Exactive HF, the mass range of the full scans was 300 to 1650 m/z and data-dependent top 10 MS2 fragmentation with higher-energy collisional dissociation (HCD) fragmentation was performed. The software XCalibur 2.2 (Thermo Fisher Scientific) was used for LC-MS method programming and data acquisition. MS/MS2 data were searched against an S. cerevisiae-specific protein database (UniProt Proteome ID UP000002311) using the software MaxQuant 1.6.0.16 [41]. The digestion mode was trypsin/P, and the maximum number of missed cleavage sites was set to three. Arg6, Arg10, Lys4, and Lys8 were defined as the medium and heavy labels in accordance with the amino acids used for cultivation of the cells. Carbamidomethylation at cysteine was set as a fixed modification, and oxidation at methionine and biotinylation of lysine were set as variable modifications. Mass tolerances of precursors and fragment ions were 4.5 ppm and 0.5 Da (CID) or 20 ppm (HCD), respectively. Re-quantification was enabled. At a minimum, two ratio counts were required for protein quantification using unique and razor peptides. False discovery rates were calculated using the revert decoy mode, and the threshold for peptide sequence matches as well as protein identifications was 0.01. MaxQuant output data were further evaluated using the Perseus software 1.6.0.7 [42].
2.6. Phenotypic Growth Tests
Yeast cells were grown to mid-log phase in YNB medium and diluted to an OD600 of 0.1. Ten-fold dilutions were prepared, and 10 or 20 µL of each dilution were dropped on yeast extract peptone dextrose (YEPD) or YNB plates, respectively. The plates were incubated for 3 days at 25 °C, 30 °C, or 37 °C. Additional compounds were added, as indicated in Figure 4. To test for haploid adhesive growth, cells were patched on 10 mM 3-amino-1,2,4-triazole (3-AT) containing YNB plates and incubated for 3 days at 30 °C. Plates were washed under a constant stream of water to test the ability of the yeast cells to adhere to the surface of the agar plate.
2.7. Accession Number
Data are available via ProteomeXchange with identifier PXD015611 [43,44].
3. Results
3.1. Expression of Rps2-BirA* in S. cerevisiae Strains of the Σ1278b Genetic Background
Our original inventory analysis of the Asc1-proxiOME using BioID revealed some expected neighboring proteins, for example, known adjacent ribosomal proteins and subunits of eIF3, as well as some unexpected ones, such as the deubiquitinase complex Ubp3-Bre5 and the RNA polymerase II degradation factor Def1. Using BioID experiments from the perspective of the Asc1-adjacent Rps2 protein, we intended first to confirm the presence of those proteins that were not expected at the head region of the 40S subunit (hr40S), and second, to study the putative Asc1-dependency of their hr40S localization. For the second aim, we compared Rps2-BioID captures from wild-type ASC1 and asc1− cells. This should reveal the impact of Asc1 on hr40S composition and/or dynamics. Rps2 was chosen as a new perspective for in vivo labeling of hr40S proteins with biotin because its C-terminus extends to this region (Figure 1A) [15]. Additionally, Rps2 was specifically biotin-labeled and captured upon Asc1-BirA* expression [18]. Also, the functionality of previously published C-terminal Rps2 fusion proteins (e.g., with GFP) encouraged us in the choice of Rps2 as BioID bait [29]. Similar to our Asc1-BioID experiments, we expressed an Rps2-BirA* fusion protein from a high-copy number plasmid in wild-type RPS2 ASC1 as well as in RPS2 asc1− cells (Figure 1A). asc1− refers to an allele encoding an asc1 mRNA with a stop codon in exon 1, abrogating translation; the pre-mRNA still contains the SNR24 gene within its intron [3]; in the following text, we refer to this allele as ASC1 deletion as it does not produce any Asc1 protein. As a negative control, we also transformed RPS2 ASC1 cells with only the unfused birA* allele on a high-copy number plasmid, giving rise to Rps2-independent background biotin-labeling activity. BirA* and Rps2-BirA* expression and the stability of the fusion protein in both strain backgrounds were confirmed by Western blot experiments using an anti-BirA antibody (Figure 1B). Overall protein biotinylation in cells grown overnight in the presence of biotin was analyzed for the respective strains using horseradish peroxidase (HRP)-coupled streptavidin on Western-blot membranes (Figure 1B). The expression of mere BirA* gave rise to significantly increased background biotinylation in comparison to Rps2-BirA* expression. Rps2-BirA* expression in asc1− cells generated a very strong additional biotinylation signal at a molecular weight of approximately 26 kDa derived from Rps3 (as shown later, Figure 2A and Figure 3).
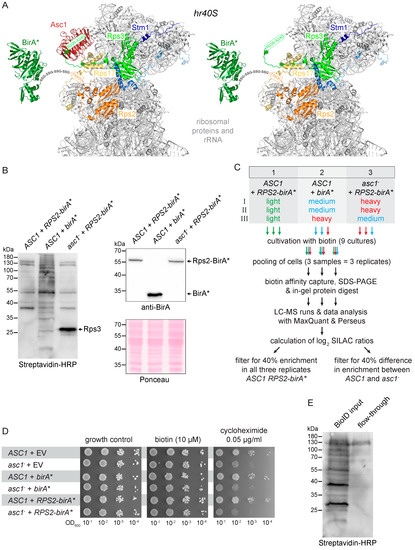
Figure 1.
Set-up of the Rps2-BioID analysis. (A) Rps2-BirA* at the hr40S in the presence and absence of Asc1 (red). The BirA* protein (dark green) is fused to the C-terminus of Rps2 (uS5, orange) with a linker sequence consisting of four repeats of Gly-Ser-Ser (GSS). The last four amino acids of Rps2 (in orange letters) were structurally not resolved and are indicated within an orange arrow. Ribosomal proteins captured with Rps2-BirA* are highlighted in different colors: Rps17 (eS17) yellow, Rps3 (uS3) light green, Stm1 blue. Biotinylated lysine residues K212 and K223 of Rps3 are indicated with green asterisks (the green arrow indicates the last 15 C-terminal amino acids of Rps3 that were structurally not resolved). The PyMOL Molecular Graphics System software was used to generate the figures from crystal structure data of the S. cerevisiae 80S ribosome and the E. coli BirA protein derived from the PDB entries 4V88 [15] and 1BIB [45], respectively. (B) An aliquot of each SILAC-labeled culture was taken prior to cell pooling for cell lysis. Extracts were applied to SDS-PAGE and proteins blotted to a membrane. Streptavidin-HRP was used to detect biotinylated proteins (left), and a BirA-specific antibody was applied to detect the BirA* and Rps2-BirA* proteins (right). The birA* and RPS2-birA* alleles were expressed from extrachromosomal high copy number plasmids in the ASC1 wild-type and asc1− background as indicated. Ponceau staining of the proteins on the membrane served as the loading control. Results are shown representatively for one of three replicates. (C) Strategy and workflow of the Biotin IDentification (BioID) experiment. Stable isotope labeling with amino acids in cell (SILAC) amino acids were used for a triple-labeling approach with three replicates (I-III), including a medium to heavy label swap. For detailed information about mass-spectrometry data acquisition (XCalibur) and analysis (MaxQuant and Perseus), see Materials and Methods and Table 3. (D) Drop dilution assays (successive 10-fold dilutions) in the presence of biotin or cycloheximide to test whether the additional expression of BirA* or Rps2-BirA* impairs colony growth. Strains transformed with the empty vector (EV) were used for growth controls. (E) Binding of biotinylated proteins on Strep-Tactin gravity flow columns. Equal volumes of the cell lysate (BioID input, applied on the Strep-Tactin column) and the flow-through were subjected to SDS-PAGE and proteins blotted to a membrane followed by detection of biotinylated proteins using streptavidin-HRP.
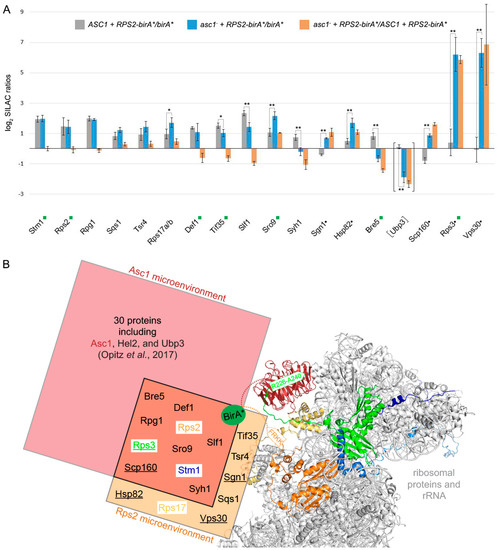
Figure 2.
BioID protein capture after Rps2-BirA*-labeling in the presence or absence of Asc1. (A) All columns represent the average of MaxQuant normalized log2 SILAC ratios of three biological replicates and are provided with the respective standard deviations. Proteins depicted in the graph were enriched with normalized log2 SILAC ratios for ASC1 RPS2-birA*/birA* greater than or equal to 0.485 (matching a minimum enrichment of approximately 40%) from either ASC1 wild-type cells (gray columns) and/or from asc1− cells (blue columns). Black dots next to protein names indicate proteins that were exclusively enriched from the asc1− cells. Orange columns represent the respective fold-changes of the SILAC protein quantification values between ASC1 wild-type and asc1− cells, indicating the degree of Asc1-dependency in their Rps2 proximal localization. The proteins are ordered from left to right according to increasing absolute differences in enrichment between ASC1 wild-type and asc1− cells (orange columns). A two-sample t-test on the ASC1 RPS2-birA*/birA* versus asc1− RPS2-birA*/birA* log2 SILAC ratios was performed, and significant changes were indicated with one or two asterisks (p-value threshold 0.05 and 0.01, respectively). A green rectangle next to the protein name indicates the identification of a biotinylated peptide for the respective protein. Although the data for Ubp3 did not pass the described filtering, they are provided for the discussion (see main text). The graph is based on the data in Table S1. (B) Comparison of the Asc1 and Rps2 microenvironments captured with BioID: The hr40S is illustrated as in Figure 1A. The biotin ligase BirA* (green sphere) was fused via a linker sequence to the C-termini of either Asc1 or Rps2 (indicated with dashed lines in red and orange, respectively). The proteinaceous microenvironment of Asc1 [18] is indicated with a red rectangle and that of Rps2 with an orange one. Proteins identified in the neighborhood of both proteins are listed in the overlap of both rectangles and are supposed to belong to a common microenvironment. Proteins only identified in the absence of Asc1 within the Rps2 microenvironment are underlined. Proximity between Asc1 or Rps2 to their neighboring proteins might also take place apart from a mature 40S ribosomal subunit. The PyMOL Molecular Graphics System software was used to generate the figure from crystal structure data of the S. cerevisiae 80S ribosome (PDB entry: 4V88) [15].
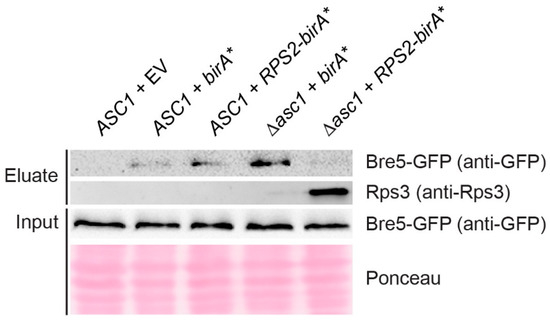
Figure 3.
Immuno-detection of Bre5 and Rps3 captured with a small-scale Rps2-BirA* BioID experiment. Rps2-BirA* BioID capture was performed in a small-scale format (see Materials and Methods for details) using strains expressing GFP-tagged Bre5. BirA* and Rps2-BirA* were expressed from high copy number plasmids in ASC1 wild-type and asc1− strains as indicated, and the ASC1 wild-type strain was additionally transformed with the empty vector (EV) as a control. Western blot experiments were performed with a GFP-antibody and an Rps3-antibody to detect protein enrichment in eluate fractions. Bre5-GFP was also detected in the total cell extracts (input). Ponceau staining of proteins served as a loading control for the input.
We next confirmed that Rps2-BirA* expression did not compromise the general growth of wild-type ASC1 or asc1− cells. Without further stress applied, the cell growth of all tested strains was similar in the absence as well as the presence of external biotin (Figure 1D). As expected, asc1− cells were sensitive to the translation inhibitor cycloheximide [10]. Expression of Rps2-BirA* as an additional Rps2 copy did not change cycloheximide sensitivity. Comparative quantitative Rps2-BioID capture experiments with strains expressing Rps2-BirA* in both ASC1 and asc1− cells (strains 1 and 2) and mere BirA* in ASC1 cells (strain 3) were performed using stable isotope labeling with amino acids in cell culture (SILAC; Figure 1C). Cellular proteins of these three yeast strains were differentially labeled with mass-specific variants of the SILAC amino acids lysine and arginine (e.g., strain 1 with light, strain 2 with medium, and strain 3 with heavy isotope amino acids incorporated) during overnight growth in the presence of 10 μM biotin. An equal number of cells from all three cultures were combined to yield one common cell pool for protein isolation and the biotin capture procedure. The quantitative Rps2-BioID experiment was performed in triplicate and was done as described previously for the Asc1-BioID in Opitz et al. (2017) [18]. The procedure applied is summarized in the Materials and Methods section. The only difference was the use of Strep-Tactin columns with 0.2 mL instead of 1 mL bed volume. As shown in Figure 1E, the smaller bed volume bound biotinylated proteins from cell extracts efficiently.
We identified 431 proteins with SILAC ratios for all three biological replicates. The ratios normalized by MaxQuant were used for data analysis. A detailed workflow for the data analysis in Perseus is provided in Table 3. Proteins that were enriched from RPS2-birA* expressing cells by more than 40% compared to the control in all three replicates were considered as Rps2 neighbors (normalized log2 SILAC ratios RPS2-birA*/birA* greater than or equal to 0.485, Figure 2A). Four proteins, Stm1, Rpg1, Sqs1, and Tsr4 were identified as Rps2 proximal proteins independent of the presence of Asc1. Seven additional proteins, Rps17 (eS17), Def1, Tif35, Slf1, Sro9, Syh1, and Bre5 were also identified in the Rps2 microenvironment but showed Asc1-dependency in their enrichment. Five proteins, Sgn1, Hsp82, Scp160, Rps3, and Vps30, were only captured with Rps2-BioID in the absence of Asc1. The identification of known neighbors of Rps2 at ribosomes, such as Rps17, Stm1, and the translation initiation factor 3 subunits a/Rpg1 and g/Tif35 [15,46,47], proves that the Rps2-BioID method is a reliable approach to uncover proximities. Furthermore, the protein Tsr4 was just recently described as a chaperone for Rps2 and interacts with its eukaryotic-specific N-terminal region [48,49]. In total, 9 of the 16 identified Rps2-neighbors were also captured with Asc1-BirA* [18], showing that we were able to monitor a common microenvironment of both proteins at the hr40S (Figure 2B).

Table 3.
Overview of MaxQuant output data evaluation with Perseus. The table lists the main steps of the Perseus analysis.
3.2. Rps2-BirA* Specifically Biotinylates Def1 and Stm1
Using Rps2-BirA*, we identified Def1 as a protein whose localization at the hr40S depends on Asc1 (Figure 2A). Upon transcriptional arrest, Def1 induces ubiquitination of the largest subunit of the stalled RNA polymerase II complex, Rbp1. A prerequisite for the transfer of Def1 from the cytoplasm to the nucleus is a substantial proteasome-mediated protein trimming through chopping off more than 200 amino acids of its Q-rich C-terminus [19]. Our previous Asc1-BioID experiment indicated the presence of the non-processed full-length Def1 version in the proximity of Asc1 [18]. At that time, we could not discriminate whether the two proteins were within proximity at or off the hr40S. The rediscovery of Def1 in the Rps2-BioID experiment here (Figure 2A) gives further evidence that Def1 actually resides at the hr40S close to Asc1 prior to its transfer into the nucleus. We previously showed that phosphorylation of Def1 at threonine 258 and serine 260 is Asc1-dependent [10]. Phosphorylation might play a role in this specific targeting of Def1.
Stm1 previously appeared as an Asc1 proximal protein [18], and it is an interactor of the translation elongation factors eEF3/Yef3 [50] and eEF2/Eft1 [51]. It resides as a putative subunit clamping factor in stalled ribosomes during glucose starvation [15], acts as a ribosome preservation factor during stationary growth [50], and has reappeared here in the Rps2-BioID experiment. The protein is homologous to the putative mammalian ortholog SERBP1 that has been described to interact with RACK1 in HeLa cells [52]. Localization of Stm1 at the hr40S does not depend on Asc1 (Figure 2A). However, its remodeling into the ribosome clamping position upon starvation might be mediated by Asc1.
3.3. Asc1 Inversely Affects Rps2-BirA*-Mediated Biotinylation of the mRNA-Binding Paralogs Sro9 and Slf1
Sro9 and Slf1 are two other proteins whose Rps2-BirA*-dependent biotinylation varied significantly in the asc1− compared to the ASC1 strain (Figure 2A). They are paralogous atypical-La motif-containing mRNA-binding proteins, and Sro9 was shown to shuttle between the nucleus and the cytoplasm in an mRNA-dependent manner [53]. Sro9 is also part of the heat-shock protein-containing HMC complex repressing the transcriptional activator heme activator protein 1 (Hap1) in the absence of heme. In the cytoplasm, Sro9 associates with translating ribosomes [54]. Our previous Asc1-BioID experiment suggested its precise location to be proximal to Asc1 [18]. The Rps2-BioID experiment now confirmed the hr40S as the site for Sro9 localization (Figure 2A). The absence of Asc1 at the hr40S even increased the capture of Sro9 as well as that of Scp160 and Sgn1 (two further mRNA-binding proteins). In contrast, the enrichment of the Sro9 paralogous protein Slf1, a key activator of translation during the oxidative stress response [55], decreased in the absence of Asc1. Asc1 seems to balance the localization of various mRNA-binding proteins at the hr40S.
3.4. Asc1 Inversely Affects Rps2-BirA*-Mediated Biotin-Enrichment of Bre5 on the One Hand and Rps3 and Vps30 on the Other Hand
Ubp3 and Bre5 together form a deubiquitinase complex and have both been identified as Asc1-proximal with our original Asc1-BioID experiment [18]. Bre5 reappeared in the Rps2-BioID experiment confirming its localization to the hr40S (Figure 2A). The capture of biotinylated Bre5 upon Rps2-BirA* expression is significantly reduced in the absence of Asc1; increased background biotinylation of Bre5 in merely BirA* expressing cells (negative control) even led to negative Rps2-BirA*-mediated enrichment values after normalization. (This is also the case for Syh1, a paralog of Smy2 that participates in COPII vesicle formation, Figure 2A) [56]. The increased biotinylation of Bre5 in the negative control was confirmed by Western blot experiments with eluates of the Rps2-BioID experiment of yeast strains expressing Bre5-GFP by use of an anti-GFP antibody. Bre5-GFP background biotinylation upon BirA* expression significantly increases in the absence of Asc1, whereas Bre5-GFP biotinylation upon Rps2-BirA* expression fades in asc1− cells (Figure 3). We showed previously that phosphorylation of Bre5 at serine 282 ceases upon depletion of Asc1 [10]. Asc1-dependent Bre5 phosphorylation might take place in a regulatory context of ribosomal subunit ubiquitination through Ubp3 to remodel ribosomes according to cellular signals. Ubp3 was not enriched from ASC1 wild-type cells in comparison to the birA* control, possibly caused by even stronger background biotinylation. However, it was significantly less captured from asc1− cells, indicating similar Asc1-dependent binding to hr40S as Bre5.
In contrast to the decreasing biotinylation and BioID capture of Bre5 in asc1− cells, the ribosomal protein Rps3 gets increasingly BioID-captured in the absence of Asc1 upon cellular Rps2-BirA* expression (Figure 2A). This finding is supported by Western blot experiments using an anti-Rps3 antibody (Figure 3). According to the crystal structure of the 80S ribosome of Ben-Shem et al. (2011) [15], Rps3 physically contacts Asc1 with its C-terminal tail. The removal of Asc1 seems to expose parts of Rps3, rendering its tail prone to Rps2-BirA*-mediated biotinylation (see also Figure 1A, right-hand side).
Vps30, a subunit of the phosphatidylinositol 3-kinase (PI3K) complexes I and II [30], was not captured upon Rps2-BirA* expression from ASC1 wild-type cells at all. It was, however, strongly enriched from asc1− cells (Figure 2A). The strong Vps30 enrichment at the hr40S equals that of Rps3 from asc1− cells. Future studies might reveal whether the exposure of the C-terminal Rps3 tail is required for Vps30 association to the hr40S and whether other subunits of the PI3K complexes locate to ribosomes to induce signaling events.
3.5. ASC1 Genetically Interacts with UBP3/BRE5 to Protect Cells Against Heat, Starvation, and Translational Stress
The increasing evidence for a functional interaction between Asc1 and the deubiquitinase Ubp3-Bre5 complex prompted us to analyze putative genetic interactions of their encoding genes. In the absence of cellular stress, single or double deletions of the genes ASC1, UBP3, and BRE5 (asc1−, Δbre5, Δubp3, asc1− Δbre5, and asc1− Δubp3 strains) do not compromise colony growth (Figure 4A, growth control in comparison to the respective wild-type strain). Growth at hyperosmotic conditions (75 mM NaCl), at elevated temperatures (37 °C instead of 30 °C), or in the presence of the translation inhibitor cycloheximide (0.05 and 0.10 μg/mL), is mildly compromised for cells with the single deletions asc1−, Δbre5, and Δubp3. The double deletions asc1− Δbre5 and asc1− Δubp3, however, lead to synergistic growth defects. Growth is almost abolished at elevated temperatures and in the presence of the translation inhibitor cycloheximide. In contrast to the growth phenotype on 37 °C, double mutation strains grew slightly better than the single deletion strains at lower temperatures (25 °C).
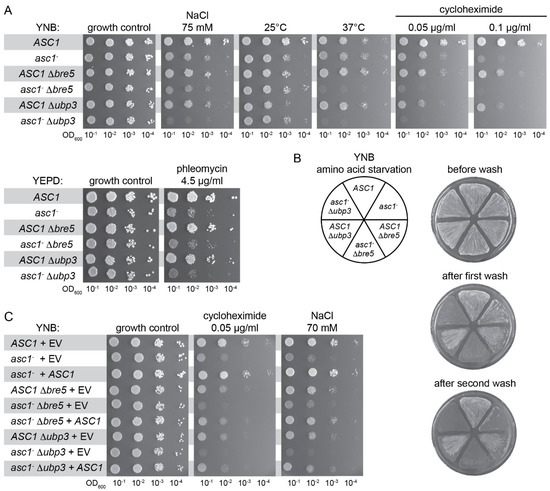
Figure 4.
Genetic interaction between ASC1 and BRE5/UBP3. S. cerevisiae ASC1, BRE5, and UBP3 single and double deletion strains were phenotypically characterized. (A) Drop dilution assays were performed to test for sensitivity of the strains to cycloheximide, phleomycin, as well as to hyperosmotic stress (NaCl), cold stress (25 °C), and heat stress (37 °C). (B) Cells were grown on 3-AT containing plates to analyze adhesive growth. The plate was photographed before and after treatment with a constant stream of water (shorter and longer wash). (C) The asc1− strain and the double deletion strains asc1− ∆bre5 and asc1− ∆ubp3 were complemented with plasmid-borne ASC1. As controls, the wild-type strain and all single and double deletion strains were transformed with the empty vector (EV). Drop dilution growth assays were performed to test for sensitivity to cycloheximide and hyperosmotic stress (NaCl).
Bilsland and colleagues described the sensitivity of Δbre5 and Δubp3 cells to the DNA damaging agent phleomycin [57]. Growth in the presence of 4.5 μg/mL phleomycin is significantly impaired for asc1− cells and to a lesser extent for Δbre5 and Δubp3 cells. Double deletion strains asc1− Δbre5 and asc1− Δubp3 do not show synergistic growth impairment and resemble the asc1− phenotype upon phleomycin treatment (Figure 4A). Adhesive cell growth of wild-type S. cerevisiae cells of the genetic Σ1278b background is generally caused by nutrient depletion, for example, starvation through prolonged growth on solid medium or in the presence of starvation-inducing agents (e.g., the histidine analog 3-aminotriazole causing amino acid starvation; shown in Figure 4B). Patches of wild-type cells remain stuck on the surface after exposing them to a stream of water (Figure 4B, ASC1 pie slice). Also, Δbre5 and Δubp3 cells stick to the surface (Figure 4B, ASC1 Δbre5 and ASC1 Δubp3 pie slices). Patches of asc1− Δbre5 and asc1− Δubp3 cells, however, are easily washed off the plates after only a short exposure time to a water stream, while the patch of asc1− cells still remains stuck at the surface (Figure 4B, first wash). A more intensive washing also removed the patches of asc1− cells from the surface in contrast to the ASC1 wild-type cells that still stick (Figure 4B, second wash). Transformation of the different asc1− strains (asc1−, asc1− Δbre5, and asc1− Δubp3) with the ASC1 gene on a centromere plasmid complemented the Asc1-dependent growth phenotypes as expected (Figure 4C, for growth in the presence of the translation inhibitor cycloheximide, 0.05 μg/mL, and growth at hyperosmotic conditions, 70 mM NaCl). In summary, the phenotypic characterization of ASC1, UBP3, and BRE5 single and double deletion strains also revealed genetic interactions that give further evidence for a cooperation of the encoded proteins at the hr40S.
4. Discussion
Protein phosphorylation and ubiquitination steer cellular decisions according to numerous intra- and extra-cellular signals. They cause remodeling of protein–protein interactions and, thus, of protein complexes and microenvironments. We have analyzed the head region of the 40S ribosomal subunit (hr40S) and consider protein remodeling there both as a prerequisite for and a consequence of ribosome activity and homeostasis. Initiation of mRNA translation, as well as ribosome stalling, might influence the protein composition and arrangement at hr40S. Now, we have analyzed this microenvironment with BioID capture experiments from two adjacent perspectives, namely the Asc1 and the Rps2 perspectives. As expected, the two neighboring viewpoints yield a similar but not identical repertoire of proximal proteins (Figure 2B). Variations in the lengths of the linker sequence between bait and biotin ligase and the occupation of different adjacent perspectives will contribute to the spatial resolution and orientation of proteins identified with BioID within a microenvironment.
The accumulation of nucleocytoplasmic-shuttling mRNA-binding proteins and of subunits of the translation initiation complex eIF3 (a/Rpg1 and g/Tif35) at the hr40S indicates that substantial cargo reception and translation initiation processes take place at this site. We provide evidence that yRACK1/Asc1 affects the positioning of the mRNA binding proteins Scp160, Sgn1, and the paralogous proteins Sro9 and Slf1. Scp160 is a well-established Asc1 interacting protein, and it was repeatedly shown that ribosome-association of Scp160 is reduced in the absence of Asc1 [58,59]. However, we made the converse observation and captured more Scp160 from the ribosomal Rps2-perspective when Asc1 was absent. An explanation might be that Asc1 shields Scp160 from biotinylation through Rps2-BirA*. Alternatively, the assays to evaluate the ribosome association of Scp160 might reflect the strength of its binding rather than its overall localization at ribosomes. The position of Scp160 at the ribosome in Asc1-deficient cells might be shifted, rendering Scp160 prone to dissociation from the ribosome during ultracentrifugation. In contrast, BioID identifies proximities without necessarily reflecting the affinity of interactions.
The mRNA-binding protein Sro9 has been described as a subunit of the heat-shock 70 protein (Hsp70)-containing complex (HMC). This complex represses the transcription factor heme activator protein 1 (Hap1) in the absence of heme [60]. Hap1, in turn, mediates oxygen sensing and heme signaling. There is a genetic link between ASC1 and oxygen-related response mechanisms: ASC1 was originally discovered in a genetic screen by Verdière and colleagues as a gene in which inactivation allowed for the growth of otherwise inviable heme-depleted hap1 mutant cells [61]. In line with this, we observed an impact of Asc1 on the ability of cells to switch from fermentative to respiratory growth [3]. In the presence of heme, enhanced association of Hsp90 with the HMC complex causes Hap1 activation [60]. Coincidently, with the increased Rps2-BioID capture of Sro9 from asc1− cells, a similar increase of the Hsp90 family chaperone Hsp82 was observed. The Sro9 paralog Slf1 has also been described as a key regulator of the oxidative stress response [55]. The results presented here indicate that cooperation of Asc1 with Sro9, Slf1, and heat shock proteins at the hr40S might account for the genetic link between ASC1 and HAP1. In mammalian cells, oxygen sensing and the transcriptional response of oxygen-dependent genes are mediated through HIF1-α, a transcription activator protein whose activity and stability are also controlled through a phosphorylation/ubiquitination-mediated interplay with heat-shock proteins and RACK1 [62]. We consider the ribosomal head region as a decisive site for Asc1/RACK1-mediated signal transfer to regulate the stability and nuclear transport of Hap1/HIF1-α.
Def1 is a CUE domain-containing protein that resides predominantly in the cytoplasm. However, it translocates into the nucleus to designate mRNA polymerase II for ubiquitination and proteasome-mediated degradation [19]. Prior to its nuclear translocation, full length Def1 obviously resides at the hr40S within the cytoplasm as observed with our Asc1-BioID experiments and now confirmed with the Rps2-BioID. The positioning of Def1 at the hr40S and its phosphorylation at threonine 258 and serine 260 are Asc1-dependent (results here and Schmitt et al., 2017 [10], respectively), and might together be important for the proteasome-mediated cropping of its Q-rich C-terminus that precedes its nuclear transport [19].
The deubiquitinase Ubp3 and its cofactor Bre5 (USP10 and G3BP1 in mammalian cells, respectively) are important factors controlling ribophagy and mitophagy [29,63,64] and were previously found to be Asc1-proximal [18]. Here, we observed the Asc1-dependent localization of Bre5 at the hr40S, as examined from the Rps2-BirA* viewpoint. We assume that the same is true for Ubp3, but strong background biotinylation in ASC1 wild-type cells expressing BirA* alone (see biotinylation within the negative control in Figure 1B, middle lane) did not lead to Rps2-BirA*-mediated Ubp3 enrichment ratios from ASC1 wild-type cells. Nevertheless, there was significant Ubp3 deprivation in asc1− cells at the hr40S (Figure 2A), leading us to cautiously claim Asc1-dependency for the Ubp3-localization as well. Beyond Asc1-dependent localization of Ubp3-Bre5 to the ribosomal hr40S, we showed evidence for the genetic interaction of Asc1 with Ubp3-Bre5 under various stress conditions. The Ubp3-Bre5 complex plays an essential role in the assembly of cytoplasmatic non-membranous ribonucleoprotein (RNP) stress granules that encompass stalled translation pre-initiation complexes [65]. A balanced mono-ubiquitination and deubiquitination of the ribosomal protein S3 (Rps3), mediated by Ubp3 and the E3 ubiquitin ligase Hel2 (ZNF598 in mammalian cells), has been demonstrated to be important for ribosome-associated quality control and autophagy processes [27]. Previously, we found Hel2 proximal to Asc1 when investigated from the Asc1-BirA* perspective [18]. However, it was not visible from the Rps2-BirA* viewpoint in this study. Rps3 gets ubiquitylated at K212 and K223 within its C-terminal tail that is physically shielded through and bound by Asc1 (Figure 1A). Also, acetylation and succinylation of K212 of Rps3 and phosphorylation of close-by sites have been described [66,67,68]. Both lysine residues are unshielded in asc1− cells leading to lysine biotinylation through Rps2-BirA* and, accordingly, to strong Rps3 biotin capture. Interaction between mRNA and Rps3 at the ribosomal mRNA entry tunnel at the 40S subunit supports both translation initiation accuracy [69] and the endonucleolytic cleavage of aberrant mRNAs during no-go decay (NGD) [25]. Ribosome collision after stalling is critical for triggering RQC activities, for example, endonucleolytic mRNA cleavage [26]. In mammalian systems, stalling is recognized by the Hel2 ortholog ZNF598 [70]. In S. cerevisiae, Hel2 was shown to bind to mRNAs at translating 40S ribosomal subunits preferentially near stop codons through the interaction with 18S rRNA and Asc1 [71]. Asc1 and Rps3 also act together in nonfunctional 18S rRNA decay (18S NRD) [72]. A Cryo-EM structure of two collided 80S ribosomes revealed that the hr40S-Asc1 microenvironment is the main physical contact site, with both Asc1 molecules directly interacting [73]. Collision might couple two 80S ribosomes via Asc1 homodimerization that, in turn, displace the C-terminal tail of Rps3 that binds Asc1 at its dimerization interface [15,74]. A displaced C-terminal Rps3 arm could then be prone to protein modifications that might be important to trigger subsequent steps in ribosome conversion or conservation. The ribosome preservation factor, Stm1, cogent BioID candidate in both Asc1- and Rps2-BioID experiments, could then be inserted into the non-translating ribosomes as a surrogate for degraded mRNA to function as a subunit clamping factor that conserves ribosomes during early starvation periods.
Severe nutrient starvation causes ribophagy, a mechanism for bulk degradation of non-translating ribosomes within the vacuole [29]. Autophagosome formation involves the generation of phosphatidylinositol 3-phosphate that is generated by phosphatidylinositol 3-kinases (PI3Ks). Our BioID experiments revealed that an integral component of the PI3K complexes I and II, Vps30, occupies a position at the hr40S proximal to Rps2 in the absence of Asc1. Mammalian RACK1 was described to promote the formation of a PI3K complex comprising the ortholog of Vps30, Beclin-1, and, thus, autophagosome induction [75]. Future studies will address the question of whether there are physiological conditions that induce a displacement of Asc1 in favor of Vps30 localization at the hr40S. This might target or activate PI3K complexes to initiate ribophagy.
There is considerable heterogeneity of ribosomes and probably of the regulatory head region of the 40S subunit as well. Depending on the cellular signals, ribosomes are supposed to be more generally or context-specifically regulated in terms of activity and homeostasis. Our Asc1- and Rps2-BioID experiments do not yet resolve the diversity at the hr40S. However, they provide a first insight into this complex microenvironment. Improved faster and smaller BioID versions will contribute to further temporally resolved protein remodeling within microenvironments [76,77]. Emerging split-BioID approaches will allow the detection of common microenvironments of proximal proteins. This method combines two chosen proximal proteins fused to inactive halves of a biotin ligase, which will only become active upon protein interaction [31]. Such context-specific split-BioID experiments, in combination with genetically engineered protein isoforms and/or specific signals/growth conditions, are planned to dissect the diversity of proteins identified and to break down the analyses of the dynamic hr40S microenvironments to specific sets of ribosomes.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4409/8/11/1384/s1, Table S1: LC-MS identification and quantification data for proteins identified in proximity of Rps2-BirA*. Data correspond to Figure 2A.
Author Contributions
Conceptualization, O.V. and K.S.; investigation, O.V. and K.S.; methodology, O.V. and K.S.; formal analysis, K.S.; validation, K.S.; data curation, O.V. and K.S.; writing—original draft preparation, O.V. and K.S.; writing—review and editing, O.V. and K.S.; visualization, K.S.; funding acquisition, O.V.
Funding
This work was supported by the DFG grants VA 352/2-2, DFG-GZ: A 630 (ZUK41/1), DFG-GZ: INST 186/1230-1 FUGG.
Acknowledgments
We thank Gerhard H. Braus for his general support. We thank Anna Lao Dudek for her technical assistance and proofreading of the manuscript. We thank our graduate students Benedikt Ni, Jonas Jennrich, and Diana Zbarsky, and especially Olga Steinbrenner, for their contribution. We thank Heike Krebber for providing the Rps3-antibody.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ferreira-Cerca, S.; Pöll, G.; Kühn, H.; Neueder, A.; Jakob, S.; Tschochner, H.; Milkereit, P. Analysis of the in vivo assembly pathway of eukaryotic 40S ribosomal proteins. Mol. Cell 2007, 28, 446–457. [Google Scholar] [CrossRef]
- Heuer, A.; Thomson, E.; Schmidt, C.; Berninghausen, O.; Becker, T.; Hurt, E.; Beckmann, R. Cryo-EM structure of a late pre-40S ribosomal subunit from Saccharomyces cerevisiae. Elife 2017, 6. [Google Scholar] [CrossRef] [PubMed]
- Rachfall, N.; Schmitt, K.; Bandau, S.; Smolinski, N.; Ehrenreich, A.; Valerius, O.; Braus, G.H. RACK1/Asc1p, a ribosomal node in cellular signaling. Mol. Cell. Proteom. 2013, 12, 87–105. [Google Scholar] [CrossRef] [PubMed]
- Valerius, O.; Kleinschmidt, M.; Rachfall, N.; Schulze, F.; López Marín, S.; Hoppert, M.; Streckfuss-Bömeke, K.; Fischer, C.; Braus, G.H. The Saccharomyces homolog of mammalian RACK1, Cpc2/Asc1p, is required for FLO11-dependent adhesive growth and dimorphism. Mol. Cell. Proteom. 2007, 6, 1968–1979. [Google Scholar] [CrossRef] [PubMed]
- Jannot, G.; Bajan, S.; Giguère, N.J.; Bouasker, S.; Banville, I.H.; Piquet, S.; Hutvagner, G.; Simard, M.J. The ribosomal protein RACK1 is required for microRNA function in both C. elegans and humans. EMBO Rep. 2011, 12, 581–586. [Google Scholar] [CrossRef]
- Li, S.; Esterberg, R.; Lachance, V.; Ren, D.; Radde-Gallwitz, K.; Chi, F.; Parent, J.L.; Fritz, A.; Chen, P. Rack1 is required for Vangl2 membrane localization and planar cell polarity signaling while attenuating canonical Wnt activity. Proc. Natl. Acad. Sci. USA 2011, 108, 2264–2269. [Google Scholar] [CrossRef] [PubMed]
- Volta, V.; Beugnet, A.; Gallo, S.; Magri, L.; Brina, D.; Pesce, E.; Calamita, P.; Sanvito, F.; Biffo, S. RACK1 depletion in a mouse model causes lethality, pigmentation deficits and reduction in protein synthesis efficiency. Cell. Mol. Life Sci. 2012, 70, 1439–1450. [Google Scholar] [CrossRef] [PubMed]
- Wehner, P.; Shnitsar, I.; Urlaub, H.; Borchers, A. RACK1 is a novel interaction partner of PTK7 that is required for neural tube closure. Development 2011, 138, 1321–1327. [Google Scholar] [CrossRef]
- Li, J.J.; Xie, D. RACK1, a versatile hub in cancer. Oncogene 2015, 34, 1890–1898. [Google Scholar] [CrossRef]
- Schmitt, K.; Smolinski, N.; Neumann, P.; Schmaul, S.; Hofer-Pretz, V.; Braus, G.H.; Valerius, O. Asc1p/RACK1 Connects Ribosomes to Eukaryotic Phosphosignaling. Mol. Cell. Biol. 2017, 37, e00279-16. [Google Scholar] [CrossRef]
- Dudley, A.M.; Janse, D.M.; Tanay, A.; Shamir, R.; Church, G.M. A global view of pleiotropy and phenotypically derived gene function in yeast. Mol. Syst. Biol. 2005, 1, 2005.0001. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Lee, I.; Moradi, E.; Hung, N.J.; Johnson, A.W.; Marcotte, E.M. Rational extension of the ribosome biogenesis pathway using network-guided genetics. PLoS Biol. 2009, 7, e1000213. [Google Scholar] [CrossRef] [PubMed]
- Tigano, M.; Ruotolo, R.; Dallabona, C.; Fontanesi, F.; Barrientos, A.; Donnini, C.; Ottonello, S. Elongator-dependent modification of cytoplasmic tRNALysUUU is required for mitochondrial function under stress conditions. Nucleic Acids Res. 2015, 43, 8368–8380. [Google Scholar] [CrossRef] [PubMed]
- Xia, L.; Jaafar, L.; Cashikar, A.; Flores-Rozas, H. Identification of genes required for protection from doxorubicin by a genome-wide screen in Saccharomyces cerevisiae. Cancer Res. 2007, 67, 11411–11418. [Google Scholar] [CrossRef] [PubMed]
- Ben-Shem, A.; Garreau de Loubresse, N.; Melnikov, S.; Jenner, L.; Yusupova, G.; Yusupov, M. The structure of the eukaryotic ribosome at 3.0 A resolution. Science 2011, 334, 1524–1529. [Google Scholar] [CrossRef] [PubMed]
- Lomakin, I.B.; Steitz, T.A. The initiation of mammalian protein synthesis and mRNA scanning mechanism. Nature 2013, 500, 307–311. [Google Scholar] [CrossRef]
- Roux, K.J.; Kim, D.I.; Raida, M.; Burke, B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 2012, 196, 801–810. [Google Scholar] [CrossRef]
- Opitz, N.; Schmitt, K.; Hofer-Pretz, V.; Neumann, B.; Krebber, H.; Braus, G.H.; Valerius, O. Capturing the Asc1p/Receptor for Activated C Kinase 1 (RACK1) Microenvironment at the Head Region of the 40S Ribosome with Quantitative BioID in Yeast. Mol. Cell. Proteom. 2017, 16, 2199–2218. [Google Scholar] [CrossRef]
- Wilson, M.D.; Harreman, M.; Taschner, M.; Reid, J.; Walker, J.; Erdjument-Bromage, H.; Tempst, P.; Svejstrup, J.Q. Proteasome-mediated processing of Def1, a critical step in the cellular response to transcription stress. Cell 2013, 154, 983–995. [Google Scholar] [CrossRef]
- Ikeuchi, K.; Izawa, T.; Inada, T. Recent Progress on the Molecular Mechanism of Quality Controls Induced by Ribosome Stalling. Front. Genet. 2019, 9, 743. [Google Scholar] [CrossRef]
- Sitron, C.S.; Park, J.H.; Brandman, O. Asc1, Hel2, and Slh1 couple translation arrest to nascent chain degradation. RNA 2017, 23, 798–810. [Google Scholar] [CrossRef] [PubMed]
- Ikeuchi, K.; Inada, T. Ribosome-associated Asc1/RACK1 is required for endonucleolytic cleavage induced by stalled ribosome at the 3′ end of nonstop mRNA. Sci. Rep. 2016, 6, 28234. [Google Scholar] [CrossRef] [PubMed]
- Kuroha, K.; Akamatsu, M.; Dimitrova, L.; Ito, T.; Kato, Y.; Shirahige, K.; Inada, T. Receptor for activated C kinase 1 stimulates nascent polypeptide-dependent translation arrest. EMBO Rep. 2010, 11, 956–961. [Google Scholar] [CrossRef] [PubMed]
- Sitron, C.S.; Brandman, O. CAT tails drive degradation of stalled polypeptides on and off the ribosome. Nat. Struct. Mol. Biol. 2019, 26, 450–459. [Google Scholar] [CrossRef] [PubMed]
- Simms, C.L.; Kim, K.Q.; Yan, L.L.; Qiu, J.; Zaher, H.S. Interactions between the mRNA and Rps3/uS3 at the entry tunnel of the ribosomal small subunit are important for no-go decay. PLoS Genet. 2018, 14, e1007818. [Google Scholar] [CrossRef]
- Simms, C.L.; Yan, L.L.; Zaher, H.S. Ribosome Collision Is Critical for Quality Control during No-Go Decay. Mol. Cell 2017, 68, 361–373. [Google Scholar] [CrossRef]
- Jung, Y.; Kim, H.D.; Yang, H.W.; Kim, H.J.; Jang, C.Y.; Kim, J. Modulating cellular balance of Rps3 mono-ubiquitination by both Hel2 E3 ligase and Ubp3 deubiquitinase regulates protein quality control. Exp. Mol. Med. 2017, 49, e390. [Google Scholar] [CrossRef]
- Sundaramoorthy, E.; Leonard, M.; Mak, R.; Liao, J.; Fulzele, A.; Bennett, E.J. ZNF598 and RACK1 Regulate Mammalian Ribosome-Associated Quality Control Function by Mediating Regulatory 40S Ribosomal Ubiquitylation. Mol. Cell 2017, 65, 751–760. [Google Scholar] [CrossRef]
- Kraft, C.; Deplazes, A.; Sohrmann, M.; Peter, M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat. Cell Biol. 2008, 10, 602–610. [Google Scholar] [CrossRef]
- Kihara, A.; Noda, T.; Ishihara, N.; Ohsumi, Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J. Cell Biol. 2001, 152, 519–530. [Google Scholar] [CrossRef]
- Schopp, I.M.; Amaya Ramirez, C.C.; Debeljak, J.; Kreibich, E.; Skribbe, M.; Wild, K.; Béthune, J. Split-BioID a conditional proteomics approach to monitor the composition of spatiotemporally defined protein complexes. Nat. Commun. 2017, 8, 15690. [Google Scholar] [CrossRef] [PubMed]
- Gibson, T.J. RACK1 research—Ships passing in the night? FEBS Lett. 2012, 586, 2787–2789. [Google Scholar] [CrossRef] [PubMed]
- Mumberg, D.; Müller, R.; Funk, M. Regulatable promoters of Saccharomyces cerevisiae: Comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994, 22, 5767–5768. [Google Scholar] [CrossRef] [PubMed]
- Gueldener, U.; Heinisch, J.; Koehler, G.J.; Voss, D.; Hegemann, J.H. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 2002, 30, e23. [Google Scholar] [CrossRef]
- Huh, W.K.; Falvo, J.V.; Gerke, L.C.; Carroll, A.S.; Howson, R.W.; Weissman, J.S.; O′Shea, E.K. Global analysis of protein localization in budding yeast. Nature 2003, 425, 686–691. [Google Scholar] [CrossRef]
- Kushnirov, V.V. Rapid and reliable protein extraction from yeast. Yeast 2000, 16, 857–860. [Google Scholar] [CrossRef]
- Wessel, D.; Flügge, U.I. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 1984, 138, 141–143. [Google Scholar] [CrossRef]
- Shevchenko, A.; Wilm, M.; Vorm, O.; Mann, M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 1996, 68, 850–858. [Google Scholar] [CrossRef]
- Rappsilber, J.; Ishihama, Y.; Mann, M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 2003, 75, 663–670. [Google Scholar] [CrossRef]
- Rappsilber, J.; Mann, M.; Ishihama, Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007, 2, 1896–1906. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Deutsch, E.W.; Csordas, A.; Sun, Z.; Jarnuczak, A.; Perez-Riverol, Y.; Ternent, T.; Campbell, D.S.; Bernal-Llinares, M.; Okuda, S.; Kawano, S.; et al. The ProteomeXchange consortium in 2017: Supporting the cultural change in proteomics public data deposition. Nucleic Acids Res. 2017, 45, D1100–D1106. [Google Scholar] [CrossRef] [PubMed]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef]
- Wilson, K.P.; Shewchuk, L.M.; Brennan, R.G.; Otsuka, A.J.; Matthews, B.W. Escherichia coli biotin holoenzyme synthetase/bio repressor crystal structure delineates the biotin- and DNA-binding domains. Proc. Natl. Acad. Sci. USA 1992, 89, 9257–9261. [Google Scholar] [CrossRef] [PubMed]
- Chiu, W.L.; Wagner, S.; Herrmannová, A.; Burela, L.; Zhang, F.; Saini, A.K.; Valásek, L.; Hinnebusch, A.G. The C-terminal region of eukaryotic translation initiation factor 3a (eIF3a) promotes mRNA recruitment, scanning, and, together with eIF3j and the eIF3b RNA recognition motif, selection of AUG start codons. Mol. Cell. Biol. 2010, 30, 4415–4434. [Google Scholar] [CrossRef]
- Cuchalová, L.; Kouba, T.; Herrmannová, A.; Dányi, I.; Chiu, W.L.; Valásek, L. The RNA recognition motif of eukaryotic translation initiation factor 3g (eIF3g) is required for resumption of scanning of posttermination ribosomes for reinitiation on GCN4 and together with eIF3i stimulates linear scanning. Mol. Cell. Biol. 2010, 30, 4671–4686. [Google Scholar] [CrossRef]
- Black, J.J.; Musalgaonkar, S.; Johnson, A.W. Tsr4 Is a Cytoplasmic Chaperone for the Ribosomal Protein Rps2 in Saccharomyces cerevisiae. Mol. Cell. Biol. 2019, 39. [Google Scholar] [CrossRef]
- Rössler, I.; Embacher, J.; Pillet, B.; Murat, G.; Liesinger, L.; Hafner, J.; Unterluggauer, J.J.; Birner-Gruenberger, R.; Kressler, D.; Pertschy, B. Tsr4 and Nap1, two novel members of the ribosomal protein chaperOME. Nucleic Acids Res. 2019, 47, 6984–7002. [Google Scholar] [CrossRef]
- Van Dyke, N.; Pickering, B.F.; Van Dyke, M.W. Stm1p alters the ribosome association of eukaryotic elongation factor 3 and affects translation elongation. Nucleic Acids Res. 2009, 37, 6116–6125. [Google Scholar] [CrossRef]
- Hayashi, H.; Nagai, R.; Abe, T.; Wada, M.; Ito, K.; Takeuchi-Tomita, N. Tight interaction of eEF2 in the presence of Stm1 on ribosome. J. Biochem. 2018, 163, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Bolger, G.B. The RNA-binding protein SERBP1 interacts selectively with the signaling protein RACK1. Cell. Signal. 2017, 35, 256–263. [Google Scholar] [CrossRef] [PubMed]
- Röther, S.; Burkert, C.; Brünger, K.M.; Mayer, A.; Kieser, A.; Strässer, K. Nucleocytoplasmic shuttling of the La motif-containing protein Sro9 might link its nuclear and cytoplasmic functions. RNA 2010, 16, 1393–1401. [Google Scholar] [CrossRef] [PubMed]
- Sobel, S.G.; Wolin, S.L. Two yeast La motif-containing proteins are RNA-binding proteins that associate with polyribosomes. Mol. Biol. Cell 1999, 10, 3849–3862. [Google Scholar] [CrossRef]
- Kershaw, C.J.; Costello, J.L.; Castelli, L.M.; Talavera, D.; Rowe, W.; Sims, P.F.; Ashe, M.P.; Hubbard, S.J.; Pavitt, G.D.; Grant, C.M. The yeast La related protein Slf1p is a key activator of translation during the oxidative stress response. PLoS Genet. 2015, 11, e1004903. [Google Scholar] [CrossRef]
- Higashio, H.; Sato, K.; Nakano, A. Smy2p participates in COPII vesicle formation through the interaction with Sec23p/Sec24p subcomplex. Traffic 2008, 9, 79–93. [Google Scholar] [CrossRef]
- Bilsland, E.; Hult, M.; Bell, S.D.; Sunnerhagen, P.; Downs, J.A. The Bre5/Ubp3 ubiquitin protease complex from budding yeast contributes to the cellular response to DNA damage. DNA Repair 2007, 6, 1471–1484. [Google Scholar] [CrossRef]
- Baum, S.; Bittins, M.; Frey, S.; Seedorf, M. Asc1p, a WD40-domain containing adaptor protein, is required for the interaction of the RNA-binding protein Scp160p with polysomes. Biochem. J. 2004, 380, 823–830. [Google Scholar] [CrossRef]
- Coyle, S.M.; Gilbert, W.V.; Doudna, J.A. Direct link between RACK1 function and localization at the ribosome in vivo. Mol. Cell. Biol. 2009, 29, 1626–1634. [Google Scholar] [CrossRef]
- Lan, C.; Lee, H.C.; Tang, S.; Zhang, L. A novel mode of chaperone action: Heme activation of Hap1 by enhanced association of Hsp90 with the repressed Hsp70-Hap1 complex. J. Biol. Chem. 2004, 279, 27607–27612. [Google Scholar] [CrossRef]
- Chantrel, Y.; Gaisne, M.; Lions, C.; Verdière, J. The transcriptional regulator Hap1p (Cyp1p) is essential for anaerobic or heme-deficient growth of Saccharomyces cerevisiae: Genetic and molecular characterization of an extragenic suppressor that encodes a WD repeat protein. Genetics 1998, 148, 559–569. [Google Scholar] [PubMed]
- Liu, Y.V.; Baek, J.H.; Zhang, H.; Diez, R.; Cole, R.N.; Semenza, G.L. RACK1 competes with HSP90 for binding to HIF-1alpha and is required for O2-independent and HSP90 inhibitor-induced degradation of HIF-1alpha. Mol. Cell 2007, 25, 207–217. [Google Scholar] [CrossRef]
- Müller, M.; Kötter, P.; Behrendt, C.; Walter, E.; Scheckhuber, C.Q.; Entian, K.D.; Reichert, A.S. Synthetic quantitative array technology identifies the Ubp3-Bre5 deubiquitinase complex as a negative regulator of mitophagy. Cell Rep. 2015, 10, 1215–1225. [Google Scholar] [CrossRef]
- Ossareh-Nazari, B.; Bonizec, M.; Cohen, M.; Dokudovskaya, S.; Delalande, F.; Schaeffer, C.; Van Dorsselaer, A.; Dargemont, C. Cdc48 and Ufd3, new partners of the ubiquitin protease Ubp3, are required for ribophagy. EMBO Rep. 2010, 11, 548–554. [Google Scholar] [CrossRef]
- Nostramo, R.; Varia, S.N.; Zhang, B.; Emerson, M.M.; Herman, P.K. The Catalytic Activity of the Ubp3 Deubiquitinating Protease Is Required for Efficient Stress Granule Assembly in Saccharomyces cerevisiae. Mol. Cell. Biol. 2015, 36, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Henriksen, P.; Wagner, S.A.; Weinert, B.T.; Sharma, S.; Bacinskaja, G.; Rehman, M.; Juffer, A.H.; Walther, T.C.; Lisby, M.; Choudhary, C. Proteome-wide analysis of lysine acetylation suggests its broad regulatory scope in Saccharomyces cerevisiae. Mol. Cell. Proteom. 2012, 11, 1510–1522. [Google Scholar] [CrossRef]
- Swaney, D.L.; Beltrao, P.; Starita, L.; Guo, A.; Rush, J.; Fields, S.; Krogan, N.J.; Villén, J. Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat. Methods 2013, 10, 676–682. [Google Scholar] [CrossRef]
- Weinert, B.T.; Scholz, C.; Wagner, S.A.; Iesmantavicius, V.; Su, D.; Daniel, J.A.; Choudhary, C. Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation. Cell Rep. 2013, 4, 842–851. [Google Scholar] [CrossRef]
- Dong, J.; Aitken, C.E.; Thakur, A.; Shin, B.S.; Lorsch, J.R.; Hinnebusch, A.G. Rps3/uS3 promotes mRNA binding at the 40S ribosome entry channel and stabilizes preinitiation complexes at start codons. Proc. Natl. Acad. Sci. USA 2017, 114, E2126–E2135. [Google Scholar] [CrossRef] [PubMed]
- Juszkiewicz, S.; Chandrasekaran, V.; Lin, Z.; Kraatz, S.; Ramakrishnan, V.; Hegde, R.S. ZNF598 Is a Quality Control Sensor of Collided Ribosomes. Mol. Cell 2018, 72, 469–481.e467. [Google Scholar] [CrossRef] [PubMed]
- Winz, M.L.; Peil, L.; Turowski, T.W.; Rappsilber, J.; Tollervey, D. Molecular interactions between Hel2 and RNA supporting ribosome-associated quality control. Nat. Commun. 2019, 10, 563. [Google Scholar] [CrossRef] [PubMed]
- Limoncelli, K.A.; Merrikh, C.N.; Moore, M.J. ASC1 and RPS3: New actors in 18S nonfunctional rRNA decay. RNA 2017, 23, 1946–1960. [Google Scholar] [CrossRef] [PubMed]
- Ikeuchi, K.; Tesina, P.; Matsuo, Y.; Sugiyama, T.; Cheng, J.; Saeki, Y.; Tanaka, K.; Becker, T.; Beckmann, R.; Inada, T. Collided ribosomes form a unique structural interface to induce Hel2-driven quality control pathways. EMBO J. 2019, 38. [Google Scholar] [CrossRef]
- Yatime, L.; Hein, K.L.; Nilsson, J.; Nissen, P. Structure of the RACK1 dimer from Saccharomyces cerevisiae. J. Mol. Biol. 2011, 411, 486–498. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, Q.; Qiu, G.; Zhou, S.; Jing, Z.; Wang, J.; Wang, W.; Cao, J.; Han, K.; Cheng, Q.; et al. RACK1 Promotes Autophagy by Enhancing the Atg14L-Beclin 1-Vps34-Vps15 Complex Formation upon Phosphorylation by AMPK. Cell Rep. 2015, 13, 1407–1417. [Google Scholar] [CrossRef]
- Branon, T.C.; Bosch, J.A.; Sanchez, A.D.; Udeshi, N.D.; Svinkina, T.; Carr, S.A.; Feldman, J.L.; Perrimon, N.; Ting, A.Y. Efficient proximity labeling in living cells and organisms with TurboID. Nat. Biotechnol. 2018, 36, 880–887. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.I.; Jensen, S.C.; Noble, K.A.; Kc, B.; Roux, K.H.; Motamedchaboki, K.; Roux, K.J. An improved smaller biotin ligase for BioID proximity labeling. Mol. Biol. Cell 2016, 27, 1188–1196. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).