Abstract
Rice, being a staple food crop for over one-third of the world’s population, has become a potential target for many dishonest traders and stakeholders for mixing with low-grade, low-cost grains/products and poorly nutritious adulterants to make a profit with the least effort. Single-nucleotide and insertion–deletion (InDel) polymorphisms have been widely used as DNA markers, not only in plant breeding but also to identify various traits in rice. Recently, next-generation sequencing (NGS) has produced sequences that allow for genome-wide detection of these molecular markers. These polymorphisms can potentially be used to develop high-accuracy polymerase chain reaction (PCR)-based markers. PCR-based techniques are rapid and successful methods to deal with the problem of adulteration at a commercial level. Here, we report the genome-wide analysis of InDel markers of 17 commercially available Chinese cultivars. In order to achieve accurate results, all samples were sequenced at approximately 30× genome coverage using Illumina HiSeq 2500™ system. An average of 10.6 GB clean reads per sample was produced and ~96.3% of the reads could be mapped to the rice genome reference IRGSP 1.0. After a series of filtering, we selected five InDel markers for PCR validation. The results revealed that these InDel markers can be used for authentication of Korean elite cultivars from the adulterants.
1. Introduction
Rice, being used in daily cuisine for over one-third of the world’s population, has become a potential target for many dishonest stakeholders and traders who mix low-grade/cost/nutrition adulterants to make a profit with the least effort. Though thousands of varieties with different commercial names are available, particular varieties have become popular and suit the tastes of consumers in a particular region [1,2]. Attempts to develop superior varieties/products always ended up with some poor-quality ones. This led to the existence of different quality standards with obvious price differences in the market [3,4]. This situation attracted dishonest traders to attempt adulteration of the genuine products to make surplus profit [5].
Adulteration in rice is possible from crop harvest until it reaches consumers, and leads to nutrition and health risks [6]. The common forms of rice prone to adulteration are brown rice, polished rice, rice flour, rice cakes, and rice bran oil. Authentication of rice cultivars is an important issue to address to protect the interests of farmers, dealers, millers, and food processors as well as to provide healthy food to consumers [7]. To this end, each country establishes quality standards for agricultural products by providing a label with complete details including country of origin and chemical composition. Many methods based on criteria such as morphological parameters (height, grain shape, size), physicochemical properties (starch composition, lipids, seed storage proteins, and alloenzymes), DNA (single-nucleotide polymorphism, insertions and deletions), proteins, and metabolites have been developed to detect the genuineness of the agricultural and animal food products [1,8,9,10,11,12].
In South Korea, rice consumption per person peaked in 1970 at 136.4 kg per year. Since the mid-1980s, rice consumption per person has declined almost every year, reaching 62.9 kg per year in 2015 [13]. In addition, the total rice cultivation area accounts for about 50 percent of total cultivated land. Since 2006, rice cultivation area has dropped slightly faster than total area, and was 47.5 percent of the total in 2015 (Korean Statistical Information System (KOSIS) database). Furthermore, South Korea is an important market for U.S and Chinese rice, which subsequently used adulterants in Korean varieties to meet demand. Next-generation sequencing (NGS) technology has been widely used in rice genomics and molecular breeding studies [14]. This has enabled researchers to perform an accurate genetic polymorphism analysis in order to identify unique molecular markers for particular rice varieties. However, the traceability of imported rice varieties needs in-depth analysis.
Hence, this study aimed at screening and selecting compatible polymorphic InDel markers through whole genome re-sequencing from 17 commercial rice cultivars from China. Our final goal is to identify accurate, sensitive and effective SNP, InDel markers to authenticate domestic Korean rice from those Chinese cultivars. Our results therefore lay the groundwork for long-term efforts to assess the purity of other Korean rice varieties.
2. Results
2.1. Genome Re-Sequencing and Analysis
We have sequenced 17 commercial rice cultivars from China using Illumina high-throughput sequencing technology. To facilitate downstream analysis, genomic DNA was isolated from the rice grain of each cultivar. High-throughput sequencing was performed using the Illumina HiSeq 2500™ system, and the resultant sequence reads were mapped to the IRGSP-1.0 using BWA [15], as described in methods Section 4.3. The sequence depths (30×) estimated from sequencing data for all reads of the 17 cultivars ranged from 19.28 (‘Su Jung Mi’) to 49.55 (‘Jang Rip Gaeng Mi’). The mapped sequence depths were lower than those of the total sequence, and ranged from 25.09 (‘Su Jung Mi’) to 31.75 (‘Jang Rip Gaeng Mi’). The highest and lowest coverages of a reference genome by input reads were observed in ‘Jung Rang’ (97.4%) and ‘Hang Dea Mi’ (96.1%), respectively (Table 1).

Table 1.
Sequence information for the 17 Chinese rice cultivars obtained by whole-genome re-sequencing analysis.
The total number of InDels detected in the 17 Chinese rice cultivars was less than the number of SNPs (Table 2). The number of InDels ranged from 64,613 for ‘Sunrice’ to 156,768 for ‘Kum Do Jean DongBuk Dea Mi’, with intermediate values for ‘Su Jung Mi’, ‘Saeng Tae Mi’, and ‘Jang Rip Gaeng Mi’. ‘Sunrice’ had the fewest InDels compared to the other cultivars.

Table 2.
Number of SNPs and InDels compared to the reference genome (IRGSP v. 1.0) detected in the 17 Chinese rice cultivars.
2.2. Distribution of SNP and InDel Markers
As a result of designing primers, more than two-thirds of the SNP and InDel regions were eliminated. Few SNP and InDel markers were uniquely detected in all the samples, so keeping these markers might influence the total number of SNP and InDel markers for PCR validation. In total, we selected 92 SNP and 278 InDel markers from the 17 rice cultivars. Here we selected only InDel markers for PCR validation in order to get accurate results. In terms of marker efficiency, the available markers to detect InDels were common to all rice cultivars but not ‘Hwayoung’, a Korean cultivar (data not shown). The number of InDel markers decreased as the number of cultivars sharing the same primer sequences decreased. Both selected SNP and InDel markers showed even distribution within the rice genome (Figure 1).
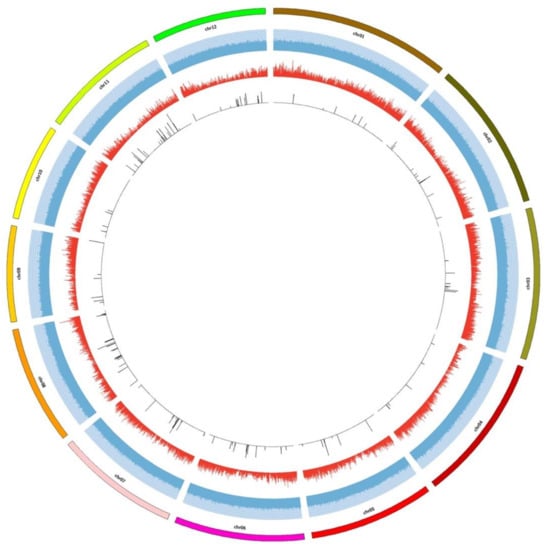
Figure 1.
An annotated Circos plot depicts the distribution pattern of the selected SNP and InDel markers. The outermost circle represents 12 chromosomes of the rice genome. Blue bars represent the GC content of the rice genome, whereas red bars represent the gene frequency. In addition, the innermost black boxes represent the SNP and InDel markers identified in the Chinese rice cultivars.
2.3. Experimental Validation of Five InDel Markers
In order to detect InDel polymorphisms by electrophoresis, we performed primer screening using PCR. As an example of validation, clear single bands were detected around the expected PCR product sizes for five markers including, KM-IND-7, KM-IND-21, KM-IND-80, KM-IND-253, and KM-IND-271 (Table 3). The unique size between the amplified fragments could be discriminated among the 17 cultivars (Figure 2; Supplementary Table S1). Our validation results shown that the markers (for 17 cultivars) with a success rate of 90% or more for PCR. In the case of KM-IND-21, polymorphism could be detected for ~80% of the rice cultivars.

Table 3.
Summary of validated InDel primers.
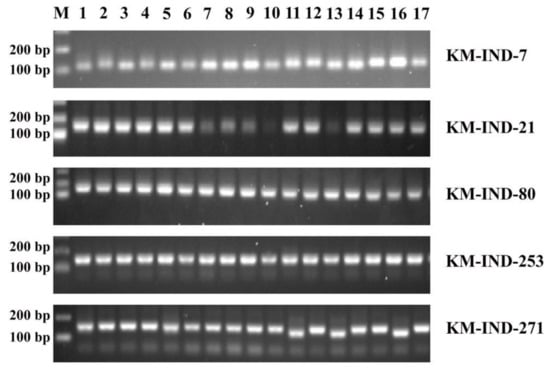
Figure 2.
Electrophoretograms for five InDel markers. M, Molecular marker (a 100-bp DNA ladder). 1–17: Chinese commercial rice cultivars (see Table 2).
3. Discussion
High accuracy is important when detecting nucleotide polymorphisms, including SNPs or InDels, using the NGS re-sequencing strategy. In the International Rice Genome Sequencing Project (2005), the entire ‘Nipponbare’ genome was sequenced through the Sanger sequencing method and thereby precise reference sequences were established. For re-sequencing with short reads, the sequencing depth affects the accuracy of polymorphism detection. Here, the sequencing depth of the mapped sequences was an average of 28.9-fold (Table 1), which is twice the average depth (~14-fold) achieved in the most current 3000 Genomes Project [16]. The results of our study suggest that this sequencing depth is sufficient to accurately detect nucleotide polymorphisms. However, it appears that there is no relationship between the genome coverage and the sequencing depth of all the rice samples (Table 1). We found that the number of polymorphic sites (SNPs and InDels) in the genomes of all the samples showed the opposite trend to the genome coverage (Table 1).
Furthermore, re-sequencing with short reads cannot physically detect InDels that are longer than the read length (100 bp) and hence we only found insertions and deletions of less than 20 bp (data not shown). InDels with a larger size (100 bp) and copy number variations (>1 kb) were undoubtedly located all over the rice genome [17]. Various sizes of InDels with different PCR amplicons were separated in both agarose and denaturing polyacrylamide gels [18,19,20]. Here, we found that InDels could be easily detected by separation on 1.8% agarose gels, and we designed the markers accordingly. Indeed, we observed a high PCR success rate for the five markers (Figure 2; Supplementary Table S1), and these markers could be used to discriminate polymorphisms by electrophoresis.
New technologies and innovative approaches are changing the way we consider and apply genetic authentication of crop cultivars. DNA markers such as InDels play a crucial role in breeding and cultivar identification. The five InDel markers we identified might deserve further study for distinguishing Korean elite cultivars from adulterants.
4. Materials and Methods
4.1. Plant Materials
We performed whole-genome re-sequencing (WGRS) of 17 commercial rice grains of Japonica populations obtained from the National Institute of Crop Science (NICS), Rural Development Administration, South Korea.
4.2. DNA Isolation and Genome Sequencing
Genomic DNA was prepared from rice grains using a ChargeSwitch® gDNA plant kit (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s protocol. Whole-genome re-sequencing of the 27 samples was performed on an Illumina HiSeq2500™ by TheragenETEX Bio Institute (Suwon, South Korea). The procedure was performed according to the standard Illumina protocol, including sample preparation and sequencing as follows: high molecular weight genomic DNA was excised from the gel and sheared using a Covaris S2 ultra sonicator system in order to get appropriate sizes and agarose gel electrophoresis was used to select fragments. Libraries with short inserts of 350–450 bp for paired-end reads were prepared using a Truseq DNA sample prep kit following the manufacturer’s protocol for Illumina. Products were quantified using the Bioanalyzer 2100 (Agilent, Santa Clara, CA, USA) and sequencing was performed by establishing a library with Illumina HiSeq2500™. To ensure high quality, low-quality reads (<20) reads with adaptor sequence and duplicated reads were filtered out, and the remaining high-quality data were used in the mapping.
4.3. Mapping of Reads to the Reference
The raw reads were subjected to quality trimming (phred quality score, <Q20) using FastQC [21] and adapter trimming was carried out by using the parameters (-O 5 and -m 32) in version 1.0 of the cutadapt software [22]. Furthermore, the clean reads were mapped to the temperate japonica Nipponbare reference genome, Os-Nipponbare-Reference-IRGSP-1.0 [23], using the Burrows–Wheeler Aligner (BWA) software [15] under the default parameters. The alignment results were then merged and indexed as BAM files [24]. Average sequencing depth and coverage were calculated using the alignment results. The mapped reads were then used to detect SNP and InDel polymorphisms.
4.4. Detection of SNPs and InDels
We used GATK tools software [25] to detect SNPs and InDels with its default parameters. In our analysis, InDels were defined as insertions or deletions the length of which was between 1 and 10 bp. The InDels falls on more than 10 cultivars are selected for further analysis.
4.5. Primer Design for Common InDel Markers
To design InDel markers for the detection of InDel polymorphisms by electrophoresis, we extracted only InDel regions with a large size (≥2 bp) and a high sequencing depth (DP, ≥5 fold) from each InDel list for the 17 cultivars. Primer pairs for the selected InDel regions were automatically designed by using a Perl script to control the Primer3 program [26]. In addition, we screened primer pairs for duplication of sequences to maintain specificity. When the sequence of a primer pair matched that of another primer pair, the corresponding pairs were eliminated because they were considered redundant. PCR product size ranged from 100 to 150 bp.
4.6. PCR Validation
To validate the marker sets, we tested the primer pairs by means of gel electrophoresis. Around 10 markers were chosen so as to cover as much of the entire genome as possible, but were randomly chosen within each portion of the genome. The InDel primer sets were validated by using those 17 Chinese cultivars used for detection of the InDel regions. Furthermore, these primer sets were PCR-amplified in approximately 10 L of reaction mixture consisting of EmeraldAmp® GT PCR Master Mix (Takara, Japan) along with 5.0 pmol of each primer, and about 120 ng of the genomic DNA template. The conditions were: an initial denaturation step for 3 min at 95 °C, then 35 cycles of 20 s at 95 °C, 30 s at 55 °C, and 30 s at 72 °C, followed by a final extension for 1 min at 72 °C. PCR products were analyzed by means of gel electrophoresis on 1.5~1.7% agarose gels in Tris/borate/EDTA buffer. After staining with ethidium bromide, the band patterns on the agarose gels were photographed under Gel Doc™ 2000 Gel Documentation System (Bio-Rad, Seoul, Korea).
Supplementary Materials
The following are available online at http://www.mdpi.com/2073-4395/8/4/36/s1.
Acknowledgments
This study was supported by grants from the “Agriculture Joint Project” program (PJ01164922017) of the Rural Development Administration, Republic of Korea.
Author Contributions
Young-Chan Cho and Dong Woo Lee conceived and designed the experiments; Kesavan Markkandan performed the experiments and wrote the manuscript; Seung-il Yoo analyzed the data.
Conflicts of Interest
The authors declare that they have no competing interests.
References
- Vemireddy, L.R.; Satyavathi, V.; Siddiq, E.; Nagaraju, J. Review of methods for the detection and quantification of adulteration of rice: Basmati as a case study. J. Food Sci. Technol. 2015, 52, 3187–3202. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.M.; Shabir, G.; Aslam, K.; Sabar, M.; Arif, M. Identification of SSR markers to find adulteration in elite basmati rice varieties. Environ. Plant Syst. 2015, 1, 4–15. [Google Scholar]
- Ghoshray, A. Asymmetric adjustment of rice export prices: The case of Thailand and Vietnam. Int. J. Appl. Econ. 2008, 5, 80–91. [Google Scholar]
- Vemireddy, L.R.; Noor, S.; Satyavathi, V.; Srividhya, A.; Kaliappan, A.; Parimala, S.; Bharathi, P.M.; Deborah, D.A.; Rao, K.S.; Shobharani, N. Discovery and mapping of genomic regions governing economically important traits of Basmati rice. BMC Plant Biol. 2015, 15, 207. [Google Scholar] [CrossRef] [PubMed]
- Colyer, A.; Macarthur, R.; Lloyd, J.; Hird, H. Comparison of calibration methods for the quantification of Basmati and non-Basmati rice using microsatellite analysis. Food Addit. Contam. 2008, 25, 1189–1194. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S. Food adulteration affecting the nutrition and health of human beings. J. Biol. Sci. Med. 2016, 1, 65–70. [Google Scholar]
- Vlachos, A.; Arvanitoyannis, I.S. A review of rice authenticity/adulteration methods and results. Crit. Rev. Food Sci. Nutr. 2008, 48, 553–598. [Google Scholar] [CrossRef] [PubMed]
- Lees, M. Food Authenticity and Traceability; Elsevier: Amsterdam, The Netherlands, 2003. [Google Scholar]
- Fontanesi, L. Genetic authentication and traceability of food products of animal origin: New developments and perspectives. Ital. J. Anim. Sci. 2009, 8 (Suppl. 2), 9–18. [Google Scholar] [CrossRef]
- Cho, K.-S.; Hong, S.-Y.; Yun, B.-K.; Won, H.-S.; Yoon, Y.-H.; Kwon, K.-B.; Mekapogu, M. Application of InDel Markers Based on the Chloroplast Genome Sequences for Authentication and Traceability of Tartary and Common Buckwheat. Czech J. Food Sci. 2017, 35, 122–130. [Google Scholar]
- Wen, J.; Zeng, L.; Sun, Y.; Chen, D.; Xu, Y.; Luo, P.; Zhao, Z.; Yu, Z.; Fan, S. Authentication and traceability of fish maw products from the market using DNA sequencing. Food Control 2015, 55, 185–189. [Google Scholar] [CrossRef]
- Chuang, H.-Y.; Lur, H.-S.; Hwu, K.-K.; Chang, M.-C. Authentication of domestic Taiwan rice varieties based on fingerprinting analysis of microsatellite DNA markers. Bot. Stud. 2011, 52, 393–405. [Google Scholar]
- Choi, S.; Dyck, J.; Childs, N. The Rice Market in South Korea; US Department of Agriculture: Washington, DC, USA, 2016. [Google Scholar]
- Chen, H.; Xie, W.; He, H.; Yu, H.; Chen, W.; Li, J.; Yu, R.; Yao, Y.; Zhang, W.; He, Y. A high-density SNP genotyping array for rice biology and molecular breeding. Mol. Plant 2014, 7, 541–553. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Li, J.-Y.; Wang, J.; Zeigler, R.S. The 3000 rice genomes project: new opportunities and challenges for future rice research. GigaScience 2014, 3, 8. [Google Scholar] [CrossRef] [PubMed]
- Feuk, L.; Carson, A.R.; Scherer, S.W. Structural variation in the human genome. Nat. Rev. Genet. 2006, 7, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.-H.; Wu, H.-P.; Wang, C.-S.; Tseng, H.-Y.; Hwu, K.-K. Genome-wide InDel marker system for application in rice breeding and mapping studies. Euphytica 2013, 192, 131–143. [Google Scholar] [CrossRef]
- Yamaki, S.; Ohyanagi, H.; Yamasaki, M.; Eiguchi, M.; Miyabayashi, T.; Kubo, T.; Kurata, N.; Nonomura, K.-I. Development of INDEL markers to discriminate all genome types rapidly in the genus Oryza. Breed. Sci. 2013, 63, 246–254. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Wen, Z.; Ma, L.; Ji, Z.; Li, X.; Yang, C. Development of 1047 insertion-deletion markers for rice genetic studies and breeding. Genet. Mol. Res. 2013, 12, 5226–5235. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S.; Krueger, F.; Seconds-Pichon, A.; Biggins, F.; Wingett, S. FastQC: A Quality Control Tool for High Throughput Sequence Data; Babraham Institute: Cambridge, UK, 2012. [Google Scholar]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Kawahara, Y.; de la Bastide, M.; Hamilton, J.P.; Kanamori, H.; McCombie, W.R.; Ouyang, S.; Schwartz, D.C.; Tanaka, T.; Wu, J.; Zhou, S. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 2013, 6, 4. [Google Scholar] [CrossRef] [PubMed]
- Barnett, D.W.; Garrison, E.K.; Quinlan, A.R.; Strömberg, M.P.; Marth, G.T. BamTools: A C++ API and toolkit for analyzing and managing BAM files. Bioinformatics 2011, 27, 1691–1692. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucl. Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).