Abstract
Recent findings of diverse populations of prion-like conformers of misfolded tau protein expand the prion concept to Alzheimer’s disease (AD) and monogenic frontotemporal lobar degeneration (FTLD)-MAPT P301L, and suggest that distinct strains of misfolded proteins drive the phenotypes and progression rates in many neurodegenerative diseases. Notable progress in the previous decades has generated many lines of proof arguing that yeast, fungal, and mammalian prions determine heritable as well as infectious traits. The extraordinary phenotypic diversity of human prion diseases arises from structurally distinct prion strains that target, at different progression speeds, variable brain structures and cells. Although human prion research presents beneficial lessons and methods to study the mechanism of strain diversity of protein-only pathogens, the fundamental molecular mechanism by which tau conformers are formed and replicate in diverse tauopathies is still poorly understood. In this review, we summarize up to date advances in identification of diverse tau conformers through biophysical and cellular experimental paradigms, and the impact of heterogeneity of pathological tau strains on personalized structure- and strain-specific therapeutic approaches in major tauopathies.
1. Phenotypic Spectra of Alzheimer’s Disease (AD) and Frontotemporal Lobar Degeneration (FTLD)
Alzheimer’s disease (AD) and Frontotemporal Lobar Degeneration (FTLD) are the prominent causes of age-related dementia [1,2,3,4,5]. Although AD is a dual proteinopathy with concurrent deposition of misfolded amyloid beta fragment and hyperphosphorylated tau aggregates, tau deposits are the best indicator of cognitive decline [6,7]. AD encompasses extraordinarily variable phenotypes and progression rates, categorized by the prevalent clinical symptomatology as an amnestic variant, posterior cortical atrophy (PCA), logopenic primary progressive aphasia (LPPA), and the frontal variant of AD [1,6,8]. Additionally, based on the neuroimaging and neuropathology data, these clinical phenotypes have been divided into typical AD, with balanced neurofibrillary tangle (NFT) depositions in the hippocampus and association cortex; limbic-predominant AD, with NFTs predominantly occurring in the hippocampus; and hippocampal-sparing AD, with counts mostly observed in the association cortex [7,9]. Moreover, approximately 10% of AD cases have a rapidly progressive form (rpAD) with an accelerated tempo of clinical decline and disease duration frequently of less than three years [10,11,12,13,14]. The sources of this clinicopathological heterogeneity including variable involvement of astroglia [15,16,17,18] are not fully understood and only ~30% of the variability can be attributed to genetic polymorphisms [19,20,21,22]. Immunohistochemistry data show that aggregated and hyperphosphorylated tau inclusions are present in glial cells in AD, FTLD-17, and other tauopathies [23,24], and in mice models as well [25,26,27], but how the intracellular trafficking in glial cells would impact propagation of different prion-like strains of tau aggregates is not known and different glia may have protective or enhancing effects [28,29,30].
Frontotemporal lobar degeneration (FTLD) is also a clinically, genetically, and pathologically diverse group of disorders and the umbrella term FTLD [31,32,33] can be divided into the behavioral variant (bvFTD) and (at least) two types of primary progressive aphasia (PPA): non-fluent variant (nfvPPA) and semantic variant (svPPA) [34]. The bvFTD accounts for nearly 60% of cases of FTLD; the language/semantic variants are less common. Intriguingly, the clinicopathological heterogeneity in the spectrum of sporadic FTD-tau disorders is also well documented in their genetic forms such as FTLD-MAPT-P301L, with a diversity of clinical manifestations observed among patients carrying the exact same mutation, and even in the same family [32,35,36].
The distribution of tau pathology in both AD and FTD-tau tends to correlate with cognitive decline, brain areas of atrophy, and the disease stage, and therefore tau misfolding and aggregation is considered to be a link to common downstream mechanisms of neurodegeneration [3,6,16,28,36,37]. Comprehensive analysis of aging brain samples indicates that the pathological changes linked to AD start early on, through the accumulation of misfolded and hyperphosphorylated tau species in isolated anatomical areas of the brain, which then spread by cell-to-cell transmission mechanisms [17,38]. The increasing evidence from cell and transgenic mice models show that various misfolded and aggregated tau strains generated in vitro or in vivo can replicate in cells [39], hasten and spread endogenous tau aggregation in recipient transgenic animals [40,41], and therefore indicate a prion-like replication phenomenon. Although aggregates of other proteins linked to a growing number of neurodegenerative diseases can be propagated from human tissues to experimental animal models or between humans [42,43], there is currently no evidence for direct infectious etiology for the human neurodegenerative disorders defined by these proteins [44]. Because of the importance of this question, the mechanism of replication and propagation of misfolded proteins deserves primary attention, and improved understanding has important implications for individualized diagnostics and therapeutic strategies.
2. Implications of Strain Concept of Human Prion Diversity for Tauopathies
Human prions cause perhaps the most heterogenous neurodegenerative disorders, which were until recently considered separate diseases; the classification was based on leading clinical symptoms or they were named after the author(s) who originally described them—sporadic and familial fatal insomnia (sFI, FFI), Creutzfeldt–Jakob disease (CJD), Gerstmann–Sträussler–Scheinker (GSS) disease, variably protease-sensitive prionopathy (VPSPR), and a spectrum of unclassified rare genetic forms [45,46,47,48,49]. Human prions are transmissible from person to person [43], to primates [50,51], and to experimental animals [52,53], and the existence of distinct prion phenotypes in the host expressing the same prion protein sequence was offered as an argument for the presence of a prion-specific nucleic acid [54,55]. However, even with frequent attempts to detect such a nucleic acid applying various approaches, and despite accumulating data contradicting the existence of a prion strain-coding polynucleotide [56,57,58,59], the existence of distinct prion strains remains a conundrum, and the most important challenge to fundamental principles of molecular biology [52,60,61]. Many lines of evidence now indicate that the extraordinary phenotypic diversity of human prion diseases arises from the structurally distinct prion strains that target variable brain structures at different progression speeds [45,46,47,48,62,63,64]. This paradigm is supported by biochemical, genetic, and animal studies [65,66,67,68,69], and lastly by a recent successful generation of the first synthetic human prions [70]. Expanding the principles developed with cloned laboratory prions, the general consensus criteria for human prion strains based on their clinicopathological and molecular characteristics are now used for differentiation, classification, and surveillance of human prion diseases in prion centers in the US and Europe (Table 1) [48,71,72,73].

Table 1.
Classification criteria of human prion strains.
3. Isoforms and Cellular Functions of Normal Human Tau Protein
Although genetic polymorphisms in APOE and a growing number of other genes are important risk factors of late onset AD, at least two features of late onset AD that cannot be explained by linkage analysis are (i) divergences between amyloid beta and tau pathological deposits and severity of clinical disease-specific manifestations [10,13,87], and (ii) the general variability of progression rates and phenotypes [6,88,89]. Early pivotal data generated with HEK reporter cells showed different patterns of tau aggregates after inoculation by tau isolates from distinct tauopathies [39] and recently even with different tau strains generated from monomeric tau in the absence of aggregated seeds [90,91]. These experiments inferred alternatively structured tau species encoding the key information for distinct phenotypes seen in cell experiments with prion-like mechanisms. However, the aggregates of fluorescently labeled tau accumulating in HEK reporter cells in response to the inoculation with conformationally different strains do not correspond to the AD tau filament assemblies [92], but rather show various morphologies and intracellular locations. These cell patterns suggest a seeding barrier effect due to the presence of mutation in the HEK cell-expressed short K18 tau fragment.
In contrast to the human prion protein (PRNP) gene that has a single open reading frame [47,48], the MAPT gene undergoes complex alternative splicing in the coding region (Figure 1). The MAPT gene is located on chromosome 17q21 and possesses 16 exons in the tau primary transcript. Unlike exons 1, 4, 5, 7, 9, 11, 12, and 13 that are constitutive, exons 2, 3, 4a, 6, 8, and 10 are subject to alternative splicing [93] (Figure 1). Overall, alternative splicing results in six tau isoforms that differ by the presence or absence of two N-terminal regions (0 N, 1 N, or 2 N) as well as conserved repeat motifs (3R or 4R) [94,95]. In the brain, tau mRNA is expressed primarily in neurons, but low expression levels of tau can be found in various types of glial cells, including astrocytes and oligodendrocytes [15,96,97]. The main function of tau as a member of the microtubule-associated proteins (MAPs) group is to bind and stabilize microtubules through the microtubule-binding domain located at the C-terminus of tau, mediate their assembly, and thus modulate vesicle/organelle transport on microtubules [98,99,100]. The N-terminal domains may influence spacing between microtubules, subcellular distribution of neuronal tau, and aggregation kinetics of tau [101,102]. The repeat motifs constitute the microtubule-binding domain of tau, and the proline-rich domain (PRD) links the C-terminal assembly domain and the N-terminal projection domain. Overall, due to its high content of polar and positively charged amino acids, tau is a water-soluble protein with a high isoelectric point. Monomeric tau lacks a stable secondary and tertiary structure and instead possesses great structural flexibility [103].
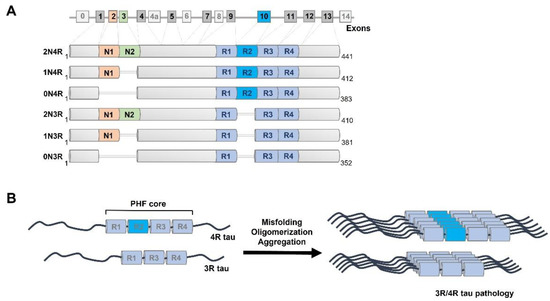
Figure 1.
Alternative splicing of MAPT gene and tau protein aggregation. (A) Alternative splicing of exons 2, 3, and 10 leads to six isoforms of tau in the adult human brain; exons 4A, 6, and 8 are exclusive to the peripheral nervous system and absent in the human brain [104]. Six isoforms of human tau occurring in the central nervous system (2N4R, 1N4R, 0N4R, 2N3R, 1N3R, 0N3R) vary by the presence or absence of N-terminal regions (N1, N2) and repeat domain 2 (R2), resulting in molecular weights ranging from 36,760 Da (0N3R) to 45,850 Da (2N4R). (B) Tau is under physiological conditions an intrinsically disordered protein that dynamically interacts with and stabilizes microtubules. Under pathological conditions, tau repeat region cores undergo conformational transition to beta sheet-rich secondary structures prone to formation of oligomers and paired helical filaments (PHF) [105] with amyloid tinctorial properties.
Neuronal tau is principally located in the axons and to a lesser degree in somatodendritic compartments such as the cell membrane [106], mitochondria [107], and nucleus [108]. Apart from important roles of tau in axonal transport [109,110], it also plays a role in nucleic acid protection [111], synaptic plasticity, and neuronal maturation [112,113,114]. Structural studies confirm that unbound tau is indeed an intrinsically disordered protein (IDP) [103] that possesses the tendency to change its unstable random conformations to more energetically favored, detergent-insoluble, partially protease-resistant protein aggregates [115] with amyloid-like tinctorial properties and high affinity for amyloid dyes-Thioflavin S, Congo Red, and their derivatives [116,117] (Figure 1). This dramatic structural transformation leads to the loss-of-function and gain-of-function outcomes that are a critical step in the pathogenesis of AD and FTLD, and in general all tau-related neurodegenerative diseases-tauopathies.
The sequence alterations in tau microtubule-binding domains that occur in mutations associated with FTLD-17 cases lead to reduced interaction with microtubules, altered ratio of 3R and 4R isoforms in favor of 4R tau, and misfolding of tau protein [118,119,120,121]. Numerous transgenic mice lines expressing mutant forms of human full-length tau were developed to investigate tau pathology at an accelerated rate. In parallel, to address the overexpression of tau linked to several mutations identified in FTDP-17 cases [120,121], the ALZ17 mice line expressing the non-mutated longest human tau 4R isoform was introduced [122]. The missense mutations such as P301L/S, W337M, and R406W result in reduced ability of tau protein to bind with microtubules and accelerated misfolding and aggregation [119,121]. These well established and extensively characterized [123,124,125,126,127,128,129,130,131,132,133] transgenic systems provide valuable tools not only for studying tau pathology caused by these mutations but also animal models for monitoring tau seeding potency and propagation of tau aggregates after intracerebral injection [132,133,134,135]. The expansion of mice models expressing various mutated/non-mutated tau isoforms has an enormous impact on understanding tau pathology initiation and spread, and several excellent reviews recently summarized the progress in this field [136,137,138,139,140].
4. Preferential Misfolding of 4R Tau Isoform in Late Onset AD and FTLD-MAPT-P301L Patients
The expression of all six tau isoforms in the adult human brain led to the assumption that neurofibrillary tangles (NFTs) are a result of random integration of different tau isoforms into paired helical filaments (PHFs) and straight filaments (SFs) in AD [141,142,143,144]. However, the sedimentation velocity separations of total brain homogenates by ultracentrifugation in sucrose gradient show that misfolded tau aggregates are, regardless of size, uniformly composed of ~80% of 4R tau and ~20% of 3R tau, even though the soluble normal tau monomers are mixtures of approximately equal proportions of 3R and 4R isoforms [10] (Figure 2). This altered distribution of 3R and 4R tau with 4-fold excess of 4R in misfolded tau aggregates indicates higher susceptibility of unfolded 4R tau monomers to conformationally convert to beta-sheet structures in AD, which correlates with seeding experiments using K18 and K19 tau constructs. Thus, 4R aggregates are likely formed with faster conversion kinetics and more rapid growth might be the mechanism explaining the 3R, 4R unequal distribution in AD tau aggregates [10]. However, whether the distinct individual tau aggregates are composed of only 4R and 3R or by both isoforms has been under debate. The recent cryoEM models of core PHFs extracted from AD brains may accommodate upstream of V306 an additional 16 amino acids, which could represent a mixture of residues 259–274 (R1) from 3R tau, or 290–305 (R2) from 4R tau [143,144], but in contrast, show 1:1 stoichiometry of R4 and R3 tau isoforms. Latest reports from Scheres’ group indicate that in vitro forming filaments of misfolded tau are exceptionally conformationally flexible and might consist of a pool of exceedingly unstable conformers with numerous structural transitions dependent on ambient conditions [104], but how to translate these in vitro experiments to aggregated and misfolded human tau samples might be mostly limited by the application of standard purification procedures. Using cell biosensors, several groups recently showed that the seeding capacity of tau present in various AD brain patients’ samples precedes the NFT formation and shows high variability between individual samples [10,145]. Cumulatively, the data imply (i) the existence of separate pathways for PHF and SFs, and (ii) large pools of misfolded tau with nonfibrillar morphology of the aggregates observed in ultracentrifugation sedimentation velocity experiments [10]. The different sizes of misfolded tau particles separated in sucrose gradients with distinct conformations corroborate this conclusion and implicate liquid-liquid phase separation (LLPS) and oligomeric states of tau (Figure 2). Notably, conformation of 3R tau in tau aggregates monitored by CDI and CSA resemble conformational profiles nearly identical to those mixed of (3R + 4R) tau [10]. To better understand the single mixed particle versus distinct particle model in nonfibrillar aggregates will require advanced techniques such as immune(cryo)EM and hydrogen exchange mass spectrometry (HX MS).
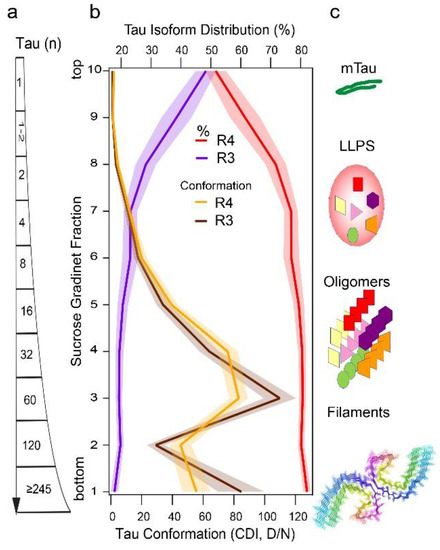
Figure 2.
Size, 3R and 4R isoform distribution, and conformation of tau particles in the hippocampus of AD cases. (a,b) Size, concentration, and conformation of insoluble misfolded 4R and 3R tau fractionated by sedimentation velocity using ultracentrifugation in sucrose gradient were analyzed simultaneously by 4R- and 3R-specific CDI. (c) The schematic representation of normal tau monomer and different structural forms of misfolded tau proteins-induced ensembles of dynamic liquid-liquid phase separation (LLPS) [146], oligomers [147], and PHF structures [105]. The composite sucrose gradient is replotted from recently published data [10] and the structure of PHF was generated by Cn3D application from published coordinates [105].
5. Conformational Diversity of Tau Isoforms in Different Phenotypes of AD, FTLD-MAPT-P301L, and TgP301L Model
The biophysical techniques that were originally developed for prions are ideally suitable to monitor the structural pattern and composition of tau directly in the brain tissue of AD cases with distinct phenotypes [46,84,85,86]. The main advantages of using these ultrasensitive methods to investigate conformational organization and properties of misfolded tau aggregates are no requirement for tau purification or in vitro amplification step before detection; they are independent of the absolute concentrations of misfolded protein particles, and thus the original characteristics of strain isolates are preserved [70,148]. While the indirect properties such as the size and protease resistance of tau aggregates and cellular or neuropathology patterns in the reporter HEK293 cells and transgenic mice [149], and posttranslational modifications (PTMs) of tau in mass spectrometry [8,147] suggested similar structure of tau in individual AD cases, the new direct biophysical and conformational data revealed evolving conformer populations driving in prion-like manner different phenotypes in individual AD [10] cases (Figure 3). This conformational paradigm is supported by the diversity of sedimentation profiles of tau aggregates (Figure 2), trypsin peptide maps, conformation-dependent immunoassay (CDI), and conformational stability assay (CSA) [10] and mirrors the data obtained with the phenotypic spectrum of FTLD-MAPT(P301L) cases and in the transgenic model of FTLD [150]. Additionally, recent advances in cryoEM provide unequivocal evidence for up to 76 distinct tau filament structures that can be generated in vitro with subtle changes in assembly conditions [104]. Cumulatively, these new findings implicate major conformational flexibility of tau in prion-like strain effects and the new hydroxyl radical footprinting approaches [76,151] to the tau strain investigation should provide unbiased structural information on tau conformers in the ensemble of all their misfolded states, particularly prefibrillar (oligomeric) forms and liquid-liquid phase separation (LLPS) droplets, and “fuzzy” coat in filamentous structures [112,146] (Figure 2).
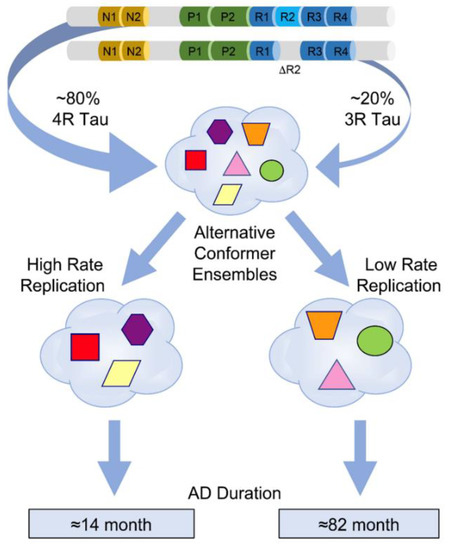
Figure 3.
Evolving ensembles of misfolded tau conformers in the pathogenesis of different progression rates of Alzheimer’s disease. Different geometric shapes and colors represent predominantly four-repeat conformers of detergent-insoluble misfolded tau. Different coexisting combinations (ensembles) of conformers corresponding to different CSA profiles are shown within the cloud outlines. The data were obtained by deconvolution of conformational stability (CSA) types seen in 43 AD cases with variable progression rates and they likely represent alternative pathways of ensemble evolution (blue arrows). Figure is reprinted from [10] with permission of the American Association for the Advancement of Science, 2022.
The deposits of hippocampal tau are considered critical in cognitive decline because they are at the crossroad in the spread of pathogenic tau aggregates, from early deposits in transentorhinal cortex (Braak stages I–II) to major projections to the hippocampus where tau pathology gradually occurs in the CA1 region (Braak II), followed by spread to the limbic structures, inferior temporal neocortex (Braak III), the amygdala and thalamus (Braak IV), and finally propagation into the neocortex (Braak V–VI) [38,40]. Notably, another predominantly 4R tau tauopathy-frontotemporal lobar degeneration (FTLD)-MAPT-P301L-also displays three distinct tau signatures in cases with different clinical phenotypes, two resembling those found in prodromal TgTauP301L mice model of FTLD [150]. As biophysical methods and HEK293 cell biosensors confirm a broad spectrum of tau strains in the mouse and human brain series, the evolution of diverse tau conformers is apparently an intrinsic feature of both tauopathies’ pathogenesis-uniallelic form of FTLD [150] and AD [10].
Another important aspect contributing to the structural complexity of tau aggregates are posttranslational modifications (PTMs). In recent studies, the correlation between seeding potency and various PTM profiles obtained by mass spectrometry of misfolded insoluble tau isolated from different clinical phenotypes has been reported [147,152]. Similar trends are also being observed at the early age of onset in familial Alzheimer’s disease (FAD) with Presenilin-1 (PSEN1) mutations [8]. These observations suggest a closer relationship between posttranslational modification and structural organization of tau during AD progression and raise the question of whether the variances in conformations are driven by distinct PTMs or vice versa. Even though the tau phosphorylation at specific sites is associated with aggregated tau burdens and is used in diagnostic and immunohistological confirmation of tauopathies [153,154,155], the causative effect of PTMs on tau misfolding and generating tau seeds competent to spread the tau pathology is debated in light of recent data that show seeding activity even in the absence of phosphorylated tau deposits [40,156,157,158]. These are important effects not only in AD and FTLD, but in other tauopathies as well, including progressive supranuclear palsy (PSP), and Pick’s disease. The interplay between PTMs and the formation of misfolded structural orders of tau may be simultaneously investigated by using high advanced approaches such as integrated cryoEM combined with conformation-sensitive mass spectrometry (HX MS) and/or synchrotron footprinting, which are applicable for both fibrillar and oligomeric forms of misfolded tau [46,70,76,159]. These approaches should also determine whether the sedimentation velocity and different trypsin peptide patterns of conformers separated by sedimentation velocity are exclusively a result of conformation and particle size, or are modified by posttranslational modifications, or Sarkosyl-resistant ligands which could be important cofactors in the tau misfolding.
6. Modelling Replication Mechanism of Tau Conformer Populations In Vitro
The important evidence for replication and propagation of misfolded tau conformers in the brain is that the misfolded protein can be amplified in vitro in a seeded reaction [70,160]; this concept was proved first with laboratory prions [148], later with human prions [71], and more recently with an increasing number of other misfolded prion-like proteins including alpha synuclein [161] and TDP-43 protein [162]. The conformational templating process for amplification of AD brain-derived tau seeds utilizes purified recombinant K18 (4R) and K19 (3R) tau substrates or their chimeras [163,164], and at the same substrate concentrations, the AD brain-derived tau seeds preferentially amplify with K18 (4R) substrate and demonstrate end point sensitivity up to 109 dilution of the brain tissue [10,165,166]. Using the tau assay, recent reports are consistently showing major interindividual variability of seeding potency of AD brain-derived tau [160,165,166] and homogenates of rapidly progressing AD cases demonstrate a significantly shorter lag phase than classical slowly progressing cases [10]. Cumulatively, these data show that in individual AD cases there is a cloud of distinct conformers of misfolded tau with various seeding potencies and that the better and more reliable predictor of seeding potency is conformation of insoluble 4R tau, which correlates with faster disease progression, than the absolute concentration of all misfolded tau seeds.
The ultracentrifugation in sucrose gradient [10], chromatographic separations [147], protease sensitivity [10], oligomer-specific antibodies [167], and lower stability of some of misfolded tau structures in conformational stability assays [10] all indicate that a significant portion of misfolded tau is in an oligomeric or prefibrillar state (Figure 2). The systematic examination of three different human prion strains has revealed that high affinity of surface domains in strain-specific prion conformers for their substrate (PrPC) results in faster replication and propagation of human prions [46,70,76]. The new biophysical data on AD tau are in agreement with this concept and point to a conformational cloud of distinct tau conformers with different interactions between misfolded and monomeric tau and affinity-driven faster replication in rapidly progressive AD [10]. The differences in these populations found in rapidly and slowly progressing AD are linked to the different replication rates in vitro in seeding assays and in cell reporters [10]. The mechanisms or aspects behind these differences are not fully understood, but the assembly of quaternary oligomer or fibrillar structures and the structure of monomeric building blocks must be thermodynamically and kinetically linked [168]. Consequently, the differences in replication slopes seen in in vitro seeding assays point to distinct mechanisms such as fibril growth from ends or from sites as theoretically suggested [169]. Additionally, the studies of prion strains of both human and animal origin imply that non-protein cofactors (phospholipids, gangliosides, negatively charged polyanions, etc.) might contribute to the diversity of misfolded strains by affecting their stability and propagation [70,159,170,171,172]. Such auxiliary cofactors in tau strains remain to be found, but they could contribute to our understanding of propagation rates of specific tau strains and their affinity to differential interactomes and distinct host cells.
7. Strain Effects of Prion-like Tau Conformers in Cell Reporters
The substitution of proline 301 with lysine (P301L) or serine (P301S) in MAPT gene is the most common mutation associated with hereditary frontotemporal dementia with parkinsonism-17, FTDP-17 [32]. This single mutation facilitates fibril formation [173,174,175] and enhances seed recruitment in cells overexpressing human mutant tau variants [40,41,133,176,177,178,179,180,181,182,183]. The recruitment of wild-type tau by P301L tau fibrils was less efficient in non-transgenic primary neurons than in P19 mice neurons [133], and an even more evident asymmetric cross-seeding barrier was repeatedly observed in other in vitro studies [184,185]. The proline substitution in tau has multiple impacts: (i) The P301L mutant has higher propensity for assembly into fibrils [134]; thus the faster incorporation of mutated tau monomers into fibrils is preferential and it was shown that tau recruitment is a time-dependent event [133]. (ii) P301L mutant tau filaments are conformationally distinct from wild-type tau filaments [185,186]. (iii) Proline 301 is located at R2, which is encoded by exon 10 and present only in 4R tau isoforms; thus this single amino acid substitution might also lead to a different ratio of tau isoforms recruitment into filamentous inclusions [33,186]. (iv) The post-translational modifications may vary between propagated as well as templated seeds of P301L-tau and wt-tau, and thus contribute to misfolding [156].
Immortalized cell lines expressing full-length tau or its truncated variants with the most frequent disease-related mutations allow the monitoring of intracellular templated amplification of misfolded tau seeds. A number of cell lines are experimentally used for this goal, namely N2a [187], HEK293 [10,39,146,177,178,188,189,190], HeLa [191], and SH-SY5Y [192]. The most frequently used are HEK293 cells that were first introduced to prion research in experiments with GFP fusion proteins of yeast Sup35p prion substrate [193]. The experiments with different tau seeds performed in vitro showed that variable aggregate morphologies can be vastly transmitted in a clonal fashion in HEK293 cell stably transfected tau protein fragments (with P301L and V337M missense mutations) fused to a YFP reporter [39,177]. The subclone (Clone 1/DS1) of these reporter cells shows various morphologies of diffuse and amorphous shapes, nuclear envelope inclusions, speckles, and threads after inoculation with conformationally different strains of tau isolated human FTLD samples and Tg mice model of FTLD(P301L) [150].
Brain extracts from transgenic mice with distinct conformational organizations of tau demonstrated considerably different seeding profiles. Interestingly, tau-enriched extracts from human brains of patients with different clinical diagnoses (behavioral variant FTLDs with/without predominant memory impairment and semantic variant of primary progressive aphasia) displayed less pronounced differences in seeding profiles using by this assay. In analogous experiments, all samples of brain-derived AD tau induce tau aggregates with speckled morphology, but notably elevated prion-like seeding activities and higher frequency of positive cells exposed at the same concentrations of tau extracted from malignant AD with rapid progression [10].
The in vitro experiments indicate an early rapid conformational transition of intracellular tau upon seeding with brain-derived FTLD tau (P301) and formation of dispersed fluorescence signals that may next be collected and relocated within processes between nearby cells [146]. The fluorescence signal of tau within a nuclear envelope and small fluorescent inclusions has all the characteristics of liquid-liquid phase separation (LLPS)—Thioflavin S signal, spherical morphology, fusion events, and may recover from photobleaching [146]. These juxtanuclear tau assemblies are apparently linked to disrupted nuclear transport and decreased cell viability. These conformation-specific and dynamic cellular effects likely represent early steps in the transition to prefibrillar (oligomeric) and fibrillar tau inclusions seen in end-stage tauopathies. These observations also suggest that using immortalized cells to perform seeding experiments, the impact of cell cycle has to be considered as distinct tau inclusion types may accumulate in different stages of the cell cycle [10,112].
8. Effects of Misfolded Tau in Neuronal Cultures
Immortalized cell lines offer fast and high throughput tools to investigate the misfolded tau seeding in a cellular environment that overexpresses tau protein; but experimental factors such as genotype of recipient cells, expression of tau fragments with P301L and V337M missense mutations (which increase the tendency to aggregate and may introduce “species barrier”-like effects due to the tau sequence mismatch), acting together, may impact the outcome and thus deserve careful consideration. Thus, using neuronal cultures is a critical next step to enhance our knowledge about the interactomes and mechanism of replication of different tau conformers. Numerous factors have to be considered in the investigation of cell-to-cell tau aggregation transmission and cell-to-cell spreading in non-overexpressed tau cell systems: (i) origin, character, and concentration of pathological tau aggregates; (ii) uptake of tau aggregates without artificial cationic lipid-mediated transfection-presence of receptors like heparan sulfate proteoglycans and LRP1 [179,194]; (iii) character of tau in recipient cells-species origin, ratio and distribution of tau isoforms; and (iv) maturation of cultures, neuronal differentiation, and cellular network to sufficiently spread tau aggregation among cells.
Human SH-SY5Y neuroblastoma, mouse N2a neuroblastoma, and catecholaminergic CAD cells among others are immortal cell lines with potential to be differentiated into neuronal subtypes expressing endogenous tau [195,196,197] (Figure 4). Overexpression of tau variants in these cells have been applied in most studies of tau pathology [187,198,199]. With the advantage of human origin, SH-SY5Y differentiate into polarized neuron-like cells with axonal sorting of human endogenous tau with both 3R, 4R isoforms [200,201] and have served as an alternative sporadic cell model for studies of pathological tau uptake, transmission and spreading [202,203].
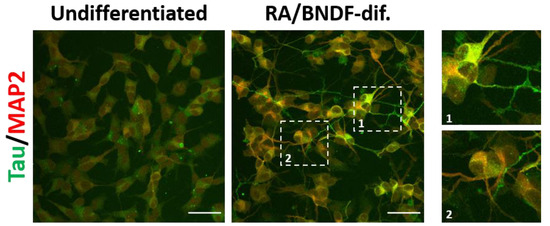
Figure 4.
Tau and MAP2 expression and localization in undifferentiated and fully (RA/BDNF) differentiated SH-SY5Y cells (cell source: ECACC). The RA/BDNF differentiation facilitates the enrichment of both microtube-associated proteins, tau and MAP2, respectively. Moreover, two distinct types of neuronal projections, associated with a polarized state of neurons, are observed in fully differentiated SH-SY5Y: axonal-like neurites enriched with tau protein and somato-dendritic compartment consisting of MAP2 (cropped images 1 and 2). Scale bars: 50 μm. Figure is modified with from [197] with permission of Elsevier, 2022.
Rodent primary neurons have been the gold standard cultures to study properties and behavior of post-mitotic fully polarized neurons with developed synaptic networking. In particular, neurons from non-transgenic mice initially express both 3R and 4R isoforms [41,204], but culture aging shifts the expression towards 4R isoforms, and thus they serve as a valuable cell model to investigate the tau misfolding propagation under conditions more resembling sporadic tauopathies. This is particularly important for investigating the cross-seeding barrier of 3R and 4R tau isoforms, which has been observed in vitro and in both reporter cells systems overexpressing tau [198,205] and primary neurons [41,206]. Overall, in vitro generated seeds from recombinant tau have a slower rate in extension of filament formation in cells than naïve tau fibrils isolated from brain material in in vitro cultures [41,181], and the distinct conformation of heparin-induced filaments and those isolated from AD brains was recently confirmed by cryo-EM studies [141]. The critical denominator of cellular effects is the structural organization of tau strains, including the size of aggregates [181,207] and only a small number of filaments with high seeding activity can trigger monomeric tau misfolding [41,176]. Several studies have shown that naïve fibrils from clinicopathologically different tauopathies have specific properties and their biological activity is likely to be associated with unique isoform composition and distinct conformational variants [206,208,209].
Over the past two decades, induced pluripotent cells (iPSCs)-derived neurons have become a valuable tool to investigate mechanisms of many neurodegenerative diseases [210,211]. There are almost limitless sources of healthy donors and patient-derived neurons that can be applied to uncover disease-specific alterations and serve as a platform to identify novel therapeutics via high throughput drug screening. Human iPSC-derived neurons grow and mature in cultures from weeks to several months and express predominantly fetal 0N3R tau isoform at early culture time-points, and only slightly shift expression to 3R, 4R representation during differentiation and maturation [212,213,214]. Importantly, neuronal progenitor cells are easily transfected with full length tau variants before differentiation including 2N4R-huTAU-P301L construct and full-length 4R tau constructs [215,216]. Although the presence of P301L mutation facilitates aggregation via in vitro generated K18-P301L fibrils, it also introduces the cross-seeding barrier between non-mutated tau and P301L variants [215,216]. Data from hiPSC-derived neurons with 3R wild-type tau and bearing P301S mutation show the cross-seeding barrier. 2N4R P301L fibrils induce robust aggregation at P301S tau hiPSC-derived neurons but not in hiPSC-derived neurons with 3R wild-type tau. However, AD brain-derived tau aggregates caused more tau aggregation in wild type hiPSC-derived neurons than in hiPSC-derived neurons with P301S mutation [217]. In a recent study of non-transfected hiPSC-derived neurons from healthy donors, full-length tau oligomers were internalized and recruited endogenous tau to induce pathological aggregation [218]. To sum up, experimental data of transmission and spreading tau pathology in neuronal cultures depends on many factors with emphasis on the origin of tau seeds and tau protein expressed in recipient neuronal cells.
9. Concluding Remarks and Future Investigations
The clinical manifestations and disease phenotypes of both sporadic AD and monogenic FTLD vary significantly among individual patients, but the mechanisms responsible for the broad spectrum of disease phenotypes have not been fully understood. One of the main factors affecting variable pathogenesis and disease progression in tauopathies seems to be related to the conformational and structural heterogeneity of misfolded tau aggregates, which show distinct behavioral and intrinsic properties such as different propagation rates, targeting various cell types, toxicity effects, and interactomes. The central role of amyloid beta due to mutations in genes of amyloid beta precursor protein APP and its endoproteases was unarguably recognized in the pathogenesis of early onset AD; however, the missing strong correlation between amyloid senile plaque deposits and severity of sporadic AD [87], and failure of attempts to therapeutically target amyloid beta-associated pathology in AD have opened more space to investigate tau protein involvement in AD [6,219]. The burden of tau aggregates, the second invariable marker of AD, correlates with cognitive decline and thus indicates the important role of tau pathology progression especially in sporadic forms of AD and FTLD. To improve our understanding of tau protein, its structural plasticity and its various misfolded and aggregated forms is the next critical direction for elucidating the disease mechanisms, and thus new targets for therapeutic interventions.
The structural evolution of tau pathological species in the familiar form of FTLD linked to MAPT P301L mutation [150] and recently published data obtained by sensitive biophysical methods reveal a broad conformational range of distinct tau conformers that accompany different phenotypes and also various progression rates in AD. Moreover, the discovery of various strains of amyloid beta that are analogous to numerous distinctive clouds of tau conformers in rpAD [11,13,220] suggest a pathogenetic association and indicate the possibility of a dual prion-like synergy, as recently suggested from experiments in vitro [221]. Additionally, the polymorphisms of the APOE gene linked to AD cases and distinct tau strains propose a imaginable parallel to circumstances noticed with human prions, where the interplay between common polymorphisms in the prion protein gene (PRNP) and variable conformational characteristics of the pathogenic prion protein leads to vastly different disease clinicopathological outcomes [47,48,64]. These observations are crucial towards more individual-based diagnostics and more precise and beneficial molecular classification of AD subtypes as well as for better understanding of mechanisms contributing to the disease progression; they thus provide essential information for exploring further AD therapeutic approaches including the necessity to consider clouds of tau misfolded conformers with extensive interindividual variability. The prion-like strain effects of misfolded tau conformers lead to a series of critical mechanistic questions relating to the role of misfolded tau aggregates in distinct phenotypes of AD and FTLD: (i) What is the molecular mechanism of replication and propagation of distinct brain-derived pathogenic aggregates, particularly prefibrillar forms of tau, in different phenotypes of AD and FTLD-tau? (ii) Which structural domains of tau control the propagation, toxicity, and evident range of pathological and clinical features in AD and FTD? (iii) What is the impact of distinct posttranslational modification on the structure of tau in different strains? (iv) What is the replication potency of different tau conformers? Answering these questions is of fundamental importance for verifying and advancing the emerging concept pointing to structurally distinct prion-like strains [89,102] of tau as a critical differentiating factor in AD and FTLD-tau development. The early data are consistent with the possibility that patients with tauopathy may have distinct molecular drivers of clinical phenotypes and emphasizes the need for personalized structure- and strain-specific therapeutic approaches.
Biosensor reporter cell lines overexpressing tau mutant variants are continuing to be an invaluable tool in investigating the capacity and diversity of various “tau seeds” to aggregate the reporter tau and have generated valuable data. However, to understand the mechanisms of tau uptake, seed-templated propagation of endogenous tau aggregation, tau pathology propagation and transmission between individual neuronal cells and alterations in molecular pathways, differentiated neuronal-like cell lines, and particularly hiPSC-derived neurons will play an irreplaceable role in improving our understanding of the origin and pathogenesis of sporadic tauopathies.
Funding
Work in the Safar lab was supported by grants from NIH (R01NS103848, 1RF1AG058267, and 1RF1AG061797) and from Alzheimer’s Association (AARF-22-918090), and the NPDPSC is funded by CDC (NU38CK00048).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors are grateful to the patients’ families for donating brain tissue, and we thank all the referring physicians and all members of the National Prion Disease Pathology Surveillance Center (NPDPSC), Cleveland, OH, for excellent technical assistance and review of clinical data.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| AD | Alzheimer’s disease |
| CDI | Conformation-Dependent Immunoassay |
| CJD | Creutzfeldt–Jakob disease |
| CSA | Conformational Stability Assay |
| FFI | fatal familial insomnia |
| FTLD | frontotemporal lobar degeneration |
| GSS | Gerstmann–Sträussler–Scheinker syndrome |
| MAPT | microtubule-associated protein tau gene |
| PrP | prion protein |
| PrPC | normal or cellular prion protein |
| PrPSc | pathogenic prion protein |
| PRNP | prion protein gene |
| CJD | sporadic Creutzfeldt–Jakob disease |
| RT QuIC | Real-Time Quaking-Induced Conversion |
| SFI | sporadic fatal insomnia |
| VPSPr | variable protease-sensitive prionopathy |
References
- Nelson, P.T.; Trojanowski, J.Q.; Abner, E.L.; Al-Janabi, O.M.; Jicha, G.A.; Schmitt, F.A.; Smith, C.D.; Fardo, D.W.; Wang, W.X.; Kryscio, R.J.; et al. “New Old Pathologies”: AD, PART, and Cerebral Age-Related TDP-43 With Sclerosis (CARTS). J. Neuropathol. Exp. Neurol. 2016, 75, 482–498. [Google Scholar] [CrossRef]
- McAleese, K.E.; Walker, L.; Erskine, D.; Thomas, A.J.; McKeith, I.G.; Attems, J. TDP-43 pathology in Alzheimer’s disease, dementia with Lewy bodies and ageing. Brain Pathol. 2017, 27, 472–479. [Google Scholar] [CrossRef]
- James, B.D.; Wilson, R.S.; Boyle, P.A.; Trojanowski, J.Q.; Bennett, D.A.; Schneider, J.A. TDP-43 stage, mixed pathologies, and clinical Alzheimer’s-type dementia. Brain 2016, 139, 2983–2993. [Google Scholar] [CrossRef] [PubMed]
- Neumann, M.; Sampathu, D.M.; Kwong, L.K.; Truax, A.C.; Micsenyi, M.C.; Chou, T.T.; Bruce, J.; Schuck, T.; Grossman, M.; Clark, C.M.; et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 2006, 314, 130–133. [Google Scholar] [CrossRef] [PubMed]
- Josephs, K.A.; Whitwell, J.L.; Weigand, S.D.; Murray, M.E.; Tosakulwong, N.; Liesinger, A.M.; Petrucelli, L.; Senjem, M.L.; Knopman, D.S.; Boeve, B.F.; et al. TDP-43 is a key player in the clinical features associated with Alzheimer’s disease. Acta Neuropathol. 2014, 127, 811–824. [Google Scholar] [CrossRef] [PubMed]
- Gallardo, G.; Holtzman, D.M. Amyloid-β and Tau at the Crossroads of Alzheimer’s Disease. Adv. Exp. Med. Biol. 2019, 1184, 187–203. [Google Scholar] [PubMed]
- Risacher, S.L.; Anderson, W.H.; Charil, A.; Castelluccio, P.F.; Shcherbinin, S.; Saykin, A.J.; Schwarz, A.J.; Weiner, M.W. Alzheimer disease brain atrophy subtypes are associated with cognition and rate of decline. Neurology 2017, 89, 2176–2186. [Google Scholar] [CrossRef]
- Sepulveda-Falla, D.; Chavez-Gutierrez, L.; Portelius, E.; Vélez, J.I.; Dujardin, S.; Barrera-Ocampo, A.; Dinkel, F.; Hagel, C.; Puig, B.; Mastronardi, C.; et al. A multifactorial model of pathology for age of onset heterogeneity in familial Alzheimer’s disease. Acta Neuropathol. 2021, 141, 217–233. [Google Scholar] [CrossRef] [PubMed]
- Murray, M.E.; Graff-Radford, N.R.; Ross, O.A.; Petersen, R.C.; Duara, R.; Dickson, D.W. Neuropathologically defined subtypes of Alzheimer’s disease with distinct clinical characteristics: A retrospective study. Lancet Neurol. 2011, 10, 785–796. [Google Scholar] [CrossRef]
- Kim, C.; Haldiman, T.; Kang, S.G.; Hromadkova, L.; Han, Z.Z.; Chen, W.; Lissemore, F.; Lerner, A.; de Silva, R.; Cohen, M.L.; et al. Distinct populations of highly potent TAU seed conformers in rapidly progressing Alzheimer’s disease. Sci. Transl. Med. 2022, 14, eabg0253. [Google Scholar] [CrossRef]
- Liu, H.; Kim, C.; Haldiman, T.; Sigurdson, C.J.; Nyström, S.; Nilsson, K.P.R.; Cohen, M.L.; Wisniewski, T.; Hammarström, P.; Safar, J.G. Distinct conformers of amyloid beta accumulate in the neocortex of patients with rapidly progressive Alzheimer’s disease. J. Biol. Chem. 2021, 297, 101267. [Google Scholar] [CrossRef] [PubMed]
- Cohen, M.; Appleby, B.; Safar, J.G. Distinct Prion-Like Strains of Amyloid Beta Implicated in Phenotypic Diversity of Alzheimer Disease. Prion 2016, 10, 9–17. [Google Scholar] [CrossRef]
- Cohen, M.L.; Kim, C.; Haldiman, T.; ElHag, M.; Mehndiratta, P.; Pichet, T.; Lissemore, F.; Shea, M.; Cohen, Y.; Chen, W.; et al. Rapidly progressive Alzheimer’s disease features distinct structures of amyloid-beta. Brain 2015, 138, 1009–1022. [Google Scholar] [CrossRef]
- Schmidt, C.; Wolff, M.; Weitz, M.; Bartlau, T.; Korth, C.; Zerr, I. Rapidly progressive Alzheimer disease. Arch Neurol. 2011, 68, 1124–1130. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, G.G. Astroglia and Tau: New Perspectives. Front. Aging Neurosci. 2020, 12, 96. [Google Scholar] [CrossRef] [PubMed]
- Rösler, T.W.; Tayaranian Marvian, A.; Brendel, M.; Nykänen, N.P.; Höllerhage, M.; Schwarz, S.C.; Hopfner, F.; Koeglsperger, T.; Respondek, G.; Schweyer, K.; et al. Four-repeat tauopathies. Prog. Neurobiol. 2019, 180, 101644. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, G.G. Molecular pathology of neurodegenerative diseases: Principles and practice. J. Clin. Pathol. 2019, 72, 725–735. [Google Scholar] [CrossRef]
- Kovacs, G.G. Invited review: Neuropathology of tauopathies: Principles and practice. Neuropathol. Appl. Neurobiol. 2015, 41, 3–23. [Google Scholar] [CrossRef] [PubMed]
- Mrdjen, D.; Fox, E.J.; Bukhari, S.A.; Montine, K.S.; Bendall, S.C.; Montine, T.J. The basis of cellular and regional vulnerability in Alzheimer’s disease. Acta Neuropathol. 2019, 138, 729–749. [Google Scholar] [CrossRef] [PubMed]
- Schellenberg, G.D.; Montine, T.J. The genetics and neuropathology of Alzheimer’s disease. Acta Neuropathol. 2012, 124, 305–323. [Google Scholar] [CrossRef]
- Selkoe, D.J. Alzheimer’s disease. Cold Spring Harb. Perspect. Biol. 2011, 3, a004457. [Google Scholar] [CrossRef] [PubMed]
- Van Cauwenberghe, C.; Van Broeckhoven, C.; Sleegers, K. The genetic landscape of Alzheimer disease: Clinical implications and perspectives. Genet. Med. 2016, 18, 421–430. [Google Scholar] [CrossRef] [PubMed]
- Iwatsubo, T.; Hasegawa, M.; Ihara, Y. Neuronal and glial tau-positive inclusions in diverse neurologic diseases share common phosphorylation characteristics. Acta Neuropathol. 1994, 88, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, I.; López-González, I.; Carmona, M.; Arregui, L.; Dalfó, E.; Torrejón-Escribano, B.; Diehl, R.; Kovacs, G.G. Glial and neuronal tau pathology in tauopathies: Characterization of disease-specific phenotypes and tau pathology progression. J. Neuropathol. Exp. Neurol. 2014, 73, 81–97. [Google Scholar] [CrossRef]
- Higuchi, M.; Ishihara, T.; Zhang, B.; Hong, M.; Andreadis, A.; Trojanowski, J.Q.; Lee, V.M.-Y. Transgenic mouse model of tauopathies with glial pathology and nervous system degeneration. Neuron 2002, 35, 433–446. [Google Scholar] [CrossRef]
- Lin, W.-L.; Lewis, J.; Yen, S.-H.; Hutton, M.; Dickson, D.W. Filamentous tau in oligodendrocytes and astrocytes of transgenic mice expressing the human tau isoform with the P301L mutation. Am. J. Pathol. 2003, 162, 213–218. [Google Scholar] [CrossRef]
- Ferrer, I.; Zelaya, M.V.; Aguiló García, M.; Carmona, M.; López-González, I.; Andrés-Benito, P.; Lidón, L.; Gavín, R.; Garcia-Esparcia, P.; Del Rio, J.A. Relevance of host tau in tau seeding and spreading in tauopathies. Brain Pathol. 2020, 30, 298–318. [Google Scholar] [CrossRef]
- Forrest, S.L.; Kril, J.J.; Kovacs, G.G. Association Between Globular Glial Tauopathies and Frontotemporal Dementia-Expanding the Spectrum of Gliocentric Disorders: A Review. JAMA Neurol. 2021, 78, 1004–1014. [Google Scholar] [CrossRef]
- Hartnell, I.J.; Blum, D.; Nicoll, J.A.; Dorothee, G.; Boche, D. Glial cells and adaptive immunity in frontotemporal dementia with tau pathology. Brain 2021, 144, 724–745. [Google Scholar] [CrossRef]
- Odfalk, K.F.; Bieniek, K.F.; Hopp, S.C. Microglia: Friend and foe in tauopathy. Prog. Neurobiol. 2022, 216, 102306. [Google Scholar] [CrossRef]
- Forrest, S.L.; Kril, J.J.; Stevens, C.H.; Kwok, J.B.; Hallupp, M.; Kim, W.S.; Huang, Y.; McGinley, C.V.; Werka, H.; Kiernan, M.C.; et al. Retiring the term FTDP-17 as MAPT mutations are genetic forms of sporadic frontotemporal tauopathies. Brain 2018, 141, 521–534. [Google Scholar] [CrossRef]
- Wszolek, Z.K.; Tsuboi, Y.; Ghetti, B.; Pickering-Brown, S.; Baba, Y.; Cheshire, W.P. Frontotemporal dementia and parkinsonism linked to chromosome 17 (FTDP-17). Orphanet J. Rare Dis. 2006, 1, 30. [Google Scholar] [CrossRef]
- Miyasaka, T.; Morishima-Kawashima, M.; Ravid, R.; Kamphorst, W.; Nagashima, K.; Ihara, Y. Selective deposition of mutant tau in the FTDP-17 brain affected by the P301L mutation. J. Neuropathol. Exp. Neurol. 2001, 60, 872–884. [Google Scholar] [CrossRef]
- Mackenzie, I.R.; Neumann, M. Molecular neuropathology of frontotemporal dementia: Insights into disease mechanisms from postmortem studies. J. Neurochem. 2016, 138 (Suppl. S1), 54–70. [Google Scholar] [CrossRef]
- Josephs, K.A.; Nelson, P.T. Unlocking the mysteries of TDP-43. Neurology 2015, 84, 870–871. [Google Scholar] [CrossRef]
- Borrego-Écija, S.; Morgado, J.; Palencia-Madrid, L.; Grau-Rivera, O.; Reñé, R.; Hernández, I.; Almenar, C.; Balasa, M.; Antonell, A.; Molinuevo, J.L.; et al. Frontotemporal Dementia Caused by the P301L Mutation in the MAPT Gene: Clinicopathological Features of 13 Cases from the Same Geographical Origin in Barcelona, Spain. Dement. Geriatr. Cogn. Disord. 2017, 44, 213–221. [Google Scholar] [CrossRef]
- Stamelou, M.; Respondek, G.; Giagkou, N.; Whitwell, J.L.; Kovacs, G.G.; Höglinger, G.U. Evolving concepts in progressive supranuclear palsy and other 4-repeat tauopathies. Nat. Rev. Neurol. 2021, 17, 601–620. [Google Scholar] [CrossRef]
- Braak, H.; Del Tredici, K. Evolutional aspects of Alzheimer’s disease pathogenesis. J. Alzheimer’s Dis. 2013, 33 (Suppl. S1), S155–S161. [Google Scholar] [CrossRef]
- Sanders, D.W.; Kaufman, S.K.; Holmes, B.B.; Diamond, M.I. Prions and Protein Assemblies that Convey Biological Information in Health and Disease. Neuron 2016, 89, 433–448. [Google Scholar] [CrossRef]
- Kaufman, S.K.; Del Tredici, K.; Thomas, T.L.; Braak, H.; Diamond, M.I. Tau seeding activity begins in the transentorhinal/entorhinal regions and anticipates phospho-tau pathology in Alzheimer’s disease and PART. Acta Neuropathol. 2018, 136, 57–67. [Google Scholar] [CrossRef]
- Guo, J.L.; Narasimhan, S.; Changolkar, L.; He, Z.; Stieber, A.; Zhang, B.; Gathagan, R.J.; Iba, M.; McBride, J.D.; Trojanowski, J.Q.; et al. Unique pathological tau conformers from Alzheimer’s brains transmit tau pathology in nontransgenic mice. J. Exp. Med. 2016, 213, 2635–2654. [Google Scholar] [CrossRef] [PubMed]
- Jaunmuktane, Z.; Mead, S.; Ellis, M.; Wadsworth, J.D.; Nicoll, A.J.; Kenny, J.; Launchbury, F.; Linehan, J.; Richard-Loendt, A.; Walker, A.S.; et al. Evidence for human transmission of amyloid-beta pathology and cerebral amyloid angiopathy. Nature 2015, 525, 247–250. [Google Scholar] [CrossRef] [PubMed]
- Cali, I.; Cohen, M.L.; Haïk, S.; Parchi, P.; Giaccone, G.; Collins, S.J.; Kofskey, D.; Wang, H.; McLean, C.A.; Brandel, J.P.; et al. Iatrogenic Creutzfeldt-Jakob disease with Amyloid-β pathology: An international study. Acta Neuropathol. Commun. 2018, 6, 5. [Google Scholar] [CrossRef] [PubMed]
- Asher, D.M.; Belay, E.; Bigio, E.; Brandner, S.; Brubaker, S.A.; Caughey, B.; Clark, B.; Damon, I.; Diamond, M.; Freund, M.; et al. Risk of Transmissibility From Neurodegenerative Disease-Associated Proteins: Experimental Knowns and Unknowns. J. Neuropathol. Exp. Neurol. 2020, 79, 1141–1146. [Google Scholar] [CrossRef] [PubMed]
- Gambetti, P.; Kong, Q.; Zou, W.; Parchi, P.; Chen, S.G. Sporadic and familial CJD: Classification and characterisation. Br. Med. Bull. 2003, 66, 213–239. [Google Scholar] [CrossRef]
- Safar, J.G.; Xiao, X.; Kabir, M.E.; Chen, S.; Kim, C.; Haldiman, T.; Cohen, Y.; Chen, W.; Cohen, M.L.; Surewicz, W.K. Structural determinants of phenotypic diversity and replication rate of human prions. PLoS Pathog. 2015, 11, e1004832. [Google Scholar] [CrossRef]
- Safar, J.G. Molecular pathogenesis of sporadic prion diseases in man. Prion 2012, 6, 108–115. [Google Scholar] [CrossRef]
- Puoti, G.; Bizzi, A.; Forloni, G.; Safar, J.G.; Tagliavini, F.; Gambetti, P. Sporadic human prion diseases: Molecular insights and diagnosis. Lancet Neurol. 2012, 11, 618–628. [Google Scholar] [CrossRef]
- Cali, I.; Cohen, I.; Blevins, J.; Castellani, R.; Al-Shekhlee, A.; Yuan, J.; Parchi, P.; Safar, J.; Zou, W.-Q.; Gambetti, P. The Co-Existence of PrPSc Type 1 and 2 in Sporadic Creutzfeldt-Jakob Disease Affects the Phenotype and PrPSc Conformation. In Journal of Neuropathology and Experimental Neurology: 2009; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2009; p. 553. [Google Scholar]
- Gibbs, C.J., Jr.; Gajdusek, D.C.; Asher, D.M.; Alpers, M.P.; Beck, E.; Daniel, P.M.; Matthews, W.B. Creutzfeldt-Jakob disease (spongiform encephalopathy): Transmission to the chimpanzee. Science 1968, 161, 388–389. [Google Scholar] [CrossRef]
- Gajdusek, D.C.; Gibbs, C.J., Jr.; Alpers, M. Experimental transmission of a kuru-like syndrome to chimpanzees. Nature 1966, 209, 794–796. [Google Scholar] [CrossRef]
- Safar, J.G.; Geschwind, M.D.; Deering, C.; Didorenko, S.; Sattavat, M.; Sanchez, H.; Serban, A.; Vey, M.; Baron, H.; Giles, K. Diagnosis of human prion disease. Proc. Natl. Acad. Sci. USA 2005, 102, 3501–3506. [Google Scholar] [CrossRef] [PubMed]
- Safar, J.G.; Giles, K.; Lessard, P.; Letessier, F.; Patel, S.; Serban, A.; DeArmond, S.J.; Prusiner, S.B. Conserved properties of human and bovine prion strains on transmission to guinea pigs. Lab. Investig. 2011, 91, 1326–1336. [Google Scholar] [CrossRef][Green Version]
- Bruce, M.E.; Dickinson, A.G. Biological evidence that the scrapie agent has an independent genome. J. Gen. Virol. 1987, 68, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, A.G.; Outram, G.W. Genetic aspects of unconventional virus infections: The basis of the virino hypothesis. In Novel Infectious Agents and the Central Nervous System. Ciba Foundation Symposium 135; Bock, G., Marsh, J., Eds.; John Wiley and Sons: Chichester, UK, 1988; pp. 63–83. [Google Scholar]
- Meyer, N.; Rosenbaum, V.; Schmidt, B.; Gilles, K.; Mirenda, C.; Groth, D.; Prusiner, S.B.; Riesner, D. Search for a putative scrapie genome in purified prion fractions reveals a paucity of nucleic acids. J. Gen. Virol. 1991, 72, 37–49. [Google Scholar] [CrossRef] [PubMed]
- Kellings, K.; Prusiner, S.B.; Riesner, D. Nucleic acids in prion preparations: Unspecific background or essential component? Philos. Trans. R. Soc. Lond. B Biol. Sci. 1994, 343, 425–430. [Google Scholar] [PubMed]
- Kellings, K.; Meyer, N.; Mirenda, C.; Prusiner, S.B.; Riesner, D. Further analysis of nucleic acids in purified scrapie prion preparations by improved return refocussing gel electrophoresis (RRGE). J. Gen. Virol. 1992, 73, 1025–1029. [Google Scholar] [CrossRef] [PubMed]
- Safar, J.G.; Kellings, K.; Serban, A.; Groth, D.; Cleaver, J.E.; Prusiner, S.B.; Riesner, D. Search for a prion-specific nucleic acid. J. Virol. 2005, 79, 10796–10806. [Google Scholar] [CrossRef]
- Prusiner, S.B. Prions (Les Prix Nobel Lecture). In Les Prix Nobel; Frängsmyr, T., Ed.; Almqvist & Wiksell International: Stockholm, Sweden, 1998; pp. 268–323. [Google Scholar]
- Weissmann, C. The state of the prion. Nat. Rev. Microbiol. 2004, 2, 861–871. [Google Scholar] [CrossRef] [PubMed]
- Sandberg, M.K.; Al-Doujaily, H.; Sharps, B.; De Oliveira, M.W.; Schmidt, C.; Richard-Londt, A.; Lyall, S.; Linehan, J.M.; Brandner, S.; Wadsworth, J.D.; et al. Prion neuropathology follows the accumulation of alternate prion protein isoforms after infective titre has peaked. Nat. Commun. 2014, 5, 4347. [Google Scholar] [CrossRef] [PubMed]
- Collinge, J.; Clarke, A.R. A general model of prion strains and their pathogenicity. Science 2007, 318, 930–936. [Google Scholar] [CrossRef] [PubMed]
- Safar, J.G. Molecular Mechanisms Encoding Quantitative and Qualitative Traits of Prion Strains. In Prions and Diseases; Zou, W.A.G.P., Ed.; Springer Verlag: New York, NY, USA, 2012; Volume 1. [Google Scholar]
- Caughey, B.; Baron, G.S.; Chesebro, B.; Jeffrey, M. Getting a grip on prions: Oligomers, amyloids, and pathological membrane interactions. Annu. Rev. Biochem. 2009, 78, 177–204. [Google Scholar] [CrossRef]
- Cobb, N.J.; Surewicz, W.K. Prion diseases and their biochemical mechanisms. Biochemistry 2009, 48, 2574–2585. [Google Scholar] [CrossRef]
- Morales, R.; Abid, K.; Soto, C. The prion strain phenomenon: Molecular basis and unprecedented features. Biochim. Biophys. Acta 2007, 1772, 681–691. [Google Scholar] [CrossRef]
- Prusiner, S.B. Molecular and genetic basis of prion diseases. In Principles of Molecular Medicine; Jameson, J.L., Ed.; Humana Press: Totowa, NJ, USA, 1998; pp. 927–939. [Google Scholar]
- Prusiner, S.B.; Legname, G.; DeArmond, S.J.; Cohen, F.E.; Safar, J.; Riesner, D.; Kaneko, K. Some strategies and methods for the study of prions. In Prion Biology and Diseases, 2nd ed.; Prusiner, S.B., Ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2004; pp. 857–920. [Google Scholar]
- Kim, C.; Xiao, X.; Chen, S.; Haldiman, T.; Smirnovas, V.; Kofskey, D.; Warren, M.; Surewicz, K.; Maurer, N.R.; Kong, Q.; et al. Artificial strain of human prions created in vitro. Nat. Commun. 2018, 9, 2166. [Google Scholar] [CrossRef]
- Foutz, A.; Appleby, B.S.; Hamlin, C.; Liu, X.; Yang, S.; Cohen, Y.; Chen, W.; Blevins, J.; Fausett, C.; Wang, H.; et al. Diagnostic and prognostic value of human prion detection in cerebrospinal fluid. Ann. Neurol. 2017, 81, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Baiardi, S.; Rossi, M.; Mammana, A.; Appleby, B.S.; Barria, M.A.; Calì, I.; Gambetti, P.; Gelpi, E.; Giese, A.; Ghetti, B.; et al. Phenotypic diversity of genetic Creutzfeldt-Jakob disease: A histo-molecular-based classification. Acta Neuropathol. 2021, 142, 707–728. [Google Scholar] [CrossRef]
- Parchi, P.; de Boni, L.; Saverioni, D.; Cohen, M.L.; Ferrer, I.; Gambetti, P.; Gelpi, E.; Giaccone, G.; Hauw, J.J.; Hoftberger, R.; et al. Consensus classification of human prion disease histotypes allows reliable identification of molecular subtypes: An inter-rater study among surveillance centres in Europe and USA. Acta Neuropathol. 2012, 124, 517–529. [Google Scholar] [CrossRef] [PubMed]
- Minikel, E.V.; Vallabh, S.M.; Lek, M.; Estrada, K.; Samocha, K.E.; Sathirapongsasuti, J.F.; McLean, C.Y.; Tung, J.Y.; Yu, L.P.; Gambetti, P.; et al. Quantifying prion disease penetrance using large population control cohorts. Sci. Transl. Med. 2016, 8, 322ra329. [Google Scholar] [CrossRef]
- Minikel, E.V.; Vallabh, S.M.; Orseth, M.C.; Brandel, J.-P.; Haïk, S.; Laplanche, J.-L.; Zerr, I.; Parchi, P.; Capellari, S.; Safar, J. Age at onset in genetic prion disease and the design of preventive clinical trials. Neurology 2019, 93, e125–e134. [Google Scholar] [CrossRef] [PubMed]
- Siddiqi, M.K.; Kim, C.; Haldiman, T.; Kacirova, M.; Wang, B.; Bohon, J.; Chance, M.R.; Kiselar, J.; Safar, J.G. Structurally distinct external solvent-exposed domains drive replication of major human prions. PLoS Pathog. 2021, 17, e1009642. [Google Scholar] [CrossRef]
- Collins, S.J.; Sanchez-Juan, P.; Masters, C.L.; Klug, G.M.; van Duijn, C.; Poleggi, A.; Pocchiari, M.; Almonti, S.; Cuadrado-Corrales, N.; de Pedro-Cuesta, J.; et al. Determinants of diagnostic investigation sensitivities across the clinical spectrum of sporadic Creutzfeldt-Jakob disease. Brain 2006, 129, 2278–2287. [Google Scholar] [CrossRef]
- Pocchiari, M.; Puopolo, M.; Croes, E.A.; Budka, H.; Gelpi, E.; Collins, S.; Lewis, V.; Sutcliffe, T.; Guilivi, A.; Delasnerie-Laupretre, N.; et al. Predictors of survival in sporadic Creutzfeldt-Jakob disease and other human transmissible spongiform encephalopathies. Brain 2004, 127, 2348–2359. [Google Scholar] [CrossRef] [PubMed]
- Telling, G.C. Transgenic mouse models of prion diseases. Methods Mol. Biol. 2008, 459, 249–263. [Google Scholar]
- Bishop, M.T.; Will, R.G.; Manson, J.C. Defining sporadic Creutzfeldt-Jakob disease strains and their transmission properties. Proc. Natl. Acad. Sci. USA 2010, 107, 12005–12010. [Google Scholar] [CrossRef]
- Prusiner, S.; Safar, J.; DeArmond, S. Bioassays of prions. Cold Spring Harb. Monogr. Ser. 2004, 41, 143–186. [Google Scholar]
- Wadsworth, J.D.; Collinge, J. Molecular pathology of human prion disease. Acta Neuropathol. 2011, 121, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Wadsworth, J.D.; Hill, A.F.; Beck, J.A.; Collinge, J. Molecular and clinical classification of human prion disease. Br. Med. Bull. 2003, 66, 241–254. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.; Haldiman, T.; Surewicz, K.; Cohen, Y.; Chen, W.; Blevins, J.; Sy, M.S.; Cohen, M.; Kong, Q.; Telling, G.C.; et al. Small Protease Sensitive Oligomers of PrP(Sc) in Distinct Human Prions Determine Conversion Rate of PrP(C). PLoS Pathog. 2012, 8, e1002835. [Google Scholar] [CrossRef]
- Safar, J.; Wille, H.; Itri, V.; Groth, D.; Serban, H.; Torchia, M.; Cohen, F.E.; Prusiner, S.B. Eight prion strains have PrPSc molecules with different conformations. Nat. Med. 1998, 4, 1157–1165. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.; Haldiman, T.; Cohen, Y.; Chen, W.; Blevins, J.; Sy, M.S.; Cohen, M.; Safar, J.G. Protease-Sensitive Conformers in Broad Spectrum of Distinct PrP Structures in Sporadic Creutzfeldt-Jakob Disease Are Indicator of Progression Rate. PLoS Pathog. 2011, 7, e1002242. [Google Scholar] [CrossRef]
- Masters, C.L.; Selkoe, D.J. Biochemistry of amyloid beta-protein and amyloid deposits in Alzheimer disease. Cold Spring Harb. Perspect. Med. 2012, 2, a006262. [Google Scholar] [CrossRef]
- Schmidt, C.; Karch, A.; Artjomova, S.; Hoeschel, M.; Zerr, I. Pre-progression rates in Alzheimer’s disease revisited. J. Alzheimers Dis. 2013, 35, 451–454. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, C.; Haik, S.; Satoh, K.; Rabano, A.; Martinez-Martin, P.; Roeber, S.; Brandel, J.P.; Calero-Lara, M.; de Pedro-Cuesta, J.; Laplanche, J.L.; et al. Rapidly progressive Alzheimer’s disease: A multicenter update. J. Alzheimers Dis. 2012, 30, 751–756. [Google Scholar] [CrossRef]
- Mirbaha, H.; Chen, D.; Morazova, O.A.; Ruff, K.M.; Sharma, A.M.; Liu, X.; Goodarzi, M.; Pappu, R.V.; Colby, D.W.; Mirzaei, H.; et al. Inert and seed-competent tau monomers suggest structural origins of aggregation. Elife 2018, 7, e36584. [Google Scholar] [CrossRef]
- Sharma, A.M.; Thomas, T.L.; Woodard, D.R.; Kashmer, O.M.; Diamond, M.I. Tau monomer encodes strains. Elife 2018, 7, e37813. [Google Scholar] [CrossRef]
- Kaniyappan, S.; Tepper, K.; Biernat, J.; Chandupatla, R.R.; Hübschmann, S.; Irsen, S.; Bicher, S.; Klatt, C.; Mandelkow, E.-M.; Mandelkow, E. FRET-based Tau seeding assay does not represent prion-like templated assembly of Tau filaments. Mol. Neurodegener. 2020, 15, 39. [Google Scholar] [CrossRef]
- Goedert, M.; Wischik, C.M.; Crowther, R.A.; Walker, J.E.; Klug, A. Cloning and sequencing of the cDNA encoding a core protein of the paired helical filament of Alzheimer disease: Identification as the microtubule-associated protein tau. Proc. Natl. Acad. Sci. USA 1988, 85, 4051–4055. [Google Scholar] [CrossRef]
- Goedert, M.; Jakes, R. Expression of separate isoforms of human tau protein: Correlation with the tau pattern in brain and effects on tubulin polymerization. EMBO J. 1990, 9, 4225–4230. [Google Scholar] [CrossRef]
- Goedert, M.; Spillantini, M.G.; Jakes, R.; Rutherford, D.; Crowther, R.A. Multiple isoforms of human microtubule-associated protein tau: Sequences and localization in neurofibrillary tangles of Alzheimer’s disease. Neuron 1989, 3, 519–526. [Google Scholar] [CrossRef]
- Muller, R.; Heinrich, M.; Heck, S.; Blohm, D.; Richter-Landsberg, C. Expression of microtubule-associated proteins MAP2 and tau in cultured rat brain oligodendrocytes. Cell Tissue Res. 1997, 288, 239–249. [Google Scholar] [CrossRef]
- Couchie, D.; Fages, C.; Bridoux, A.M.; Rolland, B.; Tardy, M.; Nunez, J. Microtubule-associated proteins and in vitro astrocyte differentiation. J. Cell Biol. 1985, 101, 2095–2103. [Google Scholar] [CrossRef] [PubMed]
- Weingarten, M.D.; Lockwood, A.H.; Hwo, S.Y.; Kirschner, M.W. A protein factor essential for microtubule assembly. Proc. Natl. Acad. Sci. USA 1975, 72, 1858–1862. [Google Scholar] [CrossRef] [PubMed]
- Binder, L.I.; Frankfurter, A.; Rebhun, L.I. The distribution of tau in the mammalian central nervous system. J. Cell Biol. 1985, 101, 1371–1378. [Google Scholar] [CrossRef] [PubMed]
- Kempf, M.; Clement, A.; Faissner, A.; Lee, G.; Brandt, R. Tau binds to the distal axon early in development of polarity in a microtubule- and microfilament-dependent manner. J. Neurosci. 1996, 16, 5583–5592. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Kanai, Y.; Cowan, N.; Hirokawa, N. Projection domains of MAP2 and tau determine spacings between microtubules in dendrites and axons. Nature 1992, 360, 674–677. [Google Scholar] [CrossRef]
- Feinstein, H.E.; Benbow, S.J.; LaPointe, N.E.; Patel, N.; Ramachandran, S.; Do, T.D.; Gaylord, M.R.; Huskey, N.E.; Dressler, N.; Korff, M. Oligomerization of the microtubule-associated protein tau is mediated by its N-terminal sequences: Implications for normal and pathological tau action. J. Neurochem. 2016, 137, 939–954. [Google Scholar] [CrossRef]
- Mukrasch, M.D.; Bibow, S.; Korukottu, J.; Jeganathan, S.; Biernat, J.; Griesinger, C.; Mandelkow, E.; Zweckstetter, M. Structural polymorphism of 441-residue tau at single residue resolution. PLoS Biol. 2009, 7, e34. [Google Scholar] [CrossRef]
- Lövestam, S.; Koh, F.A.; van Knippenberg, B.; Kotecha, A.; Murzin, A.G.; Goedert, M.; Scheres, S.H. Assembly of recombinant tau into filaments identical to those of Alzheimer’s disease and chronic traumatic encephalopathy. Elife 2022, 11, e76494. [Google Scholar] [CrossRef]
- Fitzpatrick, A.W.; Falcon, B.; He, S.; Murzin, A.G.; Murshudov, G.; Garringer, H.J.; Crowther, R.A.; Ghetti, B.; Goedert, M.; Scheres, S.H. Cryo-EM structures of tau filaments from Alzheimer’s disease. Nature 2017, 547, 185–190. [Google Scholar] [CrossRef]
- Arrasate, M.; Perez, M.; Avila, J. Tau dephosphorylation at tau-1 site correlates with its association to cell membrane. Neurochem. Res. 2000, 25, 43–50. [Google Scholar] [CrossRef]
- Correia, S.C.; Santos, R.X.; Perry, G.; Zhu, X.; Moreira, P.I.; Smith, M.A. Mitochondrial importance in Alzheimer’s, Huntington’s and Parkinson’s diseases. Adv. Exp. Med. Biol. 2012, 724, 205–221. [Google Scholar] [PubMed]
- Lu, J.; Li, T.; He, R.; Bartlett, P.F.; Götz, J. Visualizing the microtubule-associated protein tau in the nucleus. Sci. China Life Sci. 2014, 57, 422–431. [Google Scholar] [CrossRef] [PubMed]
- Cuchillo-Ibanez, I.; Seereeram, A.; Byers, H.L.; Leung, K.Y.; Ward, M.A.; Anderton, B.H.; Hanger, D.P. Phosphorylation of tau regulates its axonal transport by controlling its binding to kinesin. FASEB J. 2008, 22, 3186–3195. [Google Scholar] [CrossRef] [PubMed]
- Morris, S.L.; Tsai, M.-Y.; Aloe, S.; Bechberger, K.; König, S.; Morfini, G.; Brady, S.T. Defined tau phosphospecies differentially inhibit fast axonal transport through activation of two independent signaling pathways. Front. Mol. Neurosci. 2021, 13, 610037. [Google Scholar] [CrossRef]
- Violet, M.; Delattre, L.; Tardivel, M.; Sultan, A.; Chauderlier, A.; Caillierez, R.; Talahari, S.; Nesslany, F.; Lefebvre, B.; Bonnefoy, E.; et al. A major role for Tau in neuronal DNA and RNA protection in vivo under physiological and hyperthermic conditions. Front. Cell Neurosci. 2014, 8, 84. [Google Scholar] [CrossRef]
- Kang, S.-G.; Eskandari-Sedighi, G.; Hromadkova, L.; Safar, J.G.; Westaway, D. Cellular biology of tau diversity and pathogenic conformers. Front. Neurol. 2020, 11, 590199. [Google Scholar] [CrossRef]
- Biundo, F.; Del Prete, D.; Zhang, H.; Arancio, O.; D’Adamio, L. A role for tau in learning, memory and synaptic plasticity. Sci. Rep. 2018, 8, 3184. [Google Scholar] [CrossRef]
- Dawson, H.N.; Ferreira, A.; Eyster, M.V.; Ghoshal, N.; Binder, L.I.; Vitek, M.P. Inhibition of neuronal maturation in primary hippocampal neurons from τ deficient mice. J. Cell Sci. 2001, 114, 1179–1187. [Google Scholar] [CrossRef]
- Novak, M.; Kabat, J.; Wischik, C.M. Molecular characterization of the minimal protease resistant tau unit of the Alzheimer’s disease paired helical filament. EMBO J. 1993, 12, 365–370. [Google Scholar] [CrossRef]
- Cantarero, L.A.; Butler, J.E.; Osborne, J.W. The binding characteristics of proteins for polystyrene and their significance in solid-phase immunoassays. Anal. Biochem. 1980, 105, 375–382. [Google Scholar] [CrossRef]
- Goedert, M.; Spillantini, M.G. Ordered Assembly of Tau Protein and Neurodegeneration. Adv. Exp. Med. Biol. 2019, 1184, 3–21. [Google Scholar] [PubMed]
- D’Souza, I.; Poorkaj, P.; Hong, M.; Nochlin, D.; Lee, V.M.-Y.; Bird, T.D.; Schellenberg, G.D. Missense and silent tau gene mutations cause frontotemporal dementia with parkinsonism-chromosome 17 type, by affecting multiple alternative RNA splicing regulatory elements. Proc. Natl. Acad. Sci. USA 1999, 96, 5598–5603. [Google Scholar] [CrossRef] [PubMed]
- Combs, B.; Gamblin, T.C. FTDP-17 tau mutations induce distinct effects on aggregation and microtubule interactions. Biochemistry 2012, 51, 8597–8607. [Google Scholar] [CrossRef]
- Spillantini, M.G.; Murrell, J.R.; Goedert, M.; Farlow, M.R.; Klug, A.; Ghetti, B. Mutation in the tau gene in familial multiple system tauopathy with presenile dementia. Proc. Natl. Acad. Sci. USA 1998, 95, 7737–7741. [Google Scholar] [CrossRef] [PubMed]
- Hutton, M.; Lendon, C.L.; Rizzu, P.; Baker, M.; Froelich, S.; Houlden, H.; Pickering-Brown, S.; Chakraverty, S.; Isaacs, A.; Grover, A. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature 1998, 393, 702–705. [Google Scholar] [CrossRef]
- Probst, A.; Götz, J.; Wiederhold, K.; Tolnay, M.; Mistl, C.; Jaton, A.; Hong, M.; Ishihara, T.; Lee, V.-Y.; Trojanowski, J. Axonopathy and amyotrophy in mice transgenic for human four-repeat tau protein. Acta Neuropathol. 2000, 99, 469–481. [Google Scholar] [CrossRef]
- Lewis, J.; McGowan, E.; Rockwood, J.; Melrose, H.; Nacharaju, P.; Van Slegtenhorst, M.; Gwinn-Hardy, K.; Murphy, M.P.; Baker, M.; Yu, X. Neurofibrillary tangles, amyotrophy and progressive motor disturbance in mice expressing mutant (P301L) tau protein. Nat. Genet. 2000, 25, 402–405. [Google Scholar] [CrossRef]
- Götz, J.; Chen, F.; Barmettler, R.; Nitsch, R.M. Tau filament formation in transgenic mice expressing P301L tau. J. Biol. Chem. 2001, 276, 529–534. [Google Scholar]
- Allen, B.; Ingram, E.; Takao, M.; Smith, M.J.; Jakes, R.; Virdee, K.; Yoshida, H.; Holzer, M.; Craxton, M.; Emson, P.C. Abundant tau filaments and nonapoptotic neurodegeneration in transgenic mice expressing human P301S tau protein. J. Neurosci. 2002, 22, 9340–9351. [Google Scholar] [CrossRef]
- Bellucci, A.; Westwood, A.J.; Ingram, E.; Casamenti, F.; Goedert, M.; Spillantini, M.G. Induction of inflammatory mediators and microglial activation in mice transgenic for mutant human P301S tau protein. Am. J. Pathol. 2004, 165, 1643–1652. [Google Scholar] [CrossRef]
- Santacruz, K.; Lewis, J.; Spires, T.; Paulson, J.; Kotilinek, L.; Ingelsson, M.; Guimaraes, A.; DeTure, M.; Ramsden, M.; McGowan, E. Tau suppression in a neurodegenerative mouse model improves memory function. Science 2005, 309, 476–481. [Google Scholar] [CrossRef] [PubMed]
- Tanemura, K.; Murayama, M.; Akagi, T.; Hashikawa, T.; Tominaga, T.; Ichikawa, M.; Yamaguchi, H.; Takashima, A. Neurodegeneration with tau accumulation in a transgenic mouse expressing V337M human tau. J. Neurosci. 2002, 22, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Tatebayashi, Y.; Miyasaka, T.; Chui, D.-H.; Akagi, T.; Mishima, K.-i.; Iwasaki, K.; Fujiwara, M.; Tanemura, K.; Murayama, M.; Ishiguro, K. Tau filament formation and associative memory deficit in aged mice expressing mutant (R406W) human tau. Proc. Natl. Acad. Sci. USA 2002, 99, 13896–13901. [Google Scholar] [CrossRef] [PubMed]
- Murakami, T.; Paitel, E.; Kawarabayashi, T.; Ikeda, M.; Chishti, M.A.; Janus, C.; Matsubara, E.; Sasaki, A.; Kawarai, T.; Phinney, A.L. Cortical neuronal and glial pathology in TgTauP301L transgenic mice: Neuronal degeneration, memory disturbance, and phenotypic variation. Am. J. Pathol. 2006, 169, 1365–1375. [Google Scholar] [CrossRef]
- Yoshiyama, Y.; Higuchi, M.; Zhang, B.; Huang, S.-M.; Iwata, N.; Saido, T.C.; Maeda, J.; Suhara, T.; Trojanowski, J.Q.; Lee, V.M.-Y. Synapse loss and microglial activation precede tangles in a P301S tauopathy mouse model. Neuron 2007, 53, 337–351. [Google Scholar] [CrossRef]
- Gibbons, G.S.; Banks, R.A.; Kim, B.; Xu, H.; Changolkar, L.; Leight, S.N.; Riddle, D.M.; Li, C.; Gathagan, R.J.; Brown, H.J. GFP-mutant human tau transgenic mice develop Tauopathy following CNS injections of Alzheimer’s brain-derived pathological tau or synthetic mutant human tau fibrils. J. Neurosci. 2017, 37, 11485–11494. [Google Scholar] [CrossRef]
- Guo, J.L.; Lee, V.M. Neurofibrillary tangle-like tau pathology induced by synthetic tau fibrils in primary neurons over-expressing mutant tau. FEBS Lett. 2013, 587, 717–723. [Google Scholar] [CrossRef]
- Eskandari-Sedighi, G.; Daude, N.; Gapeshina, H.; Sanders, D.W.; Kamali-Jamil, R.; Yang, J.; Shi, B.; Wille, H.; Ghetti, B.; Diamond, M.I. The CNS in inbred transgenic models of 4-repeat Tauopathy develops consistent tau seeding capacity yet focal and diverse patterns of protein deposition. Mol. Neurodegener. 2017, 12, 72. [Google Scholar] [CrossRef]
- Martini-Stoica, H.; Cole, A.L.; Swartzlander, D.B.; Chen, F.; Wan, Y.-W.; Bajaj, L.; Bader, D.A.; Lee, V.M.; Trojanowski, J.Q.; Liu, Z. TFEB enhances astroglial uptake of extracellular tau species and reduces tau spreading. J. Exp. Med. 2018, 215, 2355–2377. [Google Scholar] [CrossRef]
- Lee, V.M.-Y.; Kenyon, T.K.; Trojanowski, J.Q. Transgenic animal models of tauopathies. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2005, 1739, 251–259. [Google Scholar] [CrossRef]
- Sanchez-Varo, R.; Mejias-Ortega, M.; Fernandez-Valenzuela, J.J.; Nuñez-Diaz, C.; Caceres-Palomo, L.; Vegas-Gomez, L.; Sanchez-Mejias, E.; Trujillo-Estrada, L.; Garcia-Leon, J.A.; Moreno-Gonzalez, I. Transgenic Mouse Models of Alzheimer’s Disease: An Integrative Analysis. Int. J. Mol. Sci. 2022, 23, 5404. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.Z.; Kang, S.-G.; Arce, L.; Westaway, D. Prion-like strain effects in tauopathies. Cell Tissue Res. 2022, 1–21, online ahead of print. [Google Scholar] [CrossRef]
- Houben, S.; Homa, M.; Yilmaz, Z.; Leroy, K.; Brion, J.-P.; Ando, K. Tau pathology and adult hippocampal neurogenesis: What tau mouse models tell us? Front. Neurol. 2021, 12, 610330. [Google Scholar] [CrossRef] [PubMed]
- Robert, A.; Schöll, M.; Vogels, T. Tau Seeding Mouse Models with Patient Brain-Derived Aggregates. Int. J. Mol. Sci. 2021, 22, 6132. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Falcon, B.; Murzin, A.G.; Fan, J.; Crowther, R.A.; Goedert, M.; Scheres, S.H.W. Heparin-induced tau filaments are polymorphic and differ from those in Alzheimer’s and Pick’s diseases. eLife 2019, 8, e43584. [Google Scholar] [CrossRef] [PubMed]
- Falcon, B.; Zivanov, J.; Zhang, W.; Murzin, A.G.; Garringer, H.J.; Vidal, R.; Crowther, R.A.; Newell, K.L.; Ghetti, B.; Goedert, M.; et al. Novel tau filament fold in chronic traumatic encephalopathy encloses hydrophobic molecules. Nature 2019, 568, 420–423. [Google Scholar] [CrossRef]
- Goedert, M. Tau filaments in neurodegenerative diseases. FEBS Lett. 2018, 592, 2383–2391. [Google Scholar] [CrossRef] [PubMed]
- Falcon, B.; Zhang, W.; Schweighauser, M.; Murzin, A.G.; Vidal, R.; Garringer, H.J.; Ghetti, B.; Scheres, S.H.W.; Goedert, M. Tau filaments from multiple cases of sporadic and inherited Alzheimer’s disease adopt a common fold. Acta Neuropathol. 2018, 136, 699–708. [Google Scholar] [CrossRef]
- Stopschinski, B.E.; Del Tredici, K.; Estill-Terpack, S.-J.; Ghebremdehin, E.; Yu, F.F.; Braak, H.; Diamond, M.I. Anatomic survey of seeding in Alzheimer’s disease brains reveals unexpected patterns. Acta Neuropathol. Commun. 2021, 9, 164. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.G.; Han, Z.Z.; Daude, N.; McNamara, E.; Wohlgemuth, S.; Molina-Porcel, L.; Safar, J.G.; Mok, S.A.; Westaway, D. Pathologic tau conformer ensembles induce dynamic, liquid-liquid phase separation events at the nuclear envelope. BMC Biol. 2021, 19, 199. [Google Scholar] [CrossRef]
- Dujardin, S.; Commins, C.; Lathuiliere, A.; Beerepoot, P.; Fernandes, A.R.; Kamath, T.V.; De Los Santos, M.B.; Klickstein, N.; Corjuc, D.L.; Corjuc, B.T.; et al. Tau molecular diversity contributes to clinical heterogeneity in Alzheimer’s disease. Nat. Med. 2020, 26, 1256–1263. [Google Scholar] [CrossRef] [PubMed]
- Atarashi, R.; Moore, R.A.; Sim, V.L.; Hughson, A.G.; Dorward, D.W.; Onwubiko, H.A.; Priola, S.A.; Caughey, B. Ultrasensitive detection of scrapie prion protein using seeded conversion of recombinant prion protein. Nat. Methods 2007, 4, 645–650. [Google Scholar] [CrossRef] [PubMed]
- Vaquer-Alicea, J.; Diamond, M.I.; Joachimiak, L.A. Tau strains shape disease. Acta Neuropathol. 2021, 142, 57–71. [Google Scholar] [CrossRef] [PubMed]
- Daude, N.; Kim, C.; Kang, S.G.; Eskandari-Sedighi, G.; Haldiman, T.; Yang, J.; Fleck, S.C.; Gomez-Cardona, E.; Han, Z.Z.; Borrego-Ecija, S.; et al. Diverse, evolving conformer populations drive distinct phenotypes in frontotemporal lobar degeneration caused by the same MAPT-P301L mutation. Acta Neuropathol. 2020, 139, 1045–1070. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Chance, M.R. Protein footprinting comes of age: Mass spectrometry for biophysical structure assessment. Mol. Cell. Proteom. 2017, 16, 706–716. [Google Scholar] [CrossRef] [PubMed]
- Wesseling, H.; Mair, W.; Kumar, M.; Schlaffner, C.N.; Tang, S.; Beerepoot, P.; Fatou, B.; Guise, A.J.; Cheng, L.; Takeda, S.; et al. Tau PTM Profiles Identify Patient Heterogeneity and Stages of Alzheimer’s Disease. Cell 2020, 183, 1699–1713.e1613. [Google Scholar] [CrossRef] [PubMed]
- Augustinack, J.C.; Schneider, A.; Mandelkow, E.-M.; Hyman, B.T. Specific tau phosphorylation sites correlate with severity of neuronal cytopathology in Alzheimer’s disease. Acta Neuropathol. 2002, 103, 26–35. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Prokop, S.; Giasson, B.I. “Don’t Phos Over Tau”: Recent developments in clinical biomarkers and therapies targeting tau phosphorylation in Alzheimer’s disease and other tauopathies. Mol. Neurodegener. 2021, 16, 37. [Google Scholar] [CrossRef] [PubMed]
- Wegmann, S.; Biernat, J.; Mandelkow, E. A current view on Tau protein phosphorylation in Alzheimer’s disease. Curr. Opin. Neurobiol. 2021, 69, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Tseng, J.H.; Ajit, A.; Tabassum, Z.; Patel, N.; Tian, X.; Chen, Y.; Prevatte, A.W.; Ling, K.; Rigo, F.; Meeker, R.B.; et al. Tau seeds are subject to aberrant modifications resulting in distinct signatures. Cell Rep. 2021, 35, 109037. [Google Scholar] [CrossRef]
- Furman, J.L.; Vaquer-Alicea, J.; White, C.L.; Cairns, N.J.; Nelson, P.T.; Diamond, M.I. Widespread tau seeding activity at early Braak stages. Acta Neuropathol. 2017, 133, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Mirbaha, H.; Chen, D.; Mullapudi, V.; Terpack, S.J.; White, C.L., III; Joachimiak, L.A.; Diamond, M.I. Seed-competent tau monomer initiates pathology in a tauopathy mouse model. J. Biol. Chem. 2022, 298, 102163. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wang, F.; Xiao, X.; Kim, C.; Bohon, J.; Kiselar, J.; Safar, J.G.; Ma, J.; Surewicz, W.K. Structural attributes of mammalian prion infectivity: Insights from studies with synthetic prions. J. Biol. Chem. 2018, 293, 18494–18503. [Google Scholar] [CrossRef]
- Kraus, A.; Saijo, E.; Metrick, M.A., 2nd; Newell, K.; Sigurdson, C.J.; Zanusso, G.; Ghetti, B.; Caughey, B. Seeding selectivity and ultrasensitive detection of tau aggregate conformers of Alzheimer disease. Acta Neuropathol. 2019, 137, 585–598. [Google Scholar] [CrossRef] [PubMed]
- Groveman, B.R.; Orru, C.D.; Hughson, A.G.; Raymond, L.D.; Zanusso, G.; Ghetti, B.; Campbell, K.J.; Safar, J.; Galasko, D.; Caughey, B. Rapid and ultra-sensitive quantitation of disease-associated alpha-synuclein seeds in brain and cerebrospinal fluid by alphaSyn RT-QuIC. Acta Neuropathol. Commun. 2018, 6, 7. [Google Scholar] [CrossRef] [PubMed]
- Babinchak, W.M.; Haider, R.; Dumm, B.K.; Sarkar, P.; Surewicz, K.; Choi, J.K.; Surewicz, W.K. The role of liquid-liquid phase separation in aggregation of the TDP-43 low-complexity domain. J. Biol. Chem. 2019, 294, 6306–6317. [Google Scholar] [CrossRef] [PubMed]
- Jeganathan, S.; von Bergen, M.; Mandelkow, E.M.; Mandelkow, E. The natively unfolded character of tau and its aggregation to Alzheimer-like paired helical filaments. Biochemistry 2008, 47, 10526–10539. [Google Scholar] [CrossRef] [PubMed]
- Daebel, V.; Chinnathambi, S.; Biernat, J.; Schwalbe, M.; Habenstein, B.; Loquet, A.; Akoury, E.; Tepper, K.; Muller, H.; Baldus, M.; et al. beta-Sheet core of tau paired helical filaments revealed by solid-state NMR. J. Am. Chem. Soc. 2012, 134, 13982–13989. [Google Scholar] [CrossRef]
- Saijo, E.; Metrick, M.A., 2nd; Koga, S.; Parchi, P.; Litvan, I.; Spina, S.; Boxer, A.; Rojas, J.C.; Galasko, D.; Kraus, A.; et al. 4-Repeat tau seeds and templating subtypes as brain and CSF biomarkers of frontotemporal lobar degeneration. Acta Neuropathol. 2020, 139, 63–77. [Google Scholar] [CrossRef] [PubMed]
- Metrick, M.A., 2nd; Ferreira, N.D.C.; Saijo, E.; Kraus, A.; Newell, K.; Zanusso, G.; Vendruscolo, M.; Ghetti, B.; Caughey, B. A single ultrasensitive assay for detection and discrimination of tau aggregates of Alzheimer and Pick diseases. Acta Neuropathol. Commun. 2020, 8, 22. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Sahara, N. Characteristics of tau oligomers. Front. Neurol. 2013, 4, 102. [Google Scholar] [CrossRef]
- Paravastu, A.K.; Leapman, R.D.; Yau, W.M.; Tycko, R. Molecular structural basis for polymorphism in Alzheimer’s beta-amyloid fibrils. Proc. Natl. Acad. Sci. USA 2008, 105, 18349–18354. [Google Scholar] [CrossRef] [PubMed]
- Michaels, T.C.T.; Šarić, A.; Habchi, J.; Chia, S.; Meisl, G.; Vendruscolo, M.; Dobson, C.M.; Knowles, T.P.J. Chemical Kinetics for Bridging Molecular Mechanisms and Macroscopic Measurements of Amyloid Fibril Formation. Annu. Rev. Phys. Chem. 2018, 69, 273–298. [Google Scholar] [CrossRef]
- Noble, G.P.; Wang, D.W.; Walsh, D.J.; Barone, J.R.; Miller, M.B.; Nishina, K.A.; Li, S.; Supattapone, S. A Structural and Functional Comparison Between Infectious and Non-Infectious Autocatalytic Recombinant PrP Conformers. PLoS Pathog. 2015, 11, e1005017. [Google Scholar] [CrossRef] [PubMed]
- Deleault, N.R.; Walsh, D.J.; Piro, J.R.; Wang, F.; Wang, X.; Ma, J.; Rees, J.R.; Supattapone, S. Cofactor molecules maintain infectious conformation and restrict strain properties in purified prions. Proc. Natl. Acad. Sci. USA 2012, 109, E1938–E1946. [Google Scholar] [CrossRef] [PubMed]
- Deleault, N.R.; Piro, J.R.; Walsh, D.J.; Wang, F.; Ma, J.; Geoghegan, J.C.; Supattapone, S. Isolation of phosphatidylethanolamine as a solitary cofactor for prion formation in the absence of nucleic acids. Proc. Natl. Acad. Sci. USA 2012, 109, 8546–8551. [Google Scholar] [CrossRef]
- Arrasate, M.; Perez, M.; Armas-Portela, R.; Avila, J. Polymerization of tau peptides into fibrillar structures. The effect of FTDP-17 mutations. FEBS Lett. 1999, 446, 199–202. [Google Scholar] [CrossRef]
- Goedert, M.; Jakes, R.; Crowther, R.A. Effects of frontotemporal dementia FTDP-17 mutations on heparin-induced assembly of tau filaments. FEBS Lett. 1999, 450, 306–311. [Google Scholar] [CrossRef]
- Von Bergen, M.; Barghorn, S.; Li, L.; Marx, A.; Biernat, J.; Mandelkow, E.M.; Mandelkow, E. Mutations of tau protein in frontotemporal dementia promote aggregation of paired helical filaments by enhancing local beta-structure. J. Biol. Chem. 2001, 276, 48165–48174. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.L.; Lee, V.M. Seeding of normal Tau by pathological Tau conformers drives pathogenesis of Alzheimer-like tangles. J. Biol. Chem. 2011, 286, 15317–15331. [Google Scholar] [CrossRef]
- Kaufman, S.K.; Sanders, D.W.; Thomas, T.L.; Ruchinskas, A.J.; Vaquer-Alicea, J.; Sharma, A.M.; Miller, T.M.; Diamond, M.I. Tau Prion Strains Dictate Patterns of Cell Pathology, Progression Rate, and Regional Vulnerability In Vivo. Neuron 2016, 92, 796–812. [Google Scholar] [CrossRef] [PubMed]
- Sanders, D.W.; Kaufman, S.K.; DeVos, S.L.; Sharma, A.M.; Mirbaha, H.; Li, A.; Barker, S.J.; Foley, A.C.; Thorpe, J.R.; Serpell, L.C.; et al. Distinct tau prion strains propagate in cells and mice and define different tauopathies. Neuron 2014, 82, 1271–1288. [Google Scholar] [CrossRef]
- Holmes, B.B.; DeVos, S.L.; Kfoury, N.; Li, M.; Jacks, R.; Yanamandra, K.; Ouidja, M.O.; Brodsky, F.M.; Marasa, J.; Bagchi, D.P.; et al. Heparan sulfate proteoglycans mediate internalization and propagation of specific proteopathic seeds. Proc. Natl. Acad. Sci. USA 2013, 110, E3138–E3147. [Google Scholar] [CrossRef] [PubMed]
- Kfoury, N.; Holmes, B.B.; Jiang, H.; Holtzman, D.M.; Diamond, M.I. Trans-cellular propagation of Tau aggregation by fibrillar species. J. Biol. Chem. 2012, 287, 19440–19451. [Google Scholar] [CrossRef]
- Falcon, B.; Cavallini, A.; Angers, R.; Glover, S.; Murray, T.K.; Barnham, L.; Jackson, S.; O’Neill, M.J.; Isaacs, A.M.; Hutton, M.L.; et al. Conformation determines the seeding potencies of native and recombinant Tau aggregates. J. Biol. Chem. 2015, 290, 1049–1065. [Google Scholar] [CrossRef]
- Stancu, I.C.; Vasconcelos, B.; Ris, L.; Wang, P.; Villers, A.; Peeraer, E.; Buist, A.; Terwel, D.; Baatsen, P.; Oyelami, T.; et al. Templated misfolding of Tau by prion-like seeding along neuronal connections impairs neuronal network function and associated behavioral outcomes in Tau transgenic mice. Acta Neuropathol. 2015, 129, 875–894. [Google Scholar] [CrossRef]
- Strang, K.H.; Croft, C.L.; Sorrentino, Z.A.; Chakrabarty, P.; Golde, T.E.; Giasson, B.I. Distinct differences in prion-like seeding and aggregation between Tau protein variants provide mechanistic insights into tauopathies. J. Biol. Chem. 2018, 293, 4579. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.L.; Buist, A.; Soares, A.; Callaerts, K.; Calafate, S.; Stevenaert, F.; Daniels, J.P.; Zoll, B.E.; Crowe, A.; Brunden, K.R.; et al. The Dynamics and Turnover of Tau Aggregates in Cultured Cells: Insights into Therapies For Tauopathies. J. Biol. Chem. 2016, 291, 13175–13193. [Google Scholar] [CrossRef] [PubMed]
- Aoyagi, H.; Hasegawa, M.; Tamaoka, A. Fibrillogenic nuclei composed of P301L mutant tau induce elongation of P301L tau but not wild-type tau. J. Biol. Chem. 2007, 282, 20309–20318. [Google Scholar] [CrossRef] [PubMed]
- Spillantini, M.G.; Crowther, R.A.; Kamphorst, W.; Heutink, P.; van Swieten, J.C. Tau pathology in two Dutch families with mutations in the microtubule-binding region of tau. Am. J. Pathol. 1998, 153, 1359–1363. [Google Scholar] [CrossRef]
- Matsumoto, G.; Matsumoto, K.; Kimura, T.; Suhara, T.; Higuchi, M.; Sahara, N.; Mori, N. Tau Fibril Formation in Cultured Cells Compatible with a Mouse Model of Tauopathy. Int. J. Mol. Sci. 2018, 19, 1497. [Google Scholar] [CrossRef] [PubMed]
- Bandyopadhyay, B.; Li, G.; Yin, H.; Kuret, J. Tau aggregation and toxicity in a cell culture model of tauopathy. J. Biol. Chem. 2007, 282, 16454–16464. [Google Scholar] [CrossRef] [PubMed]
- Woerman, A.L.; Stohr, J.; Aoyagi, A.; Rampersaud, R.; Krejciova, Z.; Watts, J.C.; Ohyama, T.; Patel, S.; Widjaja, K.; Oehler, A.; et al. Propagation of prions causing synucleinopathies in cultured cells. Proc. Natl. Acad. Sci. USA 2015, 112, E4949–E4958. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.-G.; Han, Z.Z.; Daude, N.; McNamara, E.; Wohlgemuth, S.; Safar, J.G.; Mok, S.-A.; Westaway, D. Tau conformers in FTLD-MAPT undergo liquid-liquid phase separation and perturb the nuclear envelope. bioRxiv 2020. [Google Scholar] [CrossRef]
- Li, L.; Shi, R.; Gu, J.; Tung, Y.C.; Zhou, Y.; Zhou, D.; Wu, R.; Chu, D.; Jin, N.; Deng, K.; et al. Alzheimer’s disease brain contains tau fractions with differential prion-like activities. Acta Neuropathol. Commun. 2021, 9, 28. [Google Scholar] [CrossRef]
- Löffler, T.; Flunkert, S.; Taub, N.; Schofield, E.L.; Ward, M.A.; Windisch, M.; Hutter-Paier, B. Stable mutated tau441 transfected SH-SY5Y cells as screening tool for Alzheimer’s disease drug candidates. J. Mol. Neurosci. 2012, 47, 192–203. [Google Scholar] [CrossRef][Green Version]
- Glover, J.R.; Kowal, A.S.; Schirmer, E.C.; Patino, M.M.; Liu, J.-J.; Lindquist, S. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell 1997, 89, 811–819. [Google Scholar] [CrossRef]
- Rauch, J.N.; Luna, G.; Guzman, E.; Audouard, M.; Challis, C.; Sibih, Y.E.; Leshuk, C.; Hernandez, I.; Wegmann, S.; Hyman, B.T. LRP1 is a master regulator of tau uptake and spread. Nature 2020, 580, 381–385. [Google Scholar] [CrossRef]
- Saragoni, L.; Hernández, P.; Maccioni, R.B. Differential association of tau with subsets of microtubules containing posttranslationally-modified tubulin variants in neuroblastoma cells. Neurochem. Res. 2000, 25, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Hou, L.X.E.; Aktiv, A.; Dahlström, A. Studies of the central nervous system-derived CAD cell line, a suitable model for intraneuronal transport studies? J. Neurosci. Res. 2007, 85, 2601–2609. [Google Scholar] [CrossRef]
- Hromadkova, L.; Bezdekova, D.; Pala, J.; Schedin-Weiss, S.; Tjernberg, L.O.; Hoschl, C.; Ovsepian, S.V. Brain-derived neurotrophic factor (BDNF) promotes molecular polarization and differentiation of immature neuroblastoma cells into definitive neurons. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2020, 1867, 118737. [Google Scholar] [CrossRef]
- Nonaka, T.; Watanabe, S.T.; Iwatsubo, T.; Hasegawa, M. Seeded aggregation and toxicity of {alpha}-synuclein and tau: Cellular models of neurodegenerative diseases. J. Biol. Chem. 2010, 285, 34885–34898. [Google Scholar] [CrossRef]
- Chastagner, P.; Loria, F.; Vargas, J.Y.; Tois, J.; Diamond, I.M.; Okafo, G.; Brou, C.; Zurzolo, C. Fate and propagation of endogenously formed Tau aggregates in neuronal cells. EMBO Mol. Med. 2020, 12, e12025. [Google Scholar] [CrossRef]
- Encinas, M.; Iglesias, M.; Liu, Y.; Wang, H.; Muhaisen, A.; Cena, V.; Gallego, C.; Comella, J.X. Sequential treatment of SH-SY5Y cells with retinoic acid and brain-derived neurotrophic factor gives rise to fully differentiated, neurotrophic factor-dependent, human neuron-like cells. J. Neurochem. 2000, 75, 991–1003. [Google Scholar] [CrossRef]
- Bell, M.; Zempel, H. SH-SY5Y-derived neurons: A human neuronal model system for investigating TAU sorting and neuronal subtype-specific TAU vulnerability. Rev. Neurosci. 2022, 33, 1–15. [Google Scholar] [CrossRef]
- Pollack, S.J.; Trigg, J.; Khanom, T.; Biasetti, L.; Marshall, K.E.; Al-Hilaly, Y.K.; Rickard, J.E.; Harrington, C.R.; Wischik, C.M.; Serpell, L.C. Paired helical filament-forming region of tau (297–391) influences endogenous tau protein and accumulates in acidic compartments in human neuronal cells. J. Mol. Biol. 2020, 432, 4891–4907. [Google Scholar] [CrossRef]
- Shamir, D.B.; Deng, Y.; Wu, Q.; Modak, S.; Congdon, E.E.; Sigurdsson, E.M. Dynamics of internalization and intracellular interaction of tau antibodies and human pathological tau protein in a human neuron-like model. Front. Neurol. 2020, 11, 602292. [Google Scholar] [CrossRef]
- Ferreira, A.; Lu, Q.; Orecchio, L.; Kosik, K.S. Selective phosphorylation of adult tau isoforms in mature hippocampal neurons exposed to fibrillar A beta. Mol. Cell Neurosci. 1997, 9, 220–234. [Google Scholar] [CrossRef]
- Woerman, A.L.; Aoyagi, A.; Patel, S.; Kazmi, S.A.; Lobach, I.; Grinberg, L.T.; McKee, A.C.; Seeley, W.W.; Olson, S.H.; Prusiner, S.B. Tau prions from Alzheimer’s disease and chronic traumatic encephalopathy patients propagate in cultured cells. Proc. Natl. Acad. Sci. USA 2016, 113, E8187–E8196. [Google Scholar] [CrossRef] [PubMed]
- Narasimhan, S.; Guo, J.L.; Changolkar, L.; Stieber, A.; McBride, J.D.; Silva, L.V.; He, Z.; Zhang, B.; Gathagan, R.J.; Trojanowski, J.Q.; et al. Pathological Tau Strains from Human Brains Recapitulate the Diversity of Tauopathies in Nontransgenic Mouse Brain. J. Neurosci. 2017, 37, 11406–11423. [Google Scholar] [CrossRef]
- Wu, J.W.; Herman, M.; Liu, L.; Simoes, S.; Acker, C.M.; Figueroa, H.; Steinberg, J.I.; Margittai, M.; Kayed, R.; Zurzolo, C.; et al. Small misfolded Tau species are internalized via bulk endocytosis and anterogradely and retrogradely transported in neurons. J. Biol. Chem. 2013, 288, 1856–1870. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; McBride, J.D.; Xu, H.; Changolkar, L.; Kim, S.-J.; Zhang, B.; Narasimhan, S.; Gibbons, G.S.; Guo, J.L.; Kozak, M.; et al. Transmission of tauopathy strains is independent of their isoform composition. Nat. Commun. 2020, 11, 7. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; O’Reilly, M.; Gibbons, G.S.; Changolkar, L.; McBride, J.D.; Riddle, D.M.; Zhang, B.; Stieber, A.; Nirschl, J.; Kim, S.-J. In vitro amplification of pathogenic tau conserves disease-specific bioactive characteristics. Acta Neuropathol. 2021, 141, 193–215. [Google Scholar] [CrossRef] [PubMed]
- Mungenast, A.E.; Siegert, S.; Tsai, L.-H. Modeling Alzheimer’s disease with human induced pluripotent stem (iPS) cells. Mol. Cell. Neurosci. 2016, 73, 13–31. [Google Scholar] [CrossRef]
- Barak, M.; Fedorova, V.; Pospisilova, V.; Raska, J.; Vochyanova, S.; Sedmik, J.; Hribkova, H.; Klimova, H.; Vanova, T.; Bohaciakova, D. Human iPSC-Derived Neural Models for Studying Alzheimer’s Disease: From Neural Stem Cells to Cerebral Organoids. Stem Cell Rev. Rep. 2022, 18, 792–820. [Google Scholar] [CrossRef]
- Iovino, M.; Agathou, S.; González-Rueda, A.; Del Castillo Velasco-Herrera, M.; Borroni, B.; Alberici, A.; Lynch, T.; O’Dowd, S.; Geti, I.; Gaffney, D. Early maturation and distinct tau pathology in induced pluripotent stem cell-derived neurons from patients with MAPT mutations. Brain 2015, 138, 3345–3359. [Google Scholar] [CrossRef]
- Wray, S. Modeling tau pathology in human stem cell derived neurons. Brain Pathol. 2017, 27, 525–529. [Google Scholar] [CrossRef]
- Miguel, L.; Rovelet-Lecrux, A.; Feyeux, M.; Frebourg, T.; Nassoy, P.; Campion, D.; Lecourtois, M. Detection of all adult Tau isoforms in a 3D culture model of iPSC-derived neurons. Stem Cell Res. 2019, 40, 101541. [Google Scholar] [CrossRef]
- Verheyen, A.; Diels, A.; Dijkmans, J.; Oyelami, T.; Meneghello, G.; Mertens, L.; Versweyveld, S.; Borgers, M.; Buist, A.; Peeters, P. Using human iPSC-derived neurons to model TAU aggregation. PLoS ONE 2015, 10, e0146127. [Google Scholar] [CrossRef]
- Medda, X.; Mertens, L.; Versweyveld, S.; Diels, A.; Barnham, L.; Bretteville, A.; Buist, A.; Verheyen, A.; Royaux, I.; Ebneth, A. Development of a scalable, high-throughput-compatible assay to detect tau aggregates using iPSC-derived cortical neurons maintained in a three-dimensional culture format. J. Biomol. Screen. 2016, 21, 804–815. [Google Scholar] [CrossRef]
- Manos, J.D.; Preiss, C.N.; Venkat, N.; Tamm, J.; Reinhardt, P.; Kwon, T.; Wu, J.; Winter, A.D.; Jahn, T.R.; Yanamandra, K. Uncovering specificity of endogenous TAU aggregation in a human iPSC-neuron TAU seeding model. Iscience 2022, 25, 103658. [Google Scholar] [CrossRef] [PubMed]
- Usenovic, M.; Niroomand, S.; Drolet, R.E.; Yao, L.; Gaspar, R.C.; Hatcher, N.G.; Schachter, J.; Renger, J.J.; Parmentier-Batteur, S. Internalized tau oligomers cause neurodegeneration by inducing accumulation of pathogenic tau in human neurons derived from induced pluripotent stem cells. J. Neurosci. 2015, 35, 14234–14250. [Google Scholar] [CrossRef] [PubMed]
- Cummings, J.; Lee, G.; Ritter, A.; Sabbagh, M.; Zhong, K. Alzheimer’s disease drug development pipeline: 2019. Alzheimer’s Dement. Transl. Res. Clin. Interv. 2019, 5, 272–293. [Google Scholar] [CrossRef]
- Qiang, W.; Yau, W.-M.; Lu, J.-X.; Collinge, J.; Tycko, R. Structural variation in amyloid-β fibrils from Alzheimer’s disease clinical subtypes. Nature 2017, 541, 217–221. [Google Scholar] [CrossRef]
- Aoyagi, A.; Condello, C.; Stöhr, J.; Yue, W.; Rivera, B.M.; Lee, J.C.; Woerman, A.L.; Halliday, G.; van Duinen, S.; Ingelsson, M.; et al. Aβ and tau prion-like activities decline with longevity in the Alzheimer’s disease human brain. Sci. Transl. Med. 2019, 11, eaat8462. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).