Genome-Wide Association Study Reveals Key Genetic Loci Controlling Oil Content in Soybean Seeds
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Materials and Field Trials
2.2. Determination of Soybean Seed Oil Content
2.3. Genome-Wide Association Analysis
2.4. Identification and Validation of Candidate Genes
3. Results
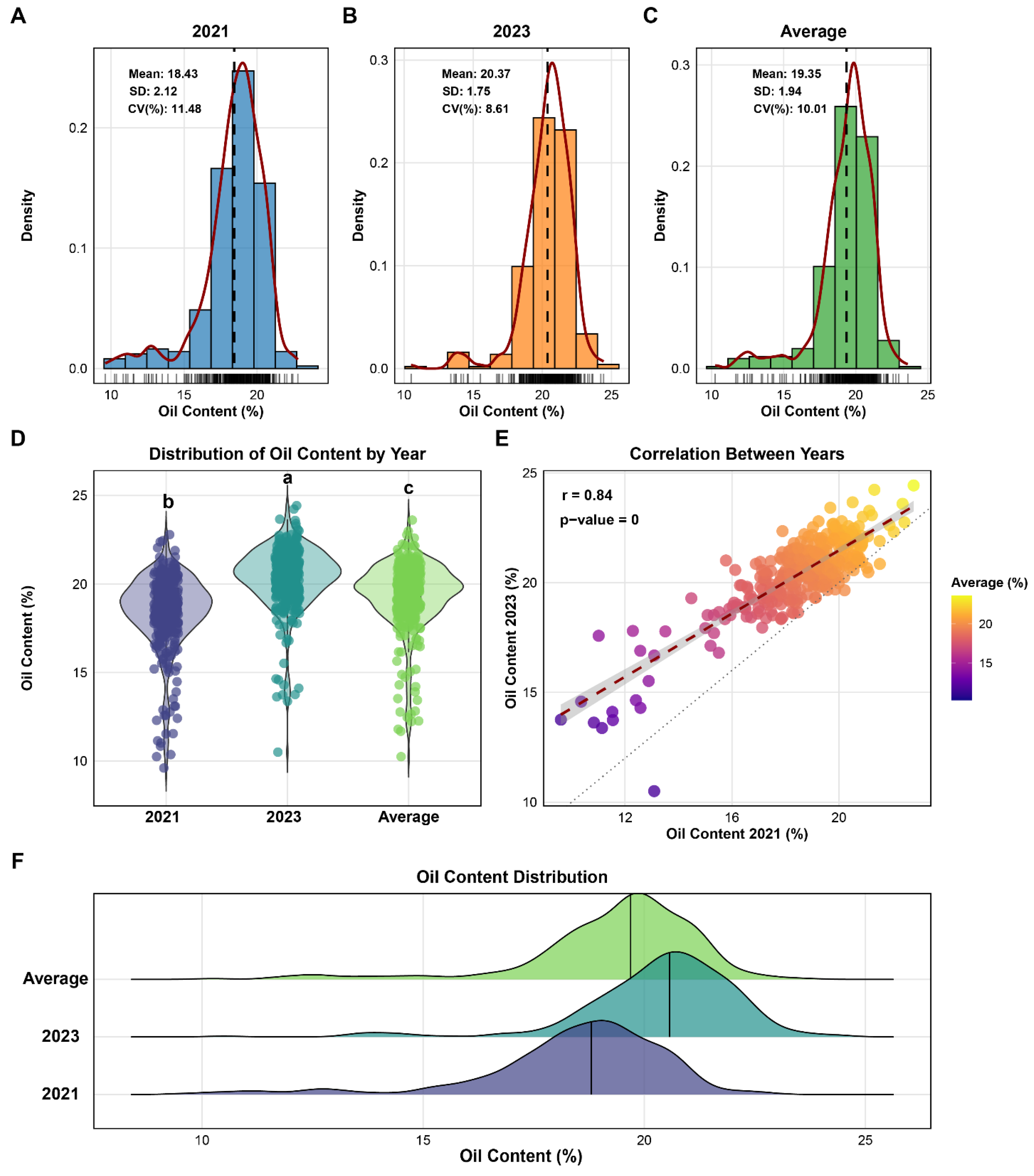
3.1. Statistical Analysis of Phenotypic Data
3.2. Soybean Oil Content Across Northern China’s Growing Regions (2021–2023)
3.3. Genome-Wide Association Analysis Results
3.4. Genome-Wide Association Analysis and Candidate Gene
3.5. Candidate Gene Screening for Association Analysis
4. Discussion
4.1. Phenotypic Variation and Environmental Effects
4.2. Population Structure and Genetic Diversity
4.3. Candidate Gene Analysis and Functional Implications
4.4. Quantitative Comparison with Prior GWASs
4.5. Breeding and Selection Implications
4.6. Study Limitations and Future Directions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, S.; Liu, S.; Wang, J.; Yokosho, K.; Zhou, B.; Yu, Y.-C.; Liu, Z.; Frommer, W.B.; Ma, J.F.; Chen, L.-Q. Simultaneous changes in seed size, oil content and protein content driven by selection of SWEET homologues during soybean domestication. Natl. Sci. Rev. 2020, 7, 1776–1786. [Google Scholar] [CrossRef] [PubMed]
- Hooker, J.C.; Smith, M.; Zapata, G.; Charette, M.; Luckert, D.; Mohr, R.M.; Daba, K.A.; Warkentin, T.D.; Hadinezhad, M.; Barlow, B. Differential gene expression provides leads to environmentally regulated soybean seed protein content. Front. Plant Sci. 2023, 14, 1260393. [Google Scholar] [CrossRef] [PubMed]
- Wilson, R.F. Seed composition. Soybeans Improv. Prod. Uses 2004, 16, 621–677. [Google Scholar]
- Carter, T.E., Jr.; Nelson, R.L.; Sneller, C.H.; Cui, Z. Genetic diversity in soybean. Soybeans Improv. Prod. Uses 2004, 16, 303–416. [Google Scholar]
- Liu, K. Chemistry and Nutritional Value of Soybean Components. In Soybeans; Springer: Boston, MA, USA, 1997; pp. 25–113. [Google Scholar] [CrossRef]
- Zhang, B.; Zhao, K.; Ren, H.; Lamlom, S.F.; Liu, X.; Wang, X.; Zhang, F.; Yuan, R.; Wang, J. Comparative study of isoflavone synthesis genes in two wild soybean varieties using transcriptomic analysis. Agriculture 2023, 13, 1164. [Google Scholar] [CrossRef]
- Liu, A.; Cheng, S.-S.; Yung, W.-S.; Li, M.-W.; Lam, H.-M. Genetic regulations of the oil and protein contents in soybean seeds and strategies for improvement. Adv. Bot. Res. 2022, 102, 259–293. [Google Scholar]
- Li, H.; Peng, Z.; Yang, X.; Wang, W.; Fu, J.; Wang, J.; Han, Y.; Chai, Y.; Guo, T.; Yang, N. Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels. Nat. Genet. 2013, 45, 43–50. [Google Scholar] [CrossRef]
- Miao, L.; Yang, S.; Zhang, K.; He, J.; Wu, C.; Ren, Y.; Gai, J.; Li, Y. Natural variation and selection in GmSWEET39 affect soybean seed oil content. New Phytol. 2020, 225, 1651–1666. [Google Scholar] [CrossRef]
- Clemente, T.E.; Cahoon, E.B. Soybean oil: Genetic approaches for modification of functionality and total content. Plant Physiol. 2009, 151, 1030–1040. [Google Scholar] [CrossRef]
- Lee, J.D.; Bilyeu, K.D.; Pantalone, V.R.; Gillen, A.M.; So, Y.S.; Shannon, J.G. Environmental stability of oleic acid concentration in seed oil for soybean lines with FAD2-1A and FAD2-1B mutant genes. Crop Sci. 2012, 52, 1290–1297. [Google Scholar] [CrossRef]
- Yang, W.; Guo, Z.; Huang, C.; Duan, L.; Chen, G.; Jiang, N.; Fang, W.; Feng, H.; Xie, W.; Lian, X. Combining high-throughput phenotyping and genome-wide association studies to reveal natural genetic variation in rice. Nat. Commun. 2014, 5, 5087. [Google Scholar] [CrossRef]
- Khan, M.R.; Rehman, N.; Inam, S.; Naeem, M.K.; Muhammad, A.; Uzair, M.; Riaz, A.; Rehman, O.U.; Muqaddas, F.; Murtaza, M. Implementation of novel genomic and biotechnological interventions for accelerated breeding of crops. In Plant Speed Breeding and High-Throughput Technologies; CRC Press: Boca Raton, FL, USA, 2024; pp. 53–81. [Google Scholar]
- Duan, Z.; Li, Q.; Wang, H.; He, X.; Zhang, M. Genetic regulatory networks of soybean seed size, oil and protein contents. Front. Plant Sci. 2023, 14, 1160418. [Google Scholar] [CrossRef]
- Yu, J.; Zhu, C.; Xuan, W.; An, H.; Tian, Y.; Wang, B.; Chi, W.; Chen, G.; Ge, Y.; Li, J. Genome-wide association studies identify OsWRKY53 as a key regulator of salt tolerance in rice. Nat. Commun. 2023, 14, 3550. [Google Scholar] [CrossRef] [PubMed]
- Liang, Q.; Chen, L.; Yang, X.; Yang, H.; Liu, S.; Kou, K.; Fan, L.; Zhang, Z.; Duan, Z.; Yuan, Y. Natural variation of Dt2 determines branching in soybean. Nat. Commun. 2022, 13, 6429. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Xu, J.; Wang, W.; Liu, X.; He, D.; Zhang, B.; Liu, B.; Lamlom, S.F.; Abdelghany, A.M.; Hong, H. Genetic architecture of shade tolerance in soybean (Glycine max L. Merr.) revealed by genome-wide association study. Crop Sci. 2025, 65, e70107. [Google Scholar] [CrossRef]
- Hwang, E.-Y.; Song, Q.; Jia, G.; Specht, J.E.; Hyten, D.L.; Costa, J.; Cregan, P.B. A genome-wide association study of seed protein and oil content in soybean. BMC Genom. 2014, 15, 1. [Google Scholar] [CrossRef]
- Cao, Y.; Li, S.; Wang, Z.; Chang, F.; Kong, J.; Gai, J.; Zhao, T. Identification of major quantitative trait loci for seed oil content in soybeans by combining linkage and genome-wide association mapping. Front. Plant Sci. 2017, 8, 1222. [Google Scholar] [CrossRef]
- Zeng, A.; Chen, P.; Korth, K.; Hancock, F.; Pereira, A.; Brye, K.; Wu, C.; Shi, A. Genome-wide association study (GWAS) of salt tolerance in worldwide soybean germplasm lines. Mol. Breed. 2017, 37, 30. [Google Scholar] [CrossRef]
- Jin, H.; Yang, X.; Zhao, H.; Song, X.; Tsvetkov, Y.D.; Wu, Y.; Gao, Q.; Zhang, R.; Zhang, J. Genetic analysis of protein content and oil content in soybean by genome-wide association study. Front. Plant Sci. 2023, 14, 1182771. [Google Scholar] [CrossRef]
- Goettel, W.; Zhang, H.; Li, Y.; Qiao, Z.; Jiang, H.; Hou, D.; Song, Q.; Pantalone, V.R.; Song, B.-H.; Yu, D. POWR1 is a domestication gene pleiotropically regulating seed quality and yield in soybean. Nat. Commun. 2022, 13, 3051. [Google Scholar] [CrossRef] [PubMed]
- Duan, Z.; Zhang, M.; Zhang, Z.; Liang, S.; Fan, L.; Yang, X.; Yuan, Y.; Pan, Y.; Zhou, G.; Liu, S. Natural allelic variation of GmST05 controlling seed size and quality in soybean. Plant Biotechnol. J. 2022, 20, 1807–1818. [Google Scholar] [CrossRef]
- Qi, Z.; Guo, C.; Li, H.; Qiu, H.; Li, H.; Jong, C.; Yu, G.; Zhang, Y.; Hu, L.; Wu, X. Natural variation in Fatty Acid 9 is a determinant of fatty acid and protein content. Plant Biotechnol. J. 2024, 22, 759–773. [Google Scholar] [CrossRef]
- Bing, L.; Peng, J.; Wu, Y.; Hu, Q.; Huang, W.; Yuan, Z.; Tang, X.; Cao, D.; Xue, Y.; Luan, X. Identification of an important QTL for seed oil content in soybean. Mol. Breed. 2023, 43, 43. [Google Scholar] [CrossRef] [PubMed]
- Gibellini, F.; Smith, T.K. The Kennedy pathway—De novo synthesis of phosphatidylethanolamine and phosphatidylcholine. IUBMB Life 2010, 62, 414–428. [Google Scholar] [CrossRef]
- Cao, J.; Li, J.-L.; Li, D.; Tobin, J.F.; Gimeno, R.E. Molecular identification of microsomal acyl-CoA: Glycerol-3-phosphate acyltransferase, a key enzyme in de novo triacylglycerol synthesis. Proc. Natl. Acad. Sci. USA 2006, 103, 19695–19700. [Google Scholar] [CrossRef]
- Zhang, D.; Zhang, H.; Hu, Z.; Chu, S.; Yu, K.; Lv, L.; Yang, Y.; Zhang, X.; Chen, X.; Kan, G. Artificial selection on GmOLEO1 contributes to the increase in seed oil during soybean domestication. PLoS Genet. 2019, 15, e1008267. [Google Scholar] [CrossRef]
- Liu, J.; Hao, W.; Liu, J.; Fan, S.; Zhao, W.; Deng, L.; Wang, X.; Hu, Z.; Hua, W.; Wang, H. A novel chimeric mitochondrial gene confers cytoplasmic effects on seed oil content in polyploid rapeseed (Brassica napus). Mol. Plant 2019, 12, 582–596. [Google Scholar] [CrossRef] [PubMed]
- Baud, S.; Wuilleme, S.; To, A.; Rochat, C.; Lepiniec, L. Role of WRINKLED1 in the transcriptional regulation of glycolytic and fatty acid biosynthetic genes in Arabidopsis. Plant J. 2009, 60, 933–947. [Google Scholar] [CrossRef]
- Baud, S.; Mendoza, M.S.; To, A.; Harscoët, E.; Lepiniec, L.; Dubreucq, B. WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J. 2007, 50, 825–838. [Google Scholar] [CrossRef] [PubMed]
- To, A.; Joubès, J.; Barthole, G.; Lécureuil, A.; Scagnelli, A.; Jasinski, S.; Lepiniec, L.; Baud, S. WRINKLED transcription factors orchestrate tissue-specific regulation of fatty acid biosynthesis in Arabidopsis. Plant Cell 2012, 24, 5007–5023. [Google Scholar] [CrossRef] [PubMed]
- Pelletier, J.M.; Kwong, R.W.; Park, S.; Le, B.H.; Baden, R.; Cagliari, A.; Hashimoto, M.; Munoz, M.D.; Fischer, R.L.; Goldberg, R.B. LEC1 sequentially regulates the transcription of genes involved in diverse developmental processes during seed development. Proc. Natl. Acad. Sci. USA 2017, 114, E6710–E6719. [Google Scholar] [CrossRef]
- Manan, S.; Ahmad, M.Z.; Zhang, G.; Chen, B.; Haq, B.U.; Yang, J.; Zhao, J. Soybean LEC2 regulates subsets of genes involved in controlling the biosynthesis and catabolism of seed storage substances and seed development. Front. Plant Sci. 2017, 8, 1604. [Google Scholar] [CrossRef]
- Lee, H.G.; Kim, H.; Suh, M.C.; Kim, H.U.; Seo, P.J. The MYB96 transcription factor regulates triacylglycerol accumulation by activating DGAT1 and PDAT1 expression in Arabidopsis seeds. Plant Cell Physiol. 2018, 59, 1432–1442. [Google Scholar] [CrossRef]
- Song, Q.-X.; Li, Q.-T.; Liu, Y.-F.; Zhang, F.-X.; Ma, B.; Zhang, W.-K.; Man, W.-Q.; Du, W.-G.; Wang, G.-D.; Chen, S.-Y. Soybean GmbZIP123 gene enhances lipid content in the seeds of transgenic Arabidopsis plants. J. Exp. Bot. 2013, 64, 4329–4341. [Google Scholar] [CrossRef]
- Cernac, A.; Benning, C. WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis. Plant J. 2004, 40, 575–585. [Google Scholar] [CrossRef]
- Ruuska, S.A.; Girke, T.; Benning, C.; Ohlrogge, J.B. Contrapuntal networks of gene expression during Arabidopsis seed filling. Plant Cell 2002, 14, 1191–1206. [Google Scholar] [CrossRef] [PubMed]
- De Vries, B.D.; Fehr, W.R.; Welke, G.A.; Dewey, R.E. Molecular analysis of mutant alleles for elevated palmitate concentration in soybean. Crop Sci. 2011, 51, 2554–2560. [Google Scholar] [CrossRef]
- Ma, W.; Kong, Q.; Mantyla, J.J.; Yang, Y.; Ohlrogge, J.B.; Benning, C. 14-3-3 protein mediates plant seed oil biosynthesis through interaction with AtWRI1. Plant J. 2016, 88, 228–235. [Google Scholar] [CrossRef]
- Li, D.; Jin, C.; Duan, S.; Zhu, Y.; Qi, S.; Liu, K.; Gao, C.; Ma, H.; Zhang, M.; Liao, Y. MYB89 transcription factor represses seed oil accumulation. Plant Physiol. 2017, 173, 1211–1225. [Google Scholar] [CrossRef] [PubMed]
- Chawla, R.; Poonia, A.; Samantara, K.; Mohapatra, S.R.; Naik, S.B.; Ashwath, M.; Djalovic, I.G.; Prasad, P.V. Green revolution to genome revolution: Driving better resilient crops against environmental instability. Front. Genet. 2023, 14, 1204585. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Lang, Z.; Zhu, J.-K. Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 2018, 19, 489–506. [Google Scholar] [CrossRef]
- Salami, M.; Heidari, B.; Batley, J.; Wang, J.; Tan, X.-L.; Richards, C.; Tan, H. Integration of genome-wide association studies, metabolomics, and transcriptomics reveals phenolic acid-and flavonoid-associated genes and their regulatory elements under drought stress in rapeseed flowers. Front. Plant Sci. 2024, 14, 1249142. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.; Daly, M.J. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Zhang, C.; Dong, S.-S.; Xu, J.-Y.; He, W.-M.; Yang, T.-L. PopLDdecay: A fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics 2019, 35, 1786–1788. [Google Scholar] [CrossRef]
- Raj, A.; Stephens, M.; Pritchard, J.K. fastSTRUCTURE: Variational inference of population structure in large SNP data sets. Genetics 2014, 197, 573–589. [Google Scholar] [CrossRef] [PubMed]
- Lipka, A.E.; Tian, F.; Wang, Q.; Peiffer, J.; Li, M.; Bradbury, P.J.; Gore, M.A.; Buckler, E.S.; Zhang, Z. GAPIT: Genome association and prediction integrated tool. Bioinformatics 2012, 28, 2397–2399. [Google Scholar] [CrossRef]
- Xin, M.; Zhang, Z.; Han, Y.; Feng, L.; Lei, Y.; Li, X.; Wu, F.; Wang, J.; Wang, Z.; Li, Y. Soybean phenological changes in response to climate warming in three northeastern provinces of China. Field Crops Res. 2023, 302, 109082. [Google Scholar] [CrossRef]
- Song, W.; Sun, S.; Wu, T.; Yang, R.; Tian, S.; Xu, C.; Jiang, B.; Yuan, S.; Hou, W.; Wu, C. Geographic distributions and the regionalization of soybean seed compositions across China. Food Res. Int. 2023, 164, 112364. [Google Scholar] [CrossRef]
- Bu, M.; Fan, W.; Li, R.; He, B.; Cui, P. Lipid metabolism and improvement in oilseed crops: Recent advances in multi-omics studies. Metabolites 2023, 13, 1170. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Wang, L.F.; Tao, J.J.; Zhang, W.K.; Chen, S.Y.; Song, Q.; Zhang, J.S. The comprehensive regulatory network in seed oil biosynthesis. J. Integr. Plant Biol. 2025, 67, 649–668. [Google Scholar] [CrossRef]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Mackay, T.F. Epistasis and quantitative traits: Using model organisms to study gene–gene interactions. Nat. Rev. Genet. 2014, 15, 22–33. [Google Scholar] [CrossRef]
- Cesari, S.; Thilliez, G.; Ribot, C.; Chalvon, V.; Michel, C.; Jauneau, A.; Rivas, S.; Alaux, L.; Kanzaki, H.; Okuyama, Y. The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell 2013, 25, 1463–1481. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zhu, J.; Wang, L.; Wang, S. Genome-wide association study identifies NBS-LRR-encoding genes related with anthracnose and common bacterial blight in the common bean. Front. Plant Sci. 2017, 8, 1398. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Nie, X.; Shen, C.; You, C.; Li, W.; Zhao, W.; Zhang, X.; Lin, Z. Population structure and genetic basis of the agronomic traits of upland cotton in China revealed by a genome-wide association study using high-density SNP s. Plant Biotechnol. J. 2017, 15, 1374–1386. [Google Scholar] [CrossRef]
- Wang, P.; Di, Q.; Liu, X.-Y. Genome-Wide association Study Identifies Candidate Genes Related to Oleic acid content of Soybean Seed. BMC Plant Biol. 2020, 20, 399. [Google Scholar] [CrossRef]
- Vuong, T.D.; Florez-Palacios, L.; Mozzoni, L.; Clubb, M.; Quigley, C.; Song, Q.; Kadam, S.; Yuan, Y.; Chang, T.F.; Mian, M.A.R.; et al. Genomic analysis and characterization of new loci associated with seed protein and oil content in soybeans. The Plant Genome. 2023, 16, e20400. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, X.; Lu, Y.; Bhusal, S.J.; Song, Q.; Cregan, P.B.; Yen, Y.; Brown, M.; Jiang, G.L. Genome-wide scan for seed composition provides insights into soybean quality improvement and the impacts of domestication and breeding. Molecular Plant. 2018, 11, 460–472. [Google Scholar] [CrossRef]
- Serson, W.R.; Gishini, M.F.S.; Stupar, R.M.; Stec, A.O.; Armstrong, P.R.; Hildebrand, D. Identification and Candidate Gene Evaluation of a Large Fast Neutron-Induced Deletion Associated with a High-Oil Phenotype in Soybean Seeds. Genes 2024, 15, 892. [Google Scholar] [CrossRef]
- Leamy, L.J.; Zhang, H.; Li, C.; Chen, C.Y.; Song, B.-H. A genome-wide association study of seed composition traits in wild soybean (Glycine soja). BMC Genom. 2017, 18, 18. [Google Scholar] [CrossRef]
- Rolling, W.R. A study of Phytophthora sojae Resistance in Soybean (Glycine max [L. Merr]) using Genome-Wide Association Analyses and Genomic Prediction; The Ohio State University: Columbus OH, USA, 2020. [Google Scholar]
- Ye, J.; Niu, X.; Yang, Y.; Wang, S.; Xu, Q.; Yuan, X.; Yu, H.; Wang, Y.; Wang, S.; Feng, Y. Divergent Hd1, Ghd7, and DTH7 alleles control heading date and yield potential of japonica rice in Northeast China. Front. Plant Sci. 2018, 9, 35. [Google Scholar] [CrossRef]
- Jiang, D.; Zhong, S.; McPeek, M.S. Retrospective binary-trait association test elucidates genetic architecture of Crohn disease. Am. J. Hum. Genet. 2016, 98, 243–255. [Google Scholar] [CrossRef]
- Wang, J.; Tang, Y.; Zhang, Z. Performing genome-wide association studies with multiple models using GAPIT. In Genome-Wide Association Studies; Springer: Berlin/Heidelberg, Germany, 2022; pp. 199–217. [Google Scholar]
- Zuo, Z.; Li, M.; Liu, D.; Li, Q.; Huang, B.; Ye, G.; Wang, J.; Tang, Y.; Zhang, Z. GWAS Procedures for Gene Mapping in Diverse Populations With Complex Structures. Bio-Protocol 2025, 15, e5284. [Google Scholar] [CrossRef] [PubMed]
- Tibbs Cortes, L.; Zhang, Z.; Yu, J. Status and prospects of genome-wide association studies in plants. Plant Genome 2021, 14, e20077. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Huang, M.; Fan, B.; Buckler, E.S.; Zhang, Z. Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLoS Genet. 2016, 12, e1005767. [Google Scholar] [CrossRef]
- Neupane, B. Systematic Comparison of GWAS Methods in Wheat: Balancing Statistical Power, False Positive Control, and Computational Efficiency. Preprints 2025. [Google Scholar] [CrossRef]
- Yu, J.; Pressoir, G.; Briggs, W.H.; Vroh Bi, I.; Yamasaki, M.; Doebley, J.F.; McMullen, M.D.; Gaut, B.S.; Nielsen, D.M.; Holland, J.B. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 2006, 38, 203–208. [Google Scholar] [CrossRef]
- Miao, C.; Yang, J.; Schnable, J.C. Optimising the identification of causal variants across varying genetic architectures in crops. Plant Biotechnol. J. 2019, 17, 893–905. [Google Scholar] [CrossRef]
- Yang, J.; Ren, Y.; Zhang, D.; Chen, X.; Huang, J.; Xu, Y.; Aucapiña, C.B.; Zhang, Y.; Miao, Y. Transcriptome-based WGCNA analysis reveals regulated metabolite fluxes between floral color and scent in Narcissus tazetta flower. Int. J. Mol. Sci. 2021, 22, 8249. [Google Scholar] [CrossRef] [PubMed]
- Ge, Z.; Liu, X.; Liu, B.; Abe, J.; Ma, F.; Kong, F. QTL mapping of soybean seed protein and oil trait. Soybean Sci. 2011, 30, 5. [Google Scholar] [CrossRef]
- Yang, Y. Fine Mapping and Candidate Gene Identification of a Soybean Seed Protein and Oil Qtl from a Wild Soybean Accession and Linkage Analysis for Whole Plant Biomass, Carbon, Nitrogen, and Seed Composition Using a RIL Mapping Population. Master’s Thesis, University of Missouri-Columbia, Columbia, MO, USA, 2021. [Google Scholar]
- Gillenwater, J.H. QTL Mapping of Seed Composition Traits and Assessment of Yield in Two Soybean Populations; North Carolina State University: Raleigh, NC, USA, 2020. [Google Scholar]



| Year | Chr | Physical Range of Significant SNP Loci | p Value | Effect | Number of Significant SNP Loci | Note | |
|---|---|---|---|---|---|---|---|
| Start | End | ||||||
| 2021 | 8 | 8,038,113 | 9,503,562 | 8.85 × 10−8 | −1.38 | 27 | Reduce oil content |
| 8 | 8,439,205 | 9,501,912 | 4.78 × 10−8 | 1.45 | 21 | Increase oil content | |
| 11 | 11,087,536 | 11,087,536 | 1.79 × 10−7 | 0.89 | 1 | Increase oil content | |
| 11 | 11,103,604 | 11,203,604 | 0.02 | −0.86 | 1 | Reduce oil content | |
| 20 | 15,764,307 | 15,764,307 | 2.80 × 10−7 | −1.01 | 1 | Reduce oil content | |
| 2023 | 1 | 10,828,034 | 10,841,167 | 1.91 × 10−7 | −1.08 | 3 | Reduce oil content |
| 1 | 10,537,841 | 10,904,178 | 2.46 × 10−7 | 1.07 | 4 | Increase oil content | |
| 8 | 9,005,247 | 9,079,037 | 1.41 × 10−9 | −1.58 | 7 | Reduce oil content | |
| 8 | 9,005,430 | 9,078,617 | 2.94 × 10−8 | 1.45 | 12 | Increase oil content | |
| 8 | 42,038,411 | 42,038,411 | 2.19 × 10−7 | 1.10 | 1 | Increase oil content | |
| 13 | 45,137,495 | 45,137,495 | 1.86 × 10−7 | −1.12 | 1 | Reduce oil content | |
| 13 | 45,140,366 | 45,163,611 | 1.56 × 10−7 | 1.13 | 2 | Increase oil content | |
| Average | 2 | 14,714,159 | 14,714,159 | 1.16 × 10−7 | −0.97 | 1 | Reduce oil content |
| 4 | 15,518,980 | 15,518,980 | 2.16 × 10−7 | −1.08 | 1 | Increase oil content | |
| 5 | 16,157,283 | 16,157,283 | 1.84 × 10−7 | 3.35 | 1 | Reduce oil content | |
| 7 | 16,529,749 | 16,529,749 | 1.12 × 10−7 | 0.94 | 1 | Increase oil content | |
| 8 | 8,432,800 | 9,501,864 | 6.65 × 10−8 | −1.32 | 13 | Reduce oil content | |
| 8 | 7,801,179 | 9,501,912 | 2.63 × 10−8 | 1.37 | 13 | Increase oil content | |
| 8 | 42,039,721 | 42,039,740 | 5.65 × 10−8 | 1.13 | 2 | Increase oil content | |
| 8 | 42,039,735 | 42,039,735 | 1.80 × 10−7 | −1.03 | 1 | Decrease oil content | |
| 11 | 33,092,710 | 33,092,710 | 2.42 × 10−7 | 1.04 | 1 | Increase oil content | |
| Year | Gene | Position (bp) | p Value | MAF | DNA Sequence Variation | Protein Sequence Variation | PROVEAN Score |
|---|---|---|---|---|---|---|---|
| 2021 | Glyma.08G110000 | 8,439,205 | 2.51 × 10−7 | 0.08 | A739C | T247P | 0.031 |
| Glyma.08G117400 | 9,074,920 | 8.62 × 10−11 | 0.05 | G682T | G228C | 1.450 | |
| Glyma.08G117600 | 9,501,436 | 6.76 × 10−12 | 0.05 | G698A | A233V | 1.233 | |
| Glyma.08G123500 | 9,501,620 | 2.36 × 10−7 | 0.06 | G2281A | V761I | −0.200 | |
| Glyma.08G123500 | 9,501,695 | 5.54 × 10−8 | 0.06 | G1650A | E550D | −0.667 | |
| Glyma.08G123500 | 9,074,920 | 2.61 × 10−7 | 0.06 | G1567C | E523Q | −0.200 | |
| Glyma.08G123500 | 9,074,920 | 2.19 × 10−7 | 0.06 | T1555C | C519R | −0.017 | |
| Glyma.08G123500 | 9,501,695 | 6.45 × 10−9 | 0.06 | A1508G | E503G | −1.633 | |
| Glyma.08G123500 | 9,501,912 | 2.03 × 10−8 | 0.06 | G1490C | L497P | −0.083 | |
| Glyma.08G123500 | 9,501,456 | 3.40 × 10−8 | 0.06 | T1488G | F496L | 0.933 | |
| Glyma.08G123500 | 9,501,556 | 1.11 × 10−7 | 0.06 | T1388A | I463N | 0.592 | |
| Glyma.08G123500 | 9,501,564 | 6.32 × 10−8 | 0.06 | T1380G | F460E | −1.092 | |
| Glyma.08G123500 | 9,501,586 | 2.39 × 10−7 | 0.06 | T1358A | V453E | 0.200 | |
| Glyma.08G123500 | 9,501,695 | 9.17 × 10−10 | 0.06 | A1249G | K417E | 0.058 | |
| Glyma.08G123500 | 9,501,776 | 3.87 × 10−8 | 0.06 | G1168A | D390N | 0.133 | |
| Glyma.08G123500 | 9,501,864 | 9.23 × 10−8 | 0.06 | C1080A | D360E | −0.033 | |
| Glyma.08G123500 | 9,501,912 | 4.54 × 10−8 | 0.06 | C1032G | N344K | 0.350 | |
| Glyma.08G123500 | 9,502,316 | 2.47 × 10−7 | 0.05 | T628G | S210A | 0.383 | |
| 2023 | Glyma.08G117400 | 9,054,741 | 4.94 × 10−12 | 0.05 | G682T | G228C | 1.450 |
| Glyma.08G117600 | 9,074,920 | 8.71 × 10−14 | 0.05 | C698T | A233V | 1.233 | |
| Average | Glyma08G117400 | 9,054,741 | 7.46 × 10−11 | 0.05 | G682T | G228C | 1.450 |
| Glyma08G117600 | 9,074,920 | 5.44 × 10−13 | 0.05 | C698T | A233V | 1.233 | |
| Glyma08G123500 | 9,500,663 | 2.60 × 10−7 | 0.06 | G2281A | V761I | −0.200 | |
| Glyma08G123500 | 9,501,454 | 1.47 × 10−7 | 0.07 | T1490C | L497P | −0.083 | |
| Glyma08G123500 | 9,501,456 | 7.61 × 10−8 | 0.06 | T1488G | F496L | 0.933 | |
| Glyma08G123500 | 9,501,695 | 3.88 × 10−9 | 0.06 | A1249G | K417E | 0.058 | |
| Glyma08G123500 | 9,501,864 | 1.66 × 10−7 | 0.07 | C1080A | D360E | −0.033 | |
| Glyma08G123500 | 9,501,912 | 1.84 × 10−7 | 0.07 | C1032G | N344K | 0.350 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Zhang, M.; Li, F.; Liu, X.; Zhang, C.; Zhang, F.; Zhao, K.; Yuan, R.; Lamlom, S.F.; Ren, H.; et al. Genome-Wide Association Study Reveals Key Genetic Loci Controlling Oil Content in Soybean Seeds. Agronomy 2025, 15, 1889. https://doi.org/10.3390/agronomy15081889
Wang X, Zhang M, Li F, Liu X, Zhang C, Zhang F, Zhao K, Yuan R, Lamlom SF, Ren H, et al. Genome-Wide Association Study Reveals Key Genetic Loci Controlling Oil Content in Soybean Seeds. Agronomy. 2025; 15(8):1889. https://doi.org/10.3390/agronomy15081889
Chicago/Turabian StyleWang, Xueyang, Min Zhang, Fuxin Li, Xiulin Liu, Chunlei Zhang, Fengyi Zhang, Kezhen Zhao, Rongqiang Yuan, Sobhi F. Lamlom, Honglei Ren, and et al. 2025. "Genome-Wide Association Study Reveals Key Genetic Loci Controlling Oil Content in Soybean Seeds" Agronomy 15, no. 8: 1889. https://doi.org/10.3390/agronomy15081889
APA StyleWang, X., Zhang, M., Li, F., Liu, X., Zhang, C., Zhang, F., Zhao, K., Yuan, R., Lamlom, S. F., Ren, H., Qiu, H., & Zhang, B. (2025). Genome-Wide Association Study Reveals Key Genetic Loci Controlling Oil Content in Soybean Seeds. Agronomy, 15(8), 1889. https://doi.org/10.3390/agronomy15081889







