Differential Sensitivity to MEK Inhibitors Highlights Distinct Entosis Mechanisms in BxPC3 and MCF7 Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Cell Culture
2.3. Cell Viability Assay
2.4. Measurement of the MEK Kinase Activity
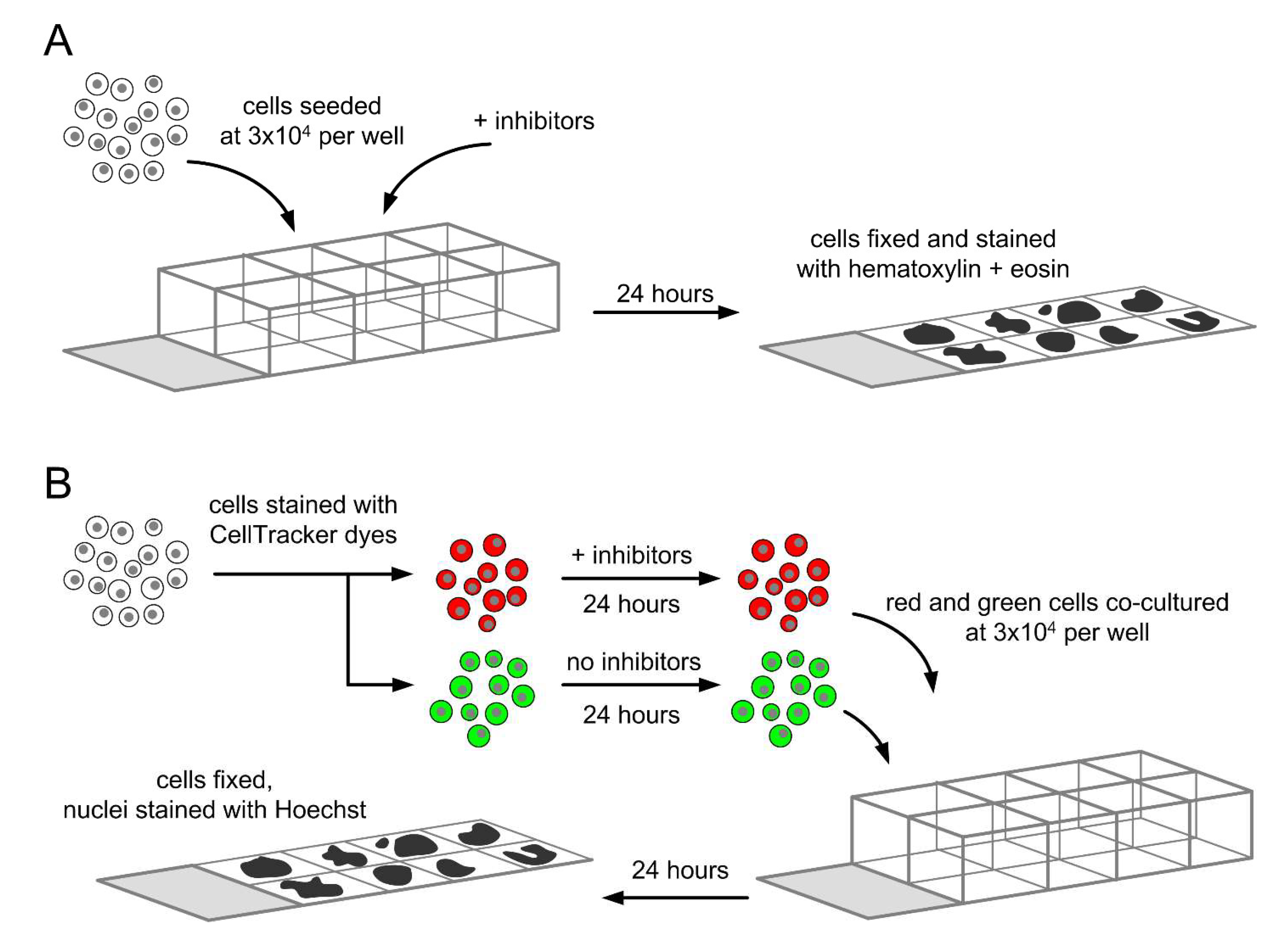
2.5. Determination of Total Entotic Index
2.6. Determination of Inner and Outer Entotic Indexes
2.7. Statistical Analysis
3. Results
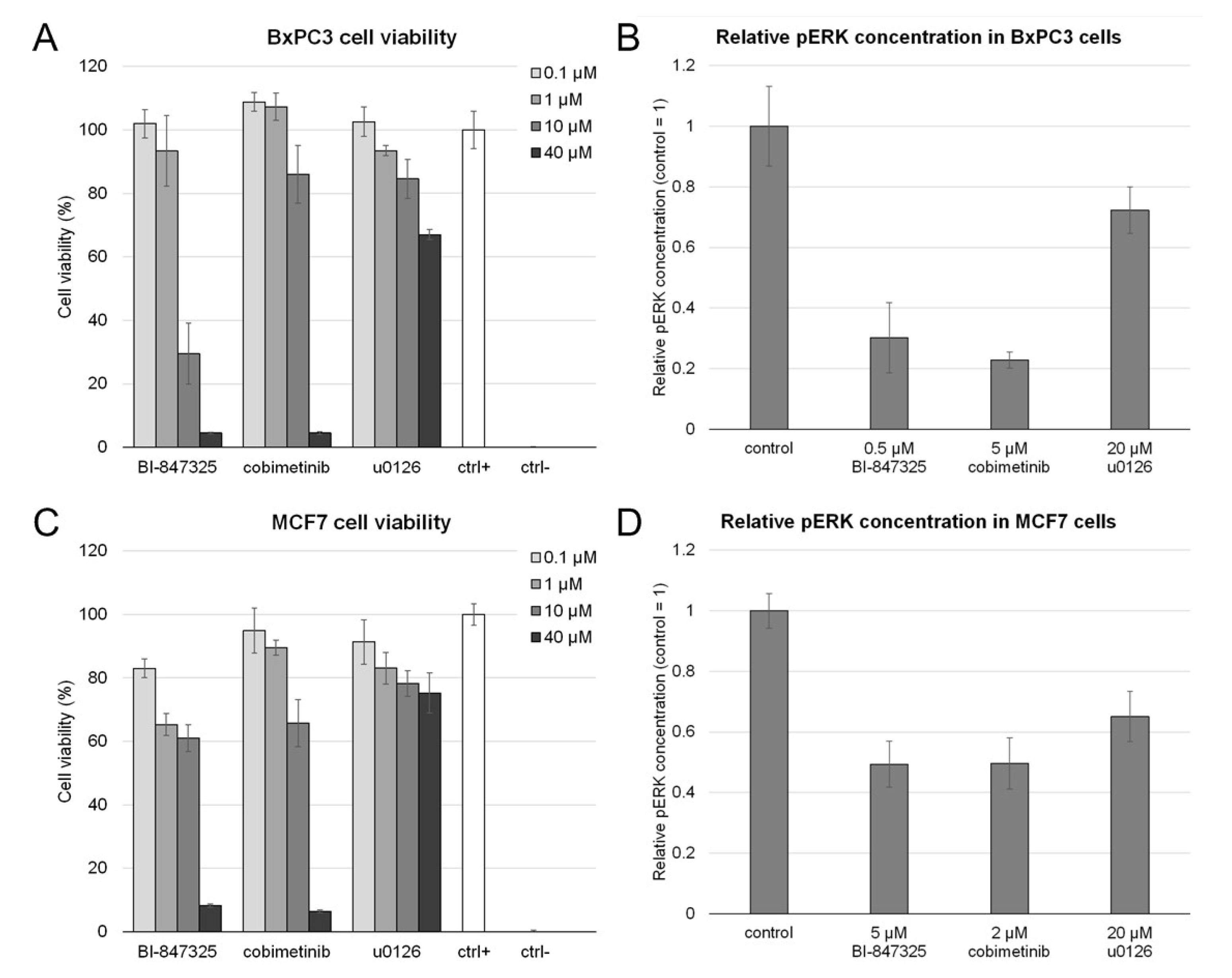
3.1. Toxicity of MEK Inhibitors and Their Effect on MEK Activity
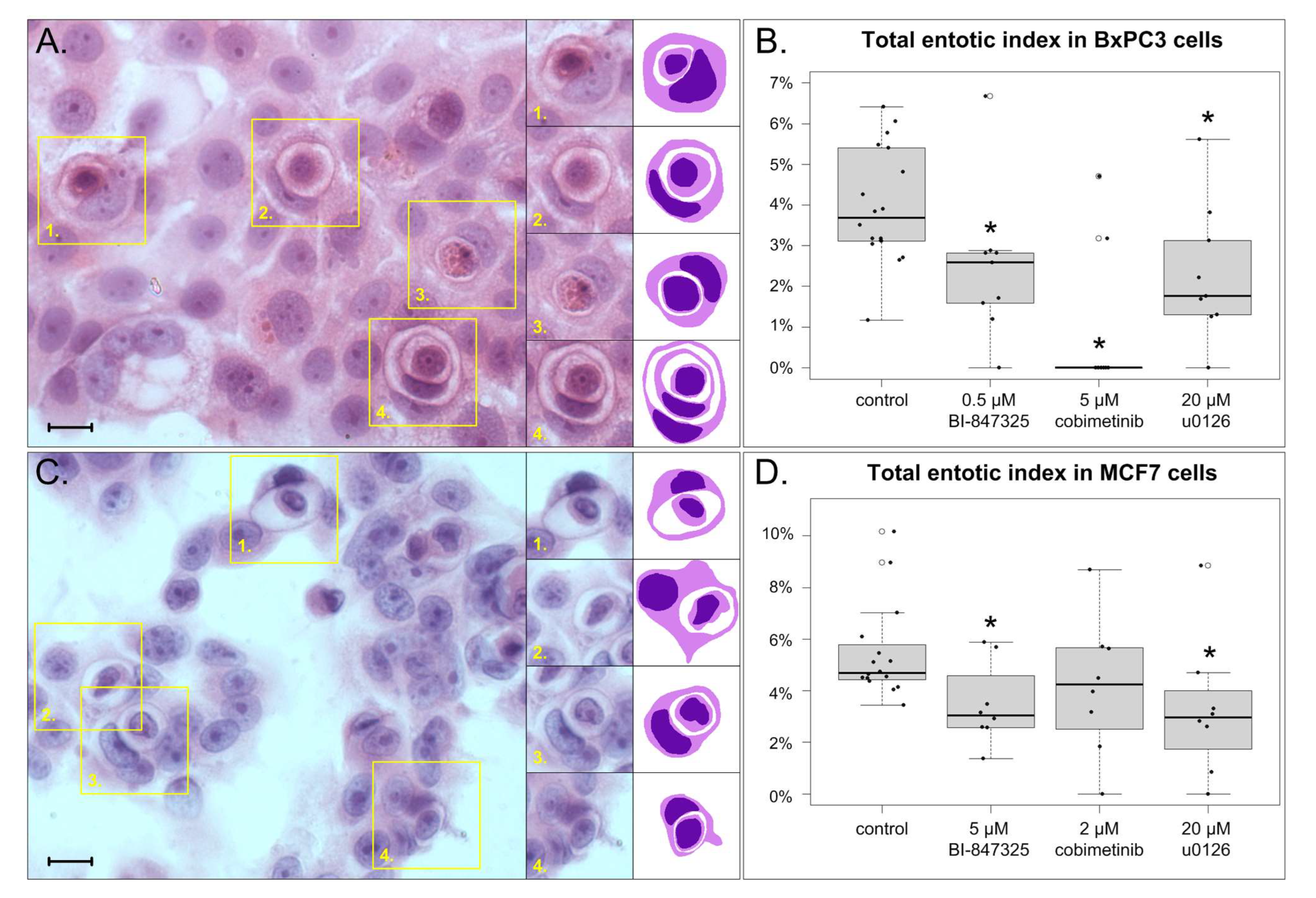
3.2. The Effect of MEK Inhibitors on Total Entotic Index
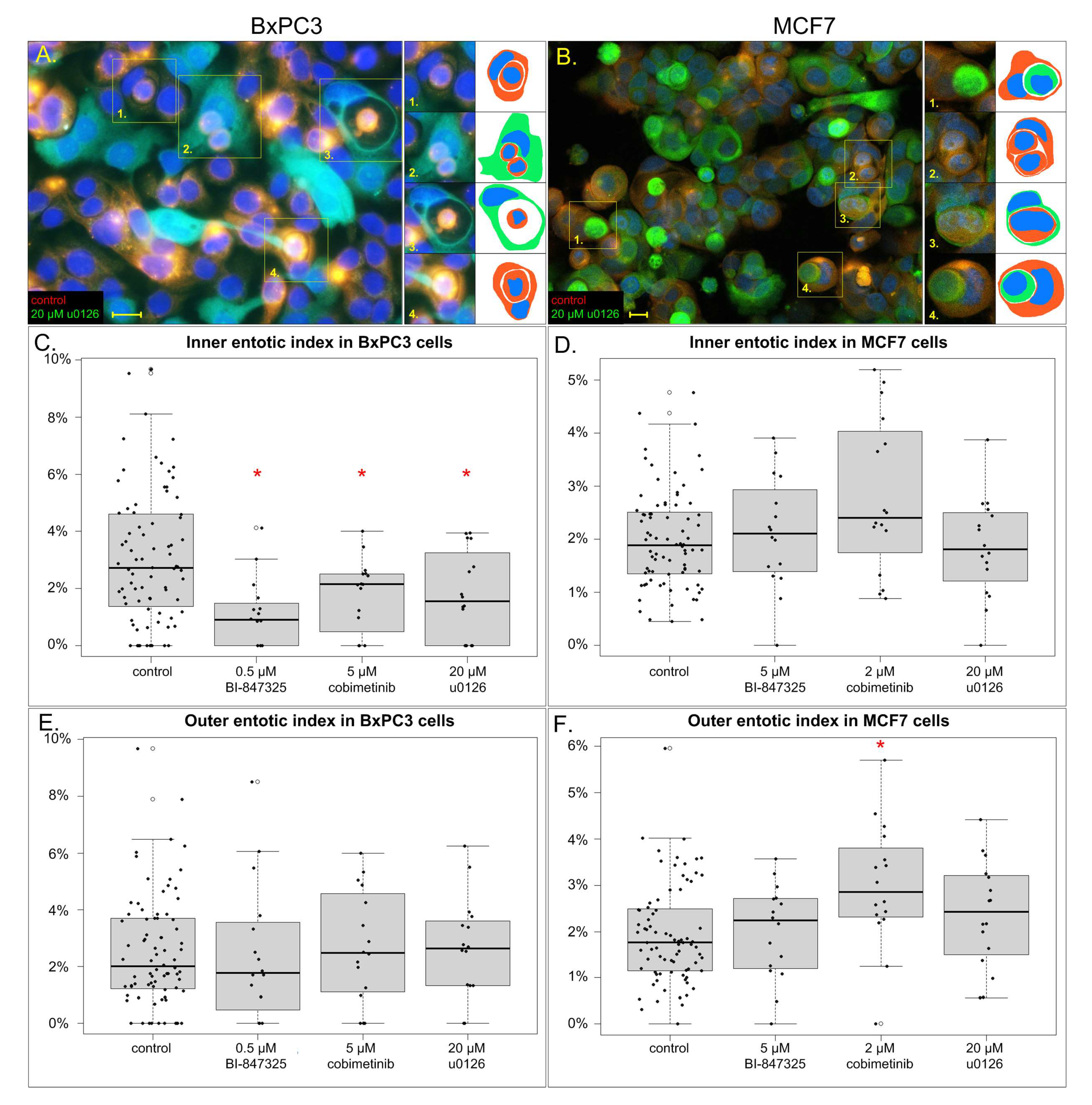
3.3. The Effect of MEK Inhibitors on Inner and Outer Entotic Indexes
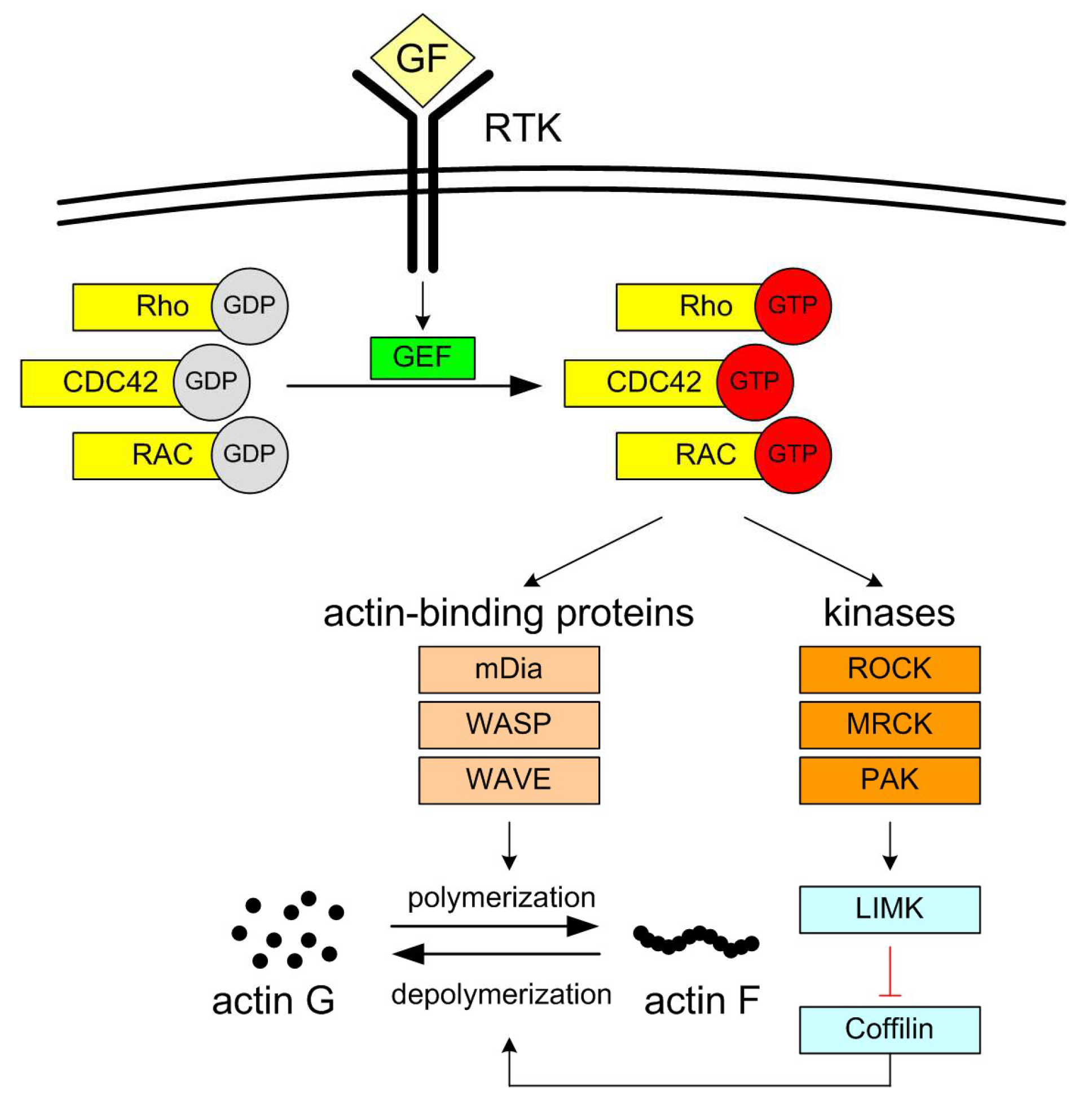
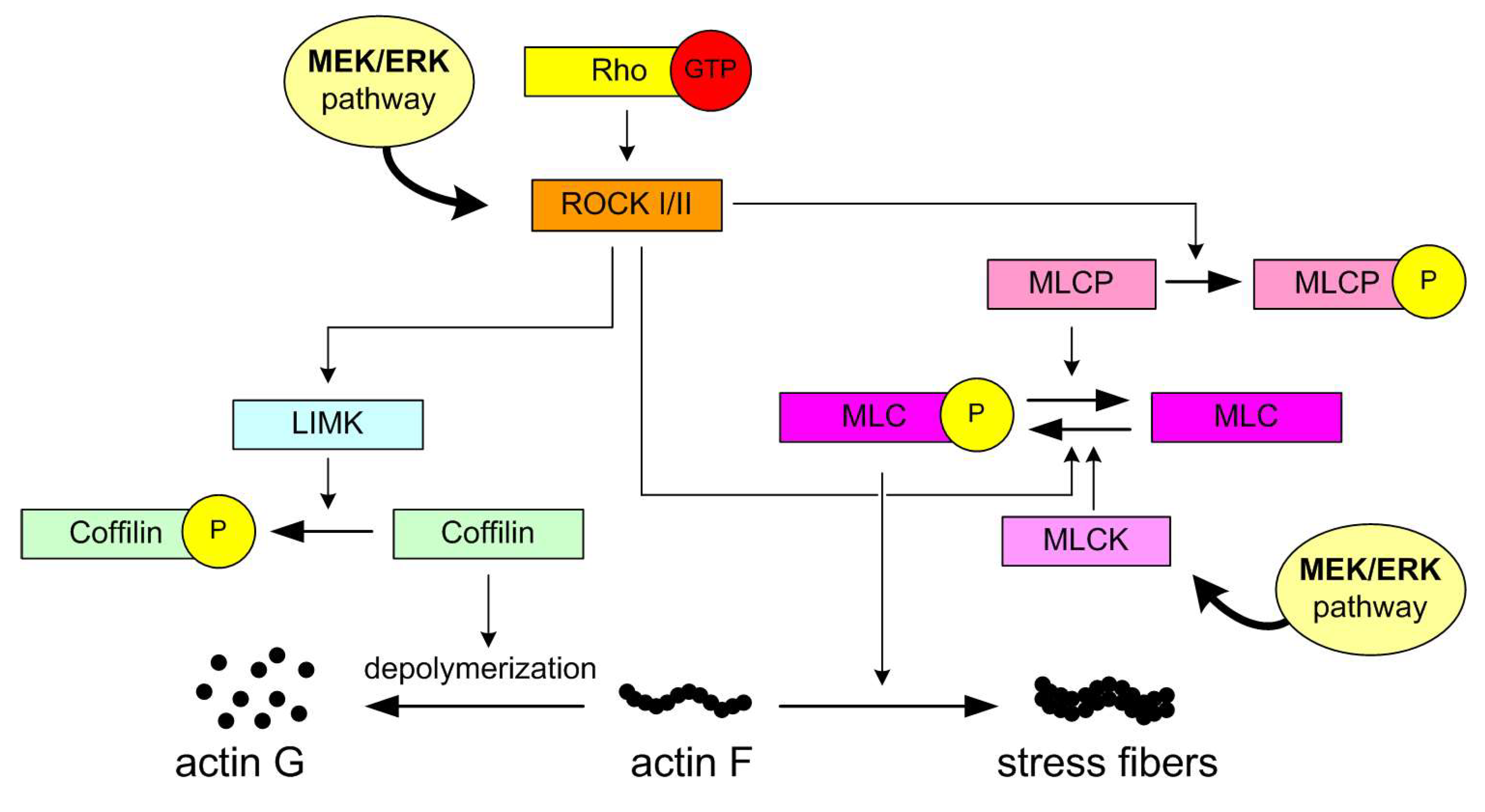
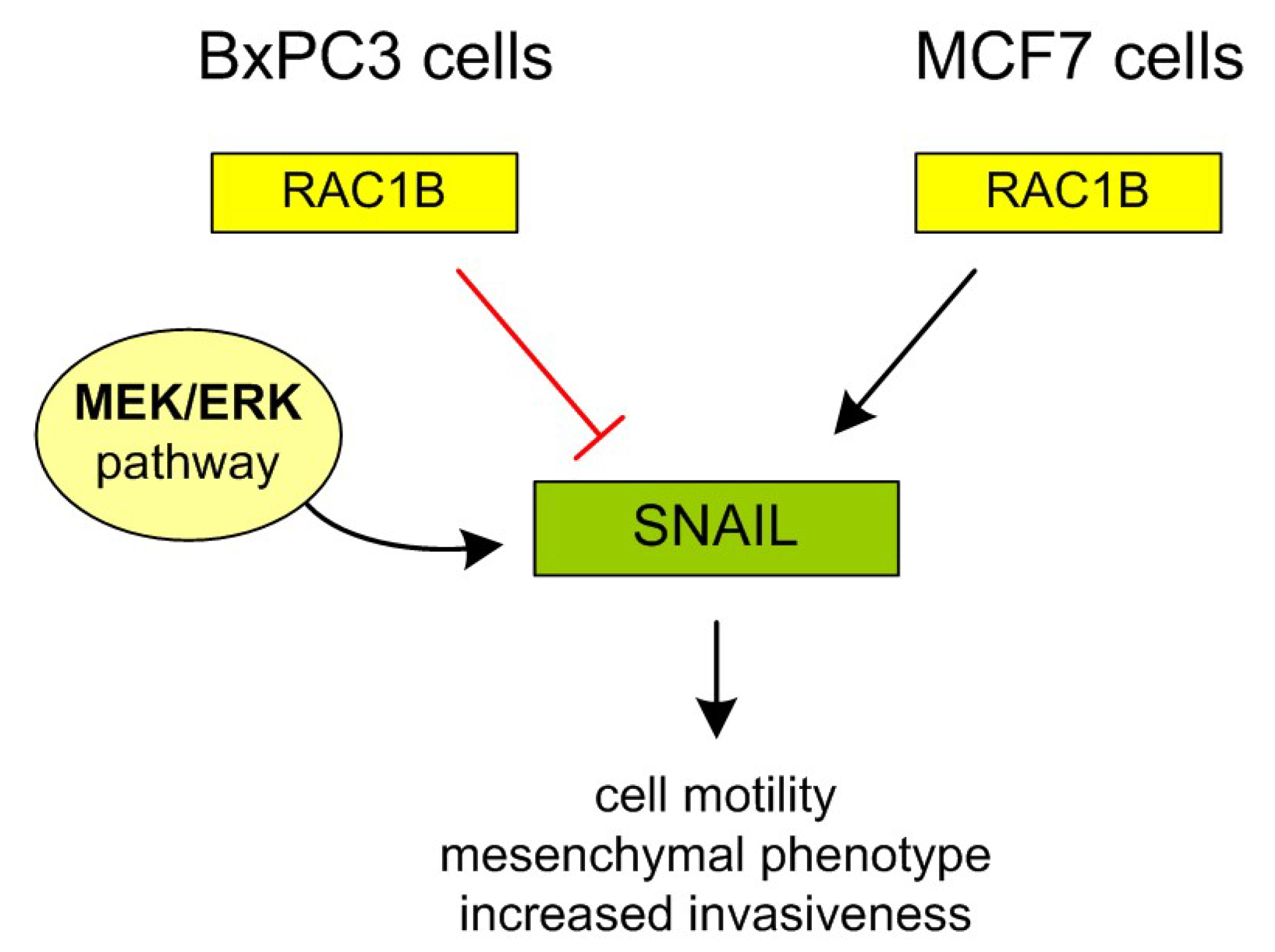
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ARHGAP22 | Rho GTPase-Activating Protein 22 |
| CIC | Cell in cell |
| CDC42 | Cell division cycle 42 |
| mDia | Protein diaphanous homolog |
| LIMK | LIM Kinase |
| MLCK | Myosin Light Chain Kinase |
| MLCP | Myosin Light Chain Phosphatase |
| MRCK | Myotonic dystrophy kinase-related Cdc42-binding kinase |
| PAK | p21 Activated Kinase |
| RAC | Ras-related C3 botulinum toxin substrate |
| ROCK | Rho-associated protein kinase |
| WASP | Wiskott–Aldrich syndrome protein, |
| WAVE | WASP family verprolin–homologous protein |
References
- Overholtzer, M.; Mailleux, A.A.; Mouneimne, G.; Normand, G.; Schnitt, S.J.; King, R.W.; Cibas, E.S.; Brugge, J.S. A nonapoptotic cell death process, entosis, that occurs by cell-in-cell invasion. Cell 2007, 131, 966–979. [Google Scholar] [CrossRef]
- Galluzzi, L.; Vitale, I.; Aaronson, S.A.; Abrams, J.M.; Adam, D.; Agostinis, P.; Alnemri, E.S.; Altucci, L.; Amelio, I.; Andrews, D.W.; et al. Molecular mechanisms of cell death: Recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018, 25, 486–541. [Google Scholar] [CrossRef]
- Liang, J.; Niu, Z.; Zhang, B.; Yu, X.; Zheng, Y.; Wang, C.; Ren, H.; Wang, M.; Ruan, B.; Qin, H.; et al. p53-dependent elimination of aneuploid mitotic offspring by entosis. Cell Death Differ. 2021, 28, 799–813. [Google Scholar] [CrossRef]
- Mlynarczuk-Bialy, I.; Dziuba, I.; Sarnecka, A.; Platos, E.; Kowalczyk, M.; Pels, K.K.; Wilczynski, G.M.; Wojcik, C.; Bialy, L.P. Entosis: From Cell Biology to Clinical Cancer Pathology. Cancers 2020, 12, 2481. [Google Scholar] [CrossRef]
- Mackay, H.L.; Moore, D.; Hall, C.; Birkbak, N.J.; Jamal-Hanjani, M.; Karim, S.A.; Phatak, V.M.; Pinon, L.; Morton, J.P.; Swanton, C.; et al. Genomic instability in mutant p53 cancer cells upon entotic engulfment. Nat. Commun. 2018, 9, 3070. [Google Scholar] [CrossRef]
- Schwegler, M.; Wirsing, A.M.; Schenker, H.M.; Ott, L.; Ries, J.M.; Buttner-Herold, M.; Fietkau, R.; Putz, F.; Distel, L.V. Prognostic Value of Homotypic Cell Internalization by Nonprofessional Phagocytic Cancer Cells. Biomed. Res. Int. 2015, 2015, 359392. [Google Scholar] [CrossRef]
- Durgan, J.; Tseng, Y.Y.; Hamann, J.C.; Domart, M.C.; Collinson, L.; Hall, A.; Overholtzer, M.; Florey, O. Mitosis can drive cell cannibalism through entosis. Elife 2017, 6, e27134. [Google Scholar] [CrossRef]
- Hamann, J.C.; Surcel, A.; Chen, R.; Teragawa, C.; Albeck, J.G.; Robinson, D.N.; Overholtzer, M. Entosis Is Induced by Glucose Starvation. Cell Rep. 2017, 20, 201–210. [Google Scholar] [CrossRef]
- Durgan, J.; Florey, O. Cancer cell cannibalism: Multiple triggers emerge for entosis. Biochim. Biophys. Acta Mol. Cell Res. 2018, 1865, 831–841. [Google Scholar] [CrossRef]
- Hinojosa, L.S.; Holst, M.; Baarlink, C.; Grosse, R. MRTF transcription and Ezrin-dependent plasma membrane blebbing are required for entotic invasion. J. Cell Biol. 2017, 216, 3087–3095. [Google Scholar] [CrossRef]
- Borensztejn, K.; Tyrna, P.; Gawel, A.M.; Dziuba, I.; Wojcik, C.; Bialy, L.P.; Mlynarczuk-Bialy, I. Classification of Cell-in-Cell Structures: Different Phenomena with Similar Appearance. Cells 2021, 10, 2569. [Google Scholar] [CrossRef]
- Tonnessen-Murray, C.A.; Frey, W.D.; Rao, S.G.; Shahbandi, A.; Ungerleider, N.A.; Olayiwola, J.O.; Murray, L.B.; Vinson, B.T.; Chrisey, D.B.; Lord, C.J.; et al. Chemotherapy-induced senescent cancer cells engulf other cells to enhance their survival. J. Cell Biol. 2019, 218, 3827–3844. [Google Scholar] [CrossRef]
- Kianfar, M.; Balcerak, A.; Chmielarczyk, M.; Tarnowski, L.; Grzybowska, E.A. Cell Death by Entosis: Triggers, Molecular Mechanisms and Clinical Significance. Int. J. Mol. Sci. 2022, 23, 4985. [Google Scholar] [CrossRef]
- Xia, P.; Zhou, J.; Song, X.; Wu, B.; Liu, X.; Li, D.; Zhang, S.; Wang, Z.; Yu, H.; Ward, T.; et al. Aurora A orchestrates entosis by regulating a dynamic MCAK-TIP150 interaction. J. Mol. Cell Biol. 2014, 6, 240–254. [Google Scholar] [CrossRef]
- Chen, R.; Ram, A.; Albeck, J.G.; Overholtzer, M. Entosis is induced by ultraviolet radiation. iScience 2021, 24, 102902. [Google Scholar] [CrossRef]
- Bozkurt, E.; Dussmann, H.; Salvucci, M.; Cavanagh, B.L.; Van Schaeybroeck, S.; Longley, D.B.; Martin, S.J.; Prehn, J.H.M. TRAIL signaling promotes entosis in colorectal cancer. J. Cell Biol. 2021, 220, e202010030. [Google Scholar] [CrossRef]
- Gaptulbarova, K.A.; Tsydenova, I.A.; Dolgasheva, D.S.; Kravtsova, E.A.; Ibragimova, M.K.; Vtorushin, S.V.; Litviakov, N.V. Mechanisms and significance of entosis for tumour growth and progression. Cell Death Discov. 2024, 10, 109. [Google Scholar] [CrossRef]
- Dziuba, I.; Gawel, A.M.; Tyrna, P.; Machtyl, J.; Olszanecka, M.; Pawlik, A.; Wojcik, C.; Bialy, L.P.; Mlynarczuk-Bialy, I. Homotypic Entosis as a Potential Novel Diagnostic Marker in Breast Cancer. Int. J. Mol. Sci. 2023, 24, 6819. [Google Scholar] [CrossRef]
- Miller, I.; Min, M.; Yang, C.; Tian, C.; Gookin, S.; Carter, D.; Spencer, S.L. Ki67 is a Graded Rather than a Binary Marker of Proliferation versus Quiescence. Cell Rep. 2018, 24, 1105–1112.e5. [Google Scholar] [CrossRef]
- Roberts, P.J.; Der, C.J. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene 2007, 26, 3291–3310. [Google Scholar] [CrossRef]
- Barbosa, R.; Acevedo, L.A.; Marmorstein, R. The MEK/ERK Network as a Therapeutic Target in Human Cancer. Mol. Cancer Res. 2021, 19, 361–374. [Google Scholar] [CrossRef]
- Song, J.; Xu, R.; Zhang, H.; Xue, X.; Ruze, R.; Chen, Y.; Yin, X.; Wang, C.; Zhao, Y. Cell-in-Cell–Mediated Entosis Reveals a Progressive Mechanism in Pancreatic Cancer. Gastroenterology 2023, 165, 1505–1521. [Google Scholar] [CrossRef]
- Kleensang, A.; Vantangoli, M.M.; Odwin-DaCosta, S.; Andersen, M.E.; Boekelheide, K.; Bouhifd, M.; Fornace, A.J., Jr.; Li, H.H.; Livi, C.B.; Madnick, S.; et al. Genetic variability in a frozen batch of MCF-7 cells invisible in routine authentication affecting cell function. Sci. Rep. 2016, 6, 28994. [Google Scholar] [CrossRef]
- Shao, T.; Zheng, Y.; Zhao, B.; Li, T.; Cheng, K.; Cai, W. Recombinant expression of different mutant K-ras gene in pancreatic cancer Bxpc-3 cells and its effects on chemotherapy sensitivity. Sci. China Life Sci. 2014, 57, 1011–1017. [Google Scholar] [CrossRef]
- Sun, Q.; Luo, T.; Ren, Y.; Florey, O.; Shirasawa, S.; Sasazuki, T.; Robinson, D.N.; Overholtzer, M. Competition between human cells by entosis. Cell Res. 2014, 24, 1299–1310. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; Foundation for Statistical Computing: Vienna, Austria, 2023. [Google Scholar]
- Druzhkova, I.; Potapov, A.; Ignatova, N.; Bugrova, M.; Shchechkin, I.; Lukina, M.; Shimolina, L.; Kolesnikova, E.; Shirmanova, M.; Zagaynova, E. Cell hiding in colorectal cancer: Correlation with response to chemotherapy in vitro and in vivo. Sci. Rep. 2024, 14, 28762. [Google Scholar] [CrossRef]
- Dziuba, I.; Gawel, A.M.; Tyrna, P.; Rybczynska, J.; Bialy, L.P.; Mlynarczuk-Bialy, I. Fate of Entosis: From the Beginning to the End in Untreated Advanced Breast Cancer. Int. J. Mol. Sci. 2023, 24, 12142. [Google Scholar] [CrossRef]
- Liu, J.; Wang, L.; Zhang, Y.; Li, S.; Sun, F.; Wang, G.; Yang, T.; Wei, D.; Guo, L.; Xiao, H. Induction of entosis in prostate cancer cells by nintedanib and its therapeutic implications. Oncol. Lett. 2019, 17, 3151–3162. [Google Scholar] [CrossRef]
- Puschel, F.; Munoz-Pinedo, C. In the Hunger Games, the Winner Takes Everything. Trends Biochem. Sci. 2017, 42, 763–764. [Google Scholar] [CrossRef][Green Version]
- Hayashi, A.; Yavas, A.; McIntyre, C.A.; Ho, Y.J.; Erakky, A.; Wong, W.; Varghese, A.M.; Melchor, J.P.; Overholtzer, M.; O’Reilly, E.M.; et al. Genetic and clinical correlates of entosis in pancreatic ductal adenocarcinoma. Mod. Pathol. 2020, 33, 1822–1831. [Google Scholar] [CrossRef]
- Kroemer, G.; Perfettini, J.L. Entosis, a key player in cancer cell competition. Cell Res. 2014, 24, 1280–1281. [Google Scholar] [CrossRef]
- Shaul, Y.D.; Seger, R. The MEK/ERK cascade: From signaling specificity to diverse functions. Biochim. Biophys. Acta 2007, 1773, 1213–1226. [Google Scholar] [CrossRef]
- Pritchard, C.A.; Hayes, L.; Wojnowski, L.; Zimmer, A.; Marais, R.M.; Norman, J.C. B-Raf acts via the ROCKII/LIMK/cofilin pathway to maintain actin stress fibers in fibroblasts. Mol. Cell. Biol. 2004, 24, 5937–5952. [Google Scholar] [CrossRef]
- Nguyen, D.H.; Catling, A.D.; Webb, D.J.; Sankovic, M.; Walker, L.A.; Somlyo, A.V.; Weber, M.J.; Gonias, S.L. Myosin light chain kinase functions downstream of Ras/ERK to promote migration of urokinase-type plasminogen activator-stimulated cells in an integrin-selective manner. J. Cell Biol. 1999, 146, 149–164. [Google Scholar] [CrossRef]
- Clayton, N.S.; Ridley, A.J. Targeting Rho GTPase Signaling Networks in Cancer. Front. Cell Dev. Biol. 2020, 8, 222. [Google Scholar] [CrossRef]
- Marcarian, H.Q.; Sivakoses, A.; Bothwell, A.L.M. Molecular Dynamics of Trogocytosis and Other Contact-Dependent Cell Trafficking Mechanisms in Tumor Pathogenesis. Cancers 2025, 17, 2268. [Google Scholar] [CrossRef]
- Matsuda, A.; Masuzawa, R.; Takahashi, K.; Takano, K.; Endo, T. MEK inhibitors and DA-Raf, a dominant-negative antagonist of the Ras-ERK pathway, prevent the migration and invasion of KRAS-mutant cancer cells. Cytoskeleton 2025, 82, 32–44. [Google Scholar] [CrossRef]
- Gong, S.; Xu, D.; Zhu, J.; Zou, F.; Peng, R. Efficacy of the MEK Inhibitor Cobimetinib and its Potential Application to Colorectal Cancer Cells. Cell. Physiol. Biochem. 2018, 47, 680–693. [Google Scholar] [CrossRef]
- Kawaguchi, K.; Igarashi, K.; Murakami, T.; Kiyuna, T.; Lwin, T.M.; Hwang, H.K.; Delong, J.C.; Clary, B.M.; Bouvet, M.; Unno, M.; et al. MEK inhibitors cobimetinib and trametinib, regressed a gemcitabine-resistant pancreatic-cancer patient-derived orthotopic xenograft (PDOX). Oncotarget 2017, 8, 47490–47496. [Google Scholar] [CrossRef]
- Samimi, H.; Haghpanah, V.; Irani, S.; Arefian, E.; Sohi, A.N.; Fallah, P.; Soleimani, M. Transcript-level regulation of MALAT1-mediated cell cycle and apoptosis genes using dual MEK/Aurora kinase inhibitor “BI-847325” on anaplastic thyroid carcinoma. Daru 2019, 27, 1–7. [Google Scholar] [CrossRef]
- Sini, P.; Gurtler, U.; Zahn, S.K.; Baumann, C.; Rudolph, D.; Baumgartinger, R.; Strauss, E.; Haslinger, C.; Tontsch-Grunt, U.; Waizenegger, I.C.; et al. Pharmacological Profile of BI 847325, an Orally Bioavailable, ATP-Competitive Inhibitor of MEK and Aurora Kinases. Mol. Cancer Ther. 2016, 15, 2388–2398. [Google Scholar] [CrossRef]
- Schoffski, P.; Aftimos, P.; Dumez, H.; Deleporte, A.; De Block, K.; Costermans, J.; Billiet, M.; Meeus, M.A.; Lee, C.; Schnell, D.; et al. A phase I study of two dosing schedules of oral BI 847325 in patients with advanced solid tumors. Cancer Chemother. Pharmacol. 2016, 77, 99–108. [Google Scholar] [CrossRef]
- Phadke, M.S.; Sini, P.; Smalley, K.S. The Novel ATP-Competitive MEK/Aurora Kinase Inhibitor BI-847325 Overcomes Acquired BRAF Inhibitor Resistance through Suppression of Mcl-1 and MEK Expression. Mol. Cancer Ther. 2015, 14, 1354–1364. [Google Scholar] [CrossRef]
- Wyckoff, J.B.; Pinner, S.E.; Gschmeissner, S.; Condeelis, J.S.; Sahai, E. ROCK- and myosin-dependent matrix deformation enables protease-independent tumor-cell invasion in vivo. Curr. Biol. 2006, 16, 1515–1523. [Google Scholar] [CrossRef]
- Sanz-Moreno, V.; Gadea, G.; Ahn, J.; Paterson, H.; Marra, P.; Pinner, S.; Sahai, E.; Marshall, C.J. Rac activation and inactivation control plasticity of tumor cell movement. Cell 2008, 135, 510–523. [Google Scholar] [CrossRef]
- Yamada, K.M.; Sixt, M. Mechanisms of 3D cell migration. Nat. Rev. Mol. Cell Biol. 2019, 20, 738–752. [Google Scholar] [CrossRef]
- Chen, F.; Gurler, S.B.; Novo, D.; Selli, C.; Alferez, D.G.; Eroglu, S.; Pavlou, K.; Zhang, J.; Sims, A.H.; Humphreys, N.E.; et al. RAC1B function is essential for breast cancer stem cell maintenance and chemoresistance of breast tumor cells. Oncogene 2023, 42, 679–692. [Google Scholar] [CrossRef]
- Zinn, R.; Otterbein, H.; Lehnert, H.; Ungefroren, H. RAC1B: A Guardian of the Epithelial Phenotype and Protector Against Epithelial-Mesenchymal Transition. Cells 2019, 8, 1569. [Google Scholar] [CrossRef]
- Du, J.; Liu, J.; Smith, B.J.; Tsao, M.S.; Cullen, J.J. Role of Rac1-dependent NADPH oxidase in the growth of pancreatic cancer. Cancer Gene Ther. 2011, 18, 135–143. [Google Scholar] [CrossRef]
- Sun, J.; Gaidosh, G.; Xu, Y.; Mookhtiar, A.; Man, N.; Cingaram, P.R.; Blumenthal, E.; Shiekhattar, R.; Goka, E.T.; Nimer, S.D.; et al. RAC1 plays an essential role in estrogen receptor alpha function in breast cancer cells. Oncogene 2021, 40, 5950–5962. [Google Scholar] [CrossRef]
- Elhasnaoui, J.; Ferrero, G.; Miano, V.; Franchitti, L.; Tarulli, I.; Coscujuela Tarrero, L.; Cutrupi, S.; De Bortoli, M. A Regulatory Axis between Epithelial Splicing Regulatory Proteins and Estrogen Receptor alpha Modulates the Alternative Transcriptome of Luminal Breast Cancer. Int. J. Mol. Sci. 2022, 23, 7835. [Google Scholar] [CrossRef]
- Xi, B.; Lai, M.; Zhang, W.; Wang, F. MicroRNA-561-5p Inhibits Cell Proliferation and Invasion by Targeting RAC1 in Pancreatic Ductal Adenocarcinoma. Ann. Clin. Lab. Sci. 2022, 52, 213–221. Available online: https://www.annclinlabsci.org/content/52/2/213.long (accessed on 22 September 2025).
- Schenker, H.; Buttner-Herold, M.; Fietkau, R.; Distel, L.V. Cell-in-cell structures are more potent predictors of outcome than senescence or apoptosis in head and neck squamous cell carcinomas. Radiat. Oncol. 2017, 12, 21. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tyrna, P.; Kostro, J.; Olszanecka, M.; Szukało, P.; Młynarczuk-Biały, I. Differential Sensitivity to MEK Inhibitors Highlights Distinct Entosis Mechanisms in BxPC3 and MCF7 Cells. Cells 2025, 14, 1500. https://doi.org/10.3390/cells14191500
Tyrna P, Kostro J, Olszanecka M, Szukało P, Młynarczuk-Biały I. Differential Sensitivity to MEK Inhibitors Highlights Distinct Entosis Mechanisms in BxPC3 and MCF7 Cells. Cells. 2025; 14(19):1500. https://doi.org/10.3390/cells14191500
Chicago/Turabian StyleTyrna, Paweł, Julia Kostro, Monika Olszanecka, Piotr Szukało, and Izabela Młynarczuk-Biały. 2025. "Differential Sensitivity to MEK Inhibitors Highlights Distinct Entosis Mechanisms in BxPC3 and MCF7 Cells" Cells 14, no. 19: 1500. https://doi.org/10.3390/cells14191500
APA StyleTyrna, P., Kostro, J., Olszanecka, M., Szukało, P., & Młynarczuk-Biały, I. (2025). Differential Sensitivity to MEK Inhibitors Highlights Distinct Entosis Mechanisms in BxPC3 and MCF7 Cells. Cells, 14(19), 1500. https://doi.org/10.3390/cells14191500






