Abstract
Lentil (Lens culinaris Medik.) is an essential legume crop providing healthy and nutritious food for people in low- to middle-income countries, worldwide. Lentil roots support symbiotic interactions with soil rhizobia species fostering nitrogen fixation; however, assemblage and diversity of the complete microbial rhizosphere community and the effect of seed genotype and origin remain largely unexplored. In this study we examined, via metagenomic analysis, the effects of seed origin on the rhizosphere’s communities in samples of the famous Greek lentil landrace, Eglouvis, derived from different local farmers and farming systems (including a Gene Bank sample), in comparison to a commercial variety. The landrace exhibited higher rhizosphere microbiome diversity compared to the commercial variety for all indexes. A core microbiome comprised of 158 taxa was present in all samples, while a greater number of unique bacterial taxa was recorded in the landrace samples compared to the commercial cultivar. Notably, landrace samples originated from organic farming had more than double the number of unique taxa compared to conventional counterparts. The study revealed a higher diversity of N2 fixers and archaea, Crenarchaeota and Thaumarchaeota, in landrace samples and particularly in those derived from organic farming, underpinning the distinct recruiting efficiency of beneficial soil microbes by the landrace.
1. Introduction
Globally, lentil (Lens culinaris Medik.) is an important food source due to its rich content of proteins, minerals, and vitamins, with yields exceeding 5 million tons per year []. Lentil local landraces are of particular interest primarily as a highly nutritious pulse crop providing high-quality, fortified products designated often as PDO (Protected Designation of Origin) and as PGI (Protected Geographical Indication). These products can foster local societies and economies and avert land degradation and desertification. In Europe alone, PDO and PGI products represent a sales value exceeding EUR 57 billion (10 × 109). Secondly, they provide a genetic reservoir of traits and genetic alleles that currently remain untapped yet could provide valuable tools to improve crop resilience and climate change adaptation to safeguard food security. Thirdly, they represent a protein-rich crop grown in low-input and rainfed systems that promote sustainability. Archeological evidence in Peloponnese, Greece, indicated that lentil cultivation dates back to 11,000 BC. Subsequently, centuries of continuous lentil cultivation have led to the development of landraces as fundamental components of Mediterranean farming systems and diet. Lentil is a self-pollinated species and it is generally accepted that several landrace characteristics are co-shaped through the continuous exposure to abiotic and biotic factors of the local environment, including poor soil fertility or exposure to pathogens and farmers’ selection to meet local needs [,]. On the other hand, breeding programs have released commercial cultivars of legumes including lentil, achieving uniformity and better yields, which are both trends of contemporary agriculture.
The famous lentil landrace Eglouvis, also known as “Faki Eglouvis”, has been cultivated in the homonymous plateau since 1700 in Lefkada Island, Greece []. The Eglouvis landrace is a microsperma-type lentil that represents an excellent paradigm of local landrace on-farm conservation, through traditional cultivation. Landraces undoubtedly represent a vital bond of tradition, history, and local identity revealing landscape distinctiveness. Landraces often guarantee higher provisioning services under non-optimal farming conditions []. Thus, understanding the intrinsic link among lentil-associated rhizosphere microbiomes and agronomic characteristics is crucial to advance food quality of plant protein crops and security, as well as soil conditions and functioning, in an environment facing climate challenges.
Nowadays, mounting evidence from research indicates that the plant genome has a central and dynamic role in shaping the composition of the microbiome [,]. Awad and coworkers have shown that the phyllosphere microbiome in grapevine cultivars is governed by the cultivar genotype in a collection of grapevine cultivars grown in the same orchard [], thus, demonstrating that the plant genotype is key player of the above-ground plant microbiome. Lentil, as a member of the Fabaceae family, maintains symbiotic associations with rhizobia species that are responsible for biological nitrogen fixation. Studies have shown that crop, genotype, and field environmental conditions contributed to the seed-associated microbial assemblage, demonstrating that the seed microbiome is also vertically transmitted in lentil, wheat, and canola crops []. Plant-associated microorganisms can be acquired indirectly from the surrounding environment or directly from the parent, in a vertical transmission pathway for microbial inheritance. Therefore, it is thought that the seed-associated microbiome may impact the development of the developing plant microbiome.
Studies have shown that legumes cultivation, particularly of pulse crops such as lentil, promotes soil fertility and enhances the productivity of sequential crops []. It is evident that plant genotype is an important factor not only from a breeding aspect, but equally for obtaining baseline information at microbial level to improve sustainable agricultural production. Lentil crops were shown to partially influence the microbial rhizosphere dynamics of sequential crops, particularly for microbial species that interceded N and P nutrient uptake [,]. The enrichment of distinct root microbes is not limited only to legumes. Wheat landraces have been shown to regulate the structure and diversity of rhizosphere microbiomes and, in turn, influence plant physiological traits []. To date, most of the lentil rhizosphere studies have been performed on commercial cultivars under various field conditions. Whether there are differences in the lentil rhizosphere microbiome among lentil cultivars and landraces under the same field and environmental conditions still remains unexplored. In a recent study it was shown that irregular high precipitation events resulted in a higher number of specific bacterial taxa identified in the rhizosphere of a lentil landrace compared to the commercial cultivars []. Nonetheless, whether a landrace maintains a distinct rhizosphere microbiome signature in comparison to modern varieties under the same field and environmental conditions remains unknown.
The so-called “rhizosphere effect” model describes the interactions and complex functions of the plant root and the surrounding soil environment [,]. The model simulates the influence sphere of the root’s local environment, depicting the interactions of plant genotype, root exudates, solutes, substrates, and microorganisms that govern nutrient uptake, cycling of resources, and plant health. Thus, the “rhizosphere effect” label was coined to describe the interplay of genotype-specific signaling molecules that support microbial proliferation and are also responsible for the formation of distinct microbial assemblages between the soil and the rhizosphere, as well as the interactions of the environment, soil type, microbiome, and plant growth and health []. It is becoming clear that the soil type and environment affect the legume rhizosphere microbiome [,]. Additionally, time of sampling and sampling compartments further impact the microbiome, often resulting in obscuring the genotype effect, which appears more profoundly in the rhizosphere microbiome of wheat genotypes []. Studies of the lentil microbiome are scarce and centered in lentil cultivars, in comparison to other legumes and crop species under normal and or erratic climate conditions [,].
Until now, research into the lentil rhizosphere microbiome has been based on bred cultivars (usually exhibiting genotypic uniformity) commonly known as modern varieties. It has been documented that lentil cultivar, generation, soil type, and their interactions are important factors controlling the lentil rhizosphere microbiome []. To obtain a further insight into the role of the genotype, seed origin, and storage conditions in the formation of the rhizosphere microbiome, we examined root microbiome communities formatted under the same field and environmental conditions in roots of the landrace Eglouvis, the seeds of which originated from different sources. The landrace Eglouvis has a polygenotypic germplasm composition [] and seeds are selected and stored by individual farmers based on their experience and diligence. Thus, samples from different farmers were included in the study to provide indications of the importance of farmer intervention. Further, crop cultivation conditions may vary among different farmers who may follow conventional or organic farming systems. One sample from organic farming was included to assess the implication of the farming system. One Gene Bank sample was included to detect storage effects under standard Gene Bank operational procedures. To evaluate the effects of the above-described factors on microbiome assemblage, we used 16S rRNA gene and ITS high-throughput amplicon sequencing and characterized the rhizosphere bacterial and fungal microbiota across Eglouvis landraces of different origins and compared its microbiomes to a commercial cultivar.
2. Materials and Methods
2.1. Plant Material
Lentil germplasm sources consisting of four Eglouvis samples and a modern commercial cultivar were used in this study. Three Eglouvis seed samples were collected from local farmers from Lefkada Island, Greece. Two of them originated from conventional farming systems and one from an organic farming system. Additionally, a conserved Gene Bank sample of Eglouvis (GRC 1015) and a commercial lentil variety Demetra were acquired from the Institute of Plant Breeding & Genetic Resources (IPB&GR), Thessaloniki, Greece (Table 1).

Table 1.
Details on the collection sites and origin, genotype, and sample source of the seeded material.
2.2. Experimental Design and Cultivation
Lentil field cultivation was carried out at IPB&GR, Thessaloniki, Greece (22°59′6.17″ E, 40°32′9.32″ N) during the growing seasons of 2020–2021. Lentils were sown directly in a randomized complete plot design (RCBD), which was part of a bigger lentil field trial (December 2020). Practically, seeds from each sample were grown scattered throughout the experimental field with the exact location and origin of each plant marked and recorded for following analyses. Lentil cultivation followed a rainfed system, without any additional fertilization or pesticide application. During cultivation, visible weeds were removed manually. Weather data including rainfall and air temperature were recorded on site. Field soil properties at 0–30 cm are summarized in Table 2.

Table 2.
Soil properties of the lentil experimental field.
2.3. Sampling
Rhizosphere soil and plant samples were collected at the mid-blooming stage (April 2021). About 10–12 random plants were carefully removed from random locations of the RCBD excluding boundary rows. Lentil plants were separated into two parts, above-ground shoot and rhizosphere. Lentil roots with the adhered soil were placed in plastic bags and quickly transported to the laboratory, while being cooled (icebox). Then rhizosphere soil was separated from roots using a soft brush. Approximately 5 g of pooled rhizosphere soil was collected in sterile tubes (Falcon 15 mL) and stored at −20 °C until further processing. Roots were rinsed clean with distilled water, and subsamples were prepared and preserved for further analyses.
2.4. Plant Morphological Traits
The above-ground plant samples were used to assess shoot biomass dry-weight (SB) and height (SH). Root samples were used to assess root length (RL), root biomass dry-weight (RB), rhizobial nodules, and arbuscular mycorrhiza fungal colonization (AMF). Specifically, rhizobia nodulations were determined microscopically. AMF colonization (counts) per plant was determined macroscopically using the ink–vinegar method, as previously described [] with some modifications. Specifically, roots were rinsed with deionized water and cleared with 10% KOH for 10 min at 80 °C. After rinsing with deionized water, roots were placed in a 1% HCl solution for 20–30 min to neutralize the pH and subsequently were stained with an ink–vinegar solution (57 mL Black Parker Quink in 1000 mL vinegar) for 5–10 min at 80 °C. Finally, roots were rinsed with deionized water and stored in 50% glycerol. Twenty-five root fragments were mounted on a microscope slide and the presence of vesicles, arbuscules, and hyphae counted at four different points along each root piece (100 intersections in total) with a light Leitz Laborlux S microscope (Ernst Leitz GmbH, Wetzlar, Germany).
2.5. DNA Extraction
DNA was extracted from soil rhizosphere samples (500 mg) using a NucleoSpin Soil kit (Macherey-Nagel, Düren, Germany) following the SL1 solution extraction protocol according to manufacturer’s instructions. DNA concentration was quantified using Qbit 4 Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA) and visualized in 1% agarose gel electrophoresis. Samples were stored in −20 °C until further analyses.
2.6. Sequencing and Bioinformatic Analyses
Library preparation and high-throughput sequencing was performed at the Sequencing Facility of the Laboratory of Genetics and Plant Breeding, School of Agriculture, Aristotle University of Thessaloniki, Greece, following a similar approach as in [] with modifications. Briefly, instead of using a Thermo 16S metagenomics kit, we amplified bacterial (V4) and fungal (ITS2) targets using the Platinum™ II Hot-Start PCR Master Mix (Thermo Fisher, USA) (see Supplementary Table S1 for primers and PCR protocols). Subsequently, equimolar concentrations of 16S and ITS2 amplicons of each sample were pooled and barcoded (Ion Xpress™ Barcode Adapters kit, Thermo Scientific, Waltham, MA, USA). Barcoded samples were quantified and pooled (Ion Library TaqMan™ Quantitation kit, Thermo Scientific, USA), resulting in an enriched library for sequencing using an Ion Studio S5 sequencing platform (Ion 520™ and Ion 530™ Kit-OT2 kit, Ion 520 chip, Ion Studio S5 Thermo Fisher, USA). Sequenced reads were demultiplexed and QC-, adaptor-, and barcode trimmed using Ion Torrent Suite prior to MG-RAST [] deposition under project number mgp98852 (https://www.mg-rast.org/linkin.cgi?project=mgp98852, accessed on 31 October 2023). Taxonomy was assigned using SILVA SSU database (version 138.1) at 97% identity (e value 10−5) using standard MG-RAST quality control (dynamic error removal (DRISEE), dynamic trimming (DYNAMICTRIM), denoising, and normalization (FASTQ-MCF)).
2.7. Statistical Analyses
Assessed lentil morphological measurements were analyzed using Multivariate Analysis of Variance (MANOVA) to detect statistically significant differences among the measured variables, according to Wilks’ Lambda test. Correlation analysis of morphological traits (shoot biomass, root biomass, shoot height, root length, root/shoot, nodules, and AMF) assessed were calculated using Pearson’s coefficient and depicted with a heatmap using the “GGally” package. The aforementioned analyses were performed in R Studio using R version 4.3.2 [].
The lentil rhizosphere microbiome was evaluated statistically for relative abundance (phylum level) and diversity indices (genus level; Chao1, Shannon (H), and Simpson (D) indices), using the phyloseq package of R Studio [] for similarity between samples (β diversity), using a Bray-Curtis dissimilarity distance matrix that was depicted with PCoA (Principal Coordinate Analysis), using the phyloseq package as mentioned above. Furthermore, the number of common species among the samples was depicted with Venn diagrams [], and rarefaction curves were calculated and depicted with the vegan package []. Illustrations were prepared using ggplot2 package []. All statistical analyses were performed in the R Studio environment. All p values < 0.05 were considered statistically significant.
2.8. Network Correlation and Analysis
Microbial co-occurrence network analysis was preformed using the Spearman’s correlation method to identify pairwise associations as previously described []. Entries that had an absolute Spearman correlation (ρ) threshold lower than 0.85 and a prevalence less than 5 were removed. This resulted in a correlation adjacency matrix of an equivalent core microbiome of 158 taxa, with 158 nodes sharing 1078 links. Network properties were calculated using the igraph package [] of the R Studio.
3. Results
3.1. Assessment of Plant Morphological Characters
The Eglouvis landrace samples exhibited, on average, 30% lower above-ground biomass compared to the commercial cultivar Demetra. Subsequently, Eglouvis samples had, on average, 35% lower shoot height than Demetra. These differences were statistically significant; Figure 1a. Interestingly, Eglouvis root length and root-to-shoot ratio were, on average, greater than those of Demetra, and those differences were statistically significant, as depicted in Figure 1a. Furthermore, Eglouvis root-related indexes, including nodules and AMF, were greater and statistically significant compared to Demetra, respectively.
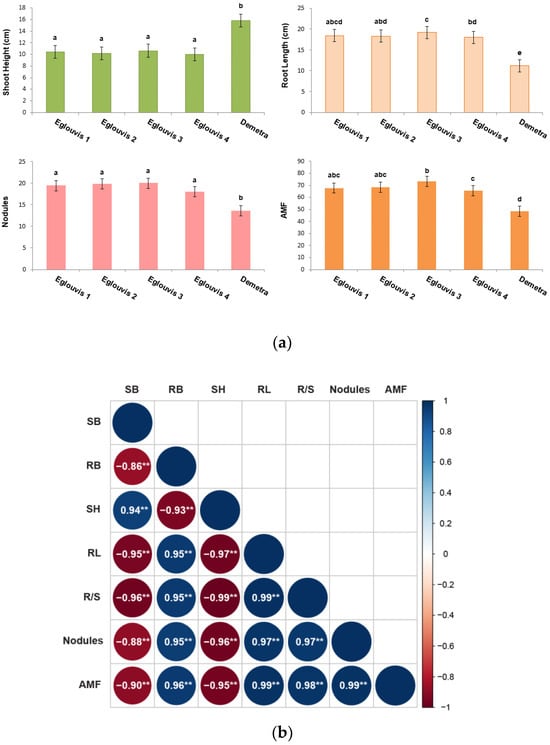
Figure 1.
Lentil plant morphological properties: shoot height (SH), shoot biomass (SB), root biomass (RB), root length (RL); root shoot ratio (R/S); nodules, arbuscular mycorrhizal fungi (AMF). (a) Barplots of shoot height, root length, nodules, and AMF. Values that share no common letter are significantly different according to Tukey’s HSD (p < 0.05). (b) Pearson’s correlation plot of the morphological measurements for segregated Eglouvis and Demetra plants (** p < 0.01).
Correlation analysis of morphological traits assessed in plants of all samples, Eglouvis 1–4 and Demetra, revealed very strong correlations among root traits. Thus, further analysis followed, from which segregating data of Eglouvis and Demetra plants unveiled a more comprehensive comparison. The highest positive correlations were observed between root length–root/shoot (0.99), root length–AMF (0.99), and AMF–nodules (0.99). Following were the correlations of root/shoot–AMF (0.98), R/S–nodules (0.97), and RL–nodules (0.97). Interestingly high correlations were also observed between RB–AMF (0.96), followed by RB–nodules (0.95), RB–R/S (0.95), and RB–RL (0.95). Shoot height was also strongly correlated with shoot biomass (0.94), indicating that root traits are key drivers of AMF and nodulation in lentil and governed by root–shoot interplay.
3.2. Sequence Analyses
3.2.1. Microbial Diversity among Genotypes
Out of 288,462 sequence reads (approx. 58,000 reads per sample), 836 different operational taxonomic units (OTUs) were identified excluding singletons. Because of ITS’ high sensitivity, several eukaryotic organisms such as plants, nematodes, and mammals were also identified. Therefore, the following phyla were excluded from the analysis: Arthropoda, Nematoda, Streptophyta, and Chordata. From the remaining OTUs (803), upon applying 97% cut-off, we used 733 OTUs from the most abundant phyla for further statistical analysis. The overall microbiome consisted of 87% bacteria, 9% eukaryota including fungi, and 4% archaea. The most abundant bacterial phyla were Actinobacteria (30%), followed by Proteobacteria (19%) and Firmicutes (18%) (Figure 2). Archaeal phyla consisted of Crenarchaeota and Thaumarchaeota, which were identified with 4502 total reads. For all samples, rarefaction curves reached a plateau after approximately 15,000 reads, indicating sufficient sequencing depth (Figure S1) for further diversity analysis of the obtained datasets. Eglouvis 3 displayed the most abundance, while Demetra, which was used as control, displayed the least. Eglouvis 1 and Eglouvis 2 displayed almost the same abundance, with almost 300 species.
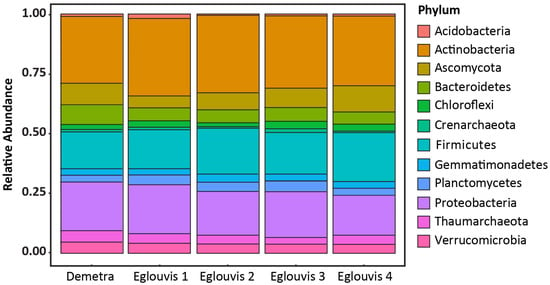
Figure 2.
Rhizosphere microbiome relative abundance at phylum level, illustrating the 12 most abundant phyla of the targeted lentil soil microbiomes.
The presence of Kabatiella microsticta, a plant pathogen also known as Aureobasidium microstictum (Bubák), was interestingly noticeable only in sample Eglouvis 4, identified by a large number of reads (12,640). Members of Kabatiella spp. belong to fungal phylum Ascomycota, which is reported to cause leaf streak on maize and ornamentals plants (Hemerocallis spp.). Recent studies reported the presence of this pathogen in China and in Norway []. Due to the extremely large number of Kabatiella spp. reads in Eglouvis 4, this pathogen was excluded from the analysis (relative abundances including Kabatiella spp. are presented in Figure S2). Furthermore, our sequencing analysis revealed several important pathogens known to infect legumes. Verticillium dahlia, which is a soil-borne pathogen that causes verticillium wilt, was detected in all samples at 7% on average. More plant pathogens were found, including Aspergillus niger (approx 0.02%), Davidiellatassiana (in Eglouvis 3; 0.01%), Myrothecium spp. (M. verrucaria, M. roridum, and M. gramineum; approx. 0.12%), and Phytophthora spp. (P. infestans, P. ramorum, P. sojae; approx. 0.16%) (Figure S3).
We observed diverse microbiomes associated with the rhizospheres of Eglouvis samples and Demetra. Among all targeted rhizosphere microbiomes, Eglouvis 3 had the greatest α diversity: 467, 4.590, and 0.979 for Chao1, Shannon, and Simpson indices, respectively (Figure 3). Eglouvis samples had, on average, greater α diversity indices than the control Demetra (Figure 3). Chi-square test analysis was conducted to assess the independence of samples, indicating that significant differences exist among relative abundances of samples at a significance level of p = 0.05. Further α diversity indices are reported in Table S2. Differences in β diversity at phylum level (Bray-Curtis) were illustrated with PCoA analysis (Figure 4). The first two PCoA axes explained approximately 73% of the total variation within the rhizosphere microbiome. Eglouvis samples (1–4) were ordinated distant to Demetra and clustering analysis separated the microbiome dataset into three groups: group a consisted of Eglouvis 3 and 4, group b of Eglouvis 1 and 2, and group c of Demetra. Furthermore, we assessed whether differences in β diversity were associated with plot distance (sampling distance) and typical physiological plant traits, including shoot and root biomass, root length, thousand seed weight, arbuscular mycorrhiza fungal colonization, and rhizobia nodules abundance, using Μantel correlation and redundancy analyses, respectively. Plot distance explained approximately 35% of the variation in rhizosphere diversity; thus this correlation was not significant (Mantel r = 0.345, p = 0.17), whereas the correlation between explanatory variables (plants traits) and response variables (rhizosphere microbiome) was strong (Mantel r = 0.81), and this association was marginally significant (p = 0.09).
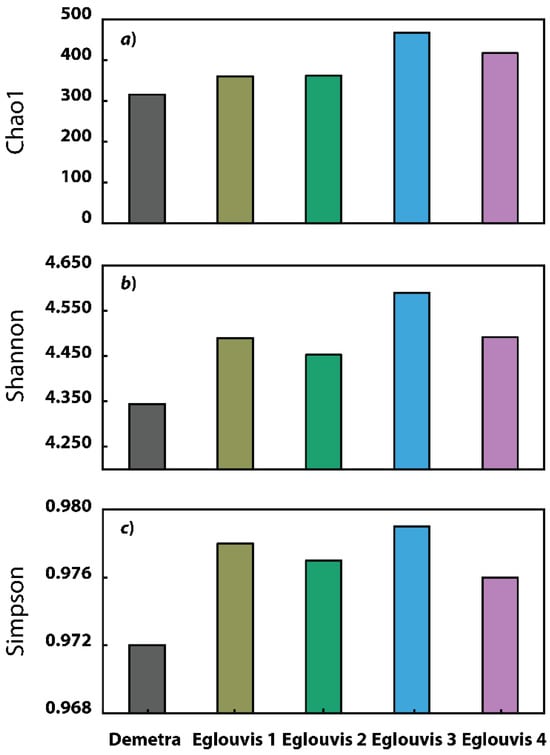
Figure 3.
Metrics of α diversity (a) Chao1, (b) Shannon, and (c) Simpson index, for the commercial cultivar (Demetra) and Eglouvis landrace (Eglouvis 1–4).
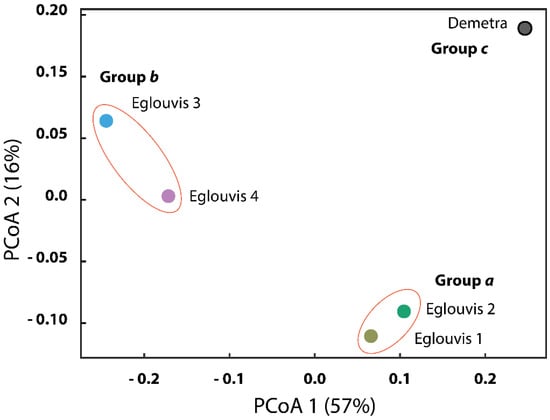
Figure 4.
PCoA of lentil genotypes constructed using the Bray-Curtis dissimilarity matrix.
Besides the clear differences in α and β diversity among the lentil Eglouvis landrace samples and the control cultivar Demetra, we mined our datasets for common species that could be part of a core microbiome (Figure 5). The Eglouvis 3 and 4 (indicated as Group a) shared ~28% of the detected OTUs and ~20% with Demetra (Group c). The Eglouvis 1 and Eglouvis 2 (comprising Group b) shared ~24% of the detected OTUs, with ~19% of them being common with Group c (Demetra). In both groups the Eglouvis samples had high portions of unique OTUs: 28% and 23% in group a, and 23% and 24% in group b, whereas Demetra had a portion of 11% unique OTUs in group a and 19% in group b. Tables of relative abundances for the two groups at phylum level are presented in the Supplementary Material (Table S3).
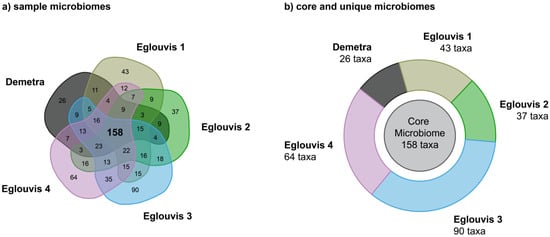
Figure 5.
Diagrams showing the relations based on microbial OTUs among samples; (a) Venn diagram of shared taxa, and (b) donut diagram of shared (core) and unique microbiomes.
3.2.2. Unique Microbial Taxa
Unique bacterial and fungal taxa were identified in higher numbers in landrace samples compared to the modern cultivar. Specifically, the unique bacterial taxa present in the Eglouvis 1–4 rhizosphere microbiome samples ranged within 38–88, with the Eglouvis 3 sample exhibiting 88 bacterial taxa, in comparison to Demetra presenting 25 (Table 3). On the other hand, far fewer fungal taxa were identified as unique in all samples, with a range of 0–2. Thus, the total number of unique taxa identified in all samples ranged within 37–90, with the Eglouvis 3 sample hosting the largest number (90) of unique taxa and Demetra the smallest (26).

Table 3.
Number of unique bacterial and fungal taxa.
Subsequently, the partitioning of major free-living N2-fixing bacterial taxa was assessed among samples at the genus level. The percentage of each N2-fixing bacterial genus detected in the rhizosphere microbiome of each sample is depicted in Figure 6. Interestingly beyond the Rhizobium and Bacillus genera present in high amounts, a number of other key nitrogen fixers were also identified, with most of them present in the landrace samples. More specifically, the commercial variety had less nitrogen-fixing taxa (942) than Eglouvis 1–4 (1433–3890), comprising nearly 8 to 15% of the sequenced taxa. A roughly two-fold greater abundance of Nitrospira, Paenibacillus, Rhodococcus, and Variovarax spp. was found in rhizosphere samples of Eglouvis 1 relative to Demetra. The abundance of Azospirillum, Flavobacterium, and Pseudomonas spp. was approximately two-fold less in Eglouvis 1 compared to that in Demetra rhizosphere samples. For Eglouvis 2, Arthrobacter, Paenibacillus, and Stenotrophomonas spp. were 50% more abundant than in Demetra. A 50% reduction was observed for taxa belonging to the genera of Nitrospira, Pseudomonas, Rhizobium, and Rhodococcus spp. when compared to Demetra rhizosphere samples. For Eglouvis 3, the abundance of Arthrobacter, Methylobacterium, Nitrospira, Paenibacillus, and Stenotrophomonas spp. was greater than 50% when compared to Demetra. Interestingly, Azotobacter spp. were identified only in the Eglouvis 3 sample. Lastly, in the rhizosphere sample of Eglouvis 4, greater abundance (≤50%) was observed for Paenibacillus, Rhodococcus, and Variovorax spp., and there was a noticeable reduction in Azospirillum, Burkholderia, Flavobacterium, and Methylobacterium spp. compared to in Demetra. The abundance of Bacillus spp. remained unchanged through all samples and comprised about 50% of the nitrogen-fixing taxa.
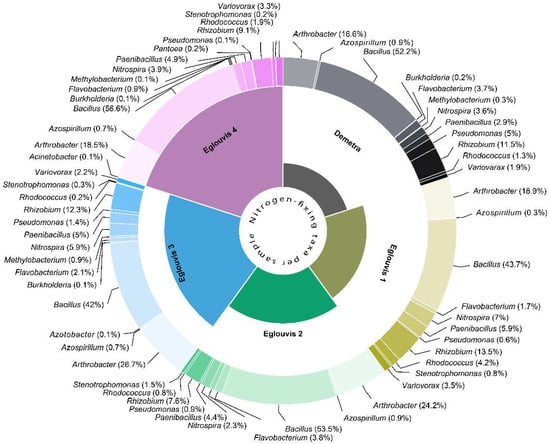
Figure 6.
Distribution and partitioning of major N2-fixing species among the five lentil rhizosphere samples identified by 16 rDNA gene sequencing. Pie sector size indicates the relative abundance of species.
3.2.3. Co-Occurrence Network Analysis
Network co-occurrence analysis indicated a linkage density of 6.8, a connectivity value of 0.086, and an average path of 3.14. Degree centrality analysis indicated that the top 10% of the key species (31/158) that were particularly important for the network hold between 18 and 20 links (Table S4). Closeness centrality indicates species that are grouped closely to each other; therefore, those nodes are considered more central; the top 10% (16/158) are listed in Table S5. Betweenness centrality is a measure of frequency for the top 10% of species (15/158) acting a “bridge” between two other nodes; betweenness ranged from 424 to 631 links (Table S6). The core microbiome co-occurrence network at the phylum level is illustrated in Figure 7.
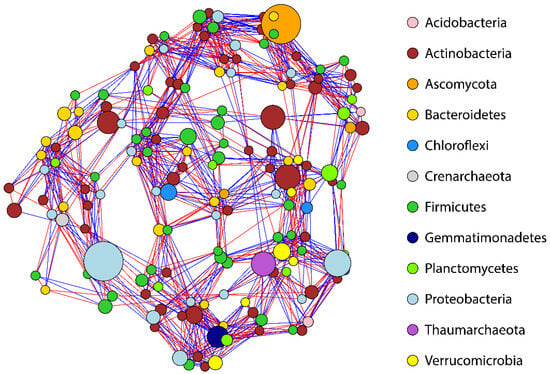
Figure 7.
The co-occurrence network of the lentil rhizosphere microbiome. Core microbiome co-occurrence network at the phylum level. Red links indicate negative and blue links indicate positive correlation. Node size indicates the average relative abundance of microbial species.
4. Discussion
Landraces are defined as dynamic populations of a cultivated plant that has a historical origin and distinct identity and lacks formal crop improvement; they are also often genetically diverse, locally adapted, and associated with traditional farming systems []. Studies of lentil landraces reported a high level of genetic diversity in Greek germplasm when compared with germplasm from other Mediterranean countries with other agro-climatic environments [,,]. The lentil Eglouvis is a landrace with a high level of genetic variation and unique genetic background that does not overlap with that of Demetra when examining individuals, or even bulk samples, using molecular markers []. As studies on the rhizosphere microbial diversity among legume species and cultivars are emerging, our understanding of how the landrace genotype may impact this diversity remains scarce. Furthermore, seeded material may act as a source of vertical transmission of the microbiome [] in modern cultivars. However, little is known about landraces and the impact of these dynamic genetic populations of genotype(s) on the rhizosphere’s microbial diversity. Thus far, studies in crops and legume species have shown that soil type, environment, and genotype may impact the rhizosphere microbiome diversity [,,,]. In our design we assessed the rhizosphere microbial community of the lentil landrace Eglouvis samples originating from three distinct systems: (a) as seed material obtained from conventional and (b) organic farming systems, and (c) a Gene Bank sample, all assessed under the same field and environmental conditions during the same cultivation period. The commercial modern cultivar Demetra was used as a reference to discern the plant genotype effect on the rhizosphere microbiome structure, thereby enabling a direct and uniform evaluation.
Studies have shown that the plant rhizosphere comprises diverse and distinct microbial communities regulating plant growth, survival in varying environmental conditions, and resistance to plant pathogens. In this study, we used the 16S rRNA gene and ITS2 regions to uncover the impact of landrace origin and farming system on the rhizosphere microbiome structure and diversity. To exclude any masking effects of environmental attributes, samples were cultivated in proximity under the same field conditions during the same cultivation period following agronomic practices of non-chemical inputs including fertilizers and pesticides. We observed significant differences in plant traits and rhizosphere microbiomes among the studied genotypes, indicating that genotype and seed origin could act as an important factor regulating plant–soil microbial interactions.
4.1. Plant Traits Impact Microbial Diversity
We found differences in commonly measured physiological traits such as root length, rhizobial nodulation, and arbuscular mycorrhizal fungal colonization. The Eglouvis landrace root length was 25% longer than the commercial cultivar in all samples assessed. Root morphology could be regulated by environmental conditions, i.e., climate and soil fertility [,,]. Under drought or nutrient-limiting, conditions lentils may develop longer root systems to scavenge available soil moisture or nutrients [,]. Considering that all lentil genotypes were cultivated under the same conditions in proximity, the landrace Eglouvis samples, which exhibited a longer root system, could withstand longer drought conditions and tolerate nutrient-poor soils. Another aspect of root morphology is linked to nitrogen fixation. Roots provide the ground plant surface for rhizobial nodulation and AMF colonization, contributing to nutritional benefits for legume growth. Thus, genotypes with longer root systems exhibit a two-fold potential, primarily for accessing relatively more soil nitrogen and secondly for hosting more rhizobial and mycorrhizal species per root length. In this study, soil nitrogen was initially low, and no nitrogen was applied during the cultivation period, following local agricultural practices. Therefore, all nitrogen used by the lentil plants is thought to come from nodules’ activity and from nitrogen uptake. Specifically, it was observed that the Eglouvis landrace possessed more rhizobia nodulation and mycorrhizal colonization than the commercial cultivar Demetra. It is suggested that root morphology may additionally influence rhizosphere microbiome, other than via root nodulation and colonization [,,]. In our study, we observed that the longer root system of the Eglouvis landrace influenced the assembly of rhizosphere microbes. A moderate correlation was found between nodules and N2-fixing taxa, indicating that other parameters may govern the rhizosphere microbial networks, which should be further explored. Thus, the influence of root morphology within landraces should be further investigated in a holistic approach, considering the temporal effects on a large set of root morphology traits.
4.2. Microbial Diversity
Research studies have demonstrated that different plant species host distinct rhizosphere microbiomes communities [,]. Changes in the rhizosphere microbiome were also evident in cropping systems under cereal–legumes rotation [,]. In previous studies, significant interactions of environment × crop and crop × generation were found [,,]. However, a considerable part of the variation remains unexplained, and it was attributed to unmeasured environmental variables or genetic attributes such as genotype/cultivar [,,,]. Recently, the role of genotype as an important factor shaping the rhizosphere microbiomes has been appreciated for legumes [], as well as for non-legumes such as wheat []. Studies showed that lentil crops select their rhizosphere microbiome from the surrounding soil, allowing endophytes to colonize roots, stems, and seeds, which in turn may inherit or re-inoculate a crop-specific microbiome. Similarly, from these and other studies [,,], we unearthed microbial taxa belonging to Actinobacteria, Proteobacteria, Firmicutes, and Chloflexi that are involved in the biological N2-fixation and nitrogen cycle (Rhizobiales, Arthrobacter, Nitrosospira spp., etc.) and in the phosphorus-cycling and organic matter decomposition process (Firmicutes and Cloroflexi spp., etc.). Proteobacteria and Bacteriodetes are classified as copiotrophic groups [] and are commonly detected in the rhizosphere []. Ascomycota prevail in fungi phyla. Similar findings were observed in the rhizosphere of legumes [,], and non-legumes [].
Lentil, as a legume species, exhibits an active close association with nitrogen-fixing bacteria, including the well-known rhizobia species, as main inhabitants of the legume nodules orchestrating the symbiotic N2 fixation. Besides the typical rhizobia inhabitants, legume nodules may harbor also other nitrogen fixers []. Recent studies of the leguminous Dalbergia odorifera have shown that non-rhizobial bacteria were detected in the host’s nodules representing 32 genera. Notably, these non-rhizobial bacteria were predominant in the N-omitted potting mix, with a relative abundance of 56–87%. Thus, it is advocated that legume nodules could be inhabited by a high diversity of non-rhizobial species, which play a critical role in nodulation and N2 fixation of the leguminous host. This diversity of nodule dwellers includes Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, and the genus Rhodococcus from the phylum Actinobacteria []. In our findings, the order Rhizobiales (Alphaproteobacteria), classes Alphaproteobacteria (including unclassified), Betaproteobacteria, and Gammaproteobacteria, and the genus Rhodococcus were most abundant in the Eglouvis samples and may influence the formation of mycorrhiza and nodules assessed in these samples compared to the commercial cultivar.
Remarkably, genus Stenotrophomonas was present only in Eglouvis samples. The genus is a Gram-negative bacteria and member of the Xanthomonadaceae family. Stenotrophomonas maltophilia was recently recognized for its plant growth-promoting rhizobacterial (PGPR) ability and bioactivities against biotic and abiotic stress in sedge species Cyperus laevigatus and wheat []. Recent studies in peanut (Arachis hypogea), a legume species, have demonstrated that S. maltophilia increased plant growth, antioxidant levels, scavenging, and stress tolerance under N2-deficit conditions, underscoring its role as an efficient PGPR for growing agricultural crops [].
Furthermore, Eglouvis samples exhibited a higher number of Variovorax spp., bacteria known for their growth-promoting effect on plants due to the presence of two enzymatic systems, nitrilase/and nitrile hydratase/amidase, which convert indole-3-acetonitrile (IAN) to the important plant hormone indole-3-acetic acid (IAA), and to the presence of nitrogen fixation ability []. The latter is further enhanced in Eglouvis samples by the presence of Azotobacter spp., a PGPR member, which is widely known for its alternative nitrogenases along with the presence of conventional nitrogenase, distinguished by the nif genes complex, as well as for its ability to release siderophores [,].
It is important to highlight the presence of archaea in the lentil rhizosphere, which is often overlooked, and specifically those archaeal groups involved in nitrogen and carbon cycling. Here, we observed members of Crenarchaeota and Thaumarchaeota in relative greater abundance in Eglouvis 3, 4, 1, and 2 than in the commercial cultivar. It is understood that archaea hold an important role in promoting plant health, but little is known about the mechanisms of such interaction [,]. It is speculated that archaea contribute to functions such as auxin production, protection against biotic and abiotic factors, interaction with fungi, and production of secondary metabolites, which protect plants against pathogens. Even though there is limited evidence in support of this theory [], emerging studies, including the current research, prompt further investigation of the active role of archaeal populations in the lentil rhizosphere, as well as that in other crops.
Inter-species variation in the host plant genome has been found to have a relatively small effect on the associated microbial communities compared with other factors such as environmental variation [,,,,]. In this work, we found that the rhizosphere bacterial communities of Eglouvis samples were divergent compared to Demetra regarding the amounts of participating N2-fixing bacteria. The observed differences in partitioning underscore a vital role of nitrogen-fixing bacteria in shaping the lentil rhizosphere that requires further investigation, as some species are present only in the Eglouvis samples, such as Acinetobacter, Azotobacter, and Stenotrophomonas, while others exhibit higher partitioning in Eglouvis samples compared to Demetra, such as Arthrobacter, Bacillus, Paenibacillus, and Rhizobium. We therefore further propose that differences in rhizosphere bacteria between the landrace and varieties could be derived from differences in recruitment (either actively or passively) of local bacteria. However, the molecular mechanisms that orchestrate the recruiting of rhizosphere microbiota, as well as how the genetic variation of the landrace governs the microbiome variation, are unknown. In this work, seeds were expected to have nominal quantities of bacteria associated with them (since reads were dominated by host DNA). Whether such differences may impact microbiota recruitment beyond the host’s exudates remains to be thoroughly characterized. Undoubtedly, differences in root architecture were noted between the landraces and the commercial cultivar, underpinning one mechanism that might differentiate their associated microbial communities; however, the implication of other mechanisms that may impart the observed differences was also reported for other crops such maize [,]. Legumes initiate plant–microbe interactions in the rhizosphere through communication pathways involving root-exudates known as flavonoids, which trigger the production of nodulation by compatible rhizobia symbionts. Mounting evidence indicates that these flavonoids not only initiate symbiosis with rhizobia, but also play a central role in shaping the legume rhizosphere community structure [,]. Whether this quorum sensing of rhizosphere microbiota assemblages is also regulated by intra-bacterial communication networks remains to be explored, as studies have shown that the lentil crop influences nutrient acquisition in subsequent crops [,]. Nutrients are generally known to be more abundant in the rhizosphere than in bulk soil. Thus, the favored development of bacterial phyla near lentil roots is detected as the result of microbial consortia of communicating patterns that are becoming apparent.
The core microbiome network in this set of data has low connectivity and only 8.6% of the possible links are realized. However, further analysis indicates key species that hold a high degree of centrality, closeness, and betweenness. At phylum level we can observe a central role in the network construction with higher relative abundance of Ascomycota and Proteobacteria, while a higher number of species (nodes) is observed in Actinobacteria and Firmicutes. Remarkably, species in these phyla play multiple roles in carbon and nitrogen cycles, and their high numbers indicate their significance in the structure of the lentil rhizosphere. Interestingly, Crenarchaeota and Thaumarchaeota species involved in nitrification constitute a single node in the network, indicating that the cluster may collapse following a possible loss of this node.
4.3. Lentil Microbial Pathogens
Although no visual symptoms were detected in the field trial, the sequencing analysis revealed potential pathogenic taxa of less than 2% relative abundance. Specifically, members of Verticillium, Aspergillus, Davidiella, Myrothecium, and Phytophthora spp. were found. This could possibly indicate that landraces have developed resistance to those pathogenic microbes or that pathogens remained dormant. The presence of pathogens within the lentil rhizobiome is not uncommon. Sequencing of lentil genotypes in Canada and India also revealed several potentially pathogenic taxa within the rhizosphere microbiome [,]. Since no visual symptoms were observed, it is important to decipher whether the presence of the identified pathogens is circumstantial or associated with certain lentil genotypes’ ability to suppress the pathogens [,].
On the other hand, the presence of Aureobasidium microstictum in the rhizosphere of the Eglouvis 4 sample obtained from Gene Bank raises important questions and concerns. Specifically, its presence highlighted an important parameter of seed storage quality that is becoming apparent with the use of new technologies such as the high-throughput sequencing (HTS) techniques []. Standard methods and protocols of seed evaluation are generally macroscopic, often aided using a stereoscope. On the other hand, most of the pathogens are transmitted through seed, while others pre-exist in the field. Seed-borne or soil-borne pathogens can survive for long periods []. Because the seeds were not disinfected, and no pesticides were applied to the field, certain pathogens could potentially have been enriched. However, the fact that the presence of the particular pathogen was only detected in the sample obtained from Gene Bank emphasizes the need for further research in deciphering the presence of seed-associated microbiomes through high-throughput sequencing.
4.4. Practical Implications
The “rhizosphere effect” is a commonly accepted model of understanding microbial assemblage and diversity within the plant’s rhizosphere. It is assumed that plants may be selecting certain microbial taxa through chemical signals to meet specific host needs, including nutrient acquisition and pathogen suppression. Thus, plants modulate and regulate the structure and composition of their microbial communities, shaping specialized ecological niches for microbial assemblages that might have positive (mutualistic), neutral (commensalistic), or deleterious (pathogenic) effects on plant fitness [].
However, to decipher the molecular mechanisms that regulate and select for microbial abundance requires further investigation. Earlier studies in the lentil rhizobiome indicated that genotype and generation are important parameters for understanding the interaction of plant–soil systems [,,]. In this study, we demonstrated that the landrace germplasm is also an important factor determining or regulating the above interaction. Furthermore, we illustrated that landrace seed origin is also essential in understanding and deciphering the complex rhizosphere effect, as the organic farming system seems to imbue the seeds with better ability to recruit beneficial bacterial taxa than the counterpart of conventional farming.
Today, most crops produced in agriculture are established by seed sowing. This process conceals that transmission of the microbiome via the seed may take place, as a driver of a root-associated microbiome in a seed-established plant crop. Studies have shown that seeds are major players of microorganisms’ vertical transmission from one plant generation to another and consequently act as a primary source of inoculum for crops []. The primary interest in this seed vertical transmission was gained by plant pathogens due to their detrimental impacts on crop yields and quality. However, seeds transmit a variety of microorganisms that represent transient colonizers of the seed’s soil habitat or are alternatively transmitted to the newly developed plantlet, influencing seedling-associated assemblages. Undoubtedly, crop fertilization and chemical inputs in agriculture crops have ensured profitable crop yields, which inevitably led to overlooking the role of seeds’ transmittal ability on soil rhizosphere-associated microbes. In this study, we showed evidence of landrace-associated attributes (seed origin, organic and conventional farming systems) affecting root microbiomes of lentil. Interestingly, a strong correlation between lentil physiological traits and the rhizosphere microbiome was observed, indicating an interplay among the root rhizobiome and landrace plant traits that remains to be elucidated.
This primary study demonstrated significant differences in rhizosphere microbial diversity that was linked to the Eglouvis landrace, under the same environment, soil type, and agricultural practices. Building upon our findings, further research is needed to dissect how the dynamic diversity enclosed in lentil landraces may affect soil attributes, including soil microbial diversity and nutrient cycles, to sustainably support soil services and function and food safety.
5. Conclusions
Metagenomic profiling of the lentil rhizosphere microbiome under the same field conditions unveiled a 30% common microbiome among samples of the landrace Eglouvis and the modern commercial cultivar Demetra, indicating the presence of a core rhizosphere for lentil plants under the same environmental conditions. Remarkably, distinct differences unveiled for all diversity indices among samples of the Eglouvis landrace and the commercial cultivar Demetra support the hypothesis that a landrace sustains higher microbiota diversity, consistent with its dynamic genetic diversity, compared to a modern improved cultivar.
Within the landrace, it was evident that the origin of the sample affects microbiome diversity, suggesting an important role of the farmer practices and farming system conditions. Higher microbiota diversity was unveiled in the landrace sample derived from organic farming, suggesting that organic cultivation could be a fundamental tool for conservation of microbial genetic resources, expansion of valuable soil microbiota, and crop fortification, especially in soils degraded due to intensive monoculture and heavy fertilization of contemporary agriculture. A Gene Bank sample that was included in the study displayed high numbers of a plant pathogen, suggesting that special care should be taken in Gene Bank standard operational procedures to avoid seed contamination.
As soil biodiversity and agriculture sustainability are closely intertwined with food safety, the study results underscored key drivers of soil × landrace interactions, highlighting their vital role in biodiversity, in both landraces as a dynamic genetic pool and in the soil microbial assemblages, for maintaining water and nutrient availability for plants in a changing climate.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/agronomy13122910/s1, Figure S1: Rarefaction curves of lentil samples; Figure S2: Relative abundances of lentil samples for the 12 most abundant phyla including pathogen Kabatiella microsticta; Figure S3: Relative abundances of lentil samples for the pathogens at genus level; Figure S4: Relative abundances of lentil samples for N2-fixation bacteria at family level; Table S1: Amplicon primers, PCR reagents, and PCR cycling program; Table S2: Metrics of α diversity among the studied genotypes; Eglouvis landrace (Eglouvis 1–4) and the commercial cultivar Demetra; Table S3: Relative abundances of common taxa at phylum level for (a) Demetra, Eglouvis 1, and Eglouvis 2, and (b) Demetra, Eglouvis 3, and Eglouvis 4; Table S4: Top 10% of key species (31/158) after degree centrality analysis; Table S5: Top 10% (16/158) key species after closeness centrality analysis; Table S6: Top 10% of key species (15/158) after betweenness centrality.
Author Contributions
Conceptualization, P.V.M.; Methodology, A.G., G.G., A.N.P. and P.V.M.; Project Administration: P.V.M.; Investigation and formal analysis: A.G., G.G. and P.V.M.; Funding acquisition: P.V.M.; Writing—original draft preparation, A.G. and G.G.; Writing—review and editing, A.N.P. and P.V.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
Data supporting reported results can be found, under project number mgp98852 under the following https://www.mg-rast.org/linkin.cgi?project=mgp98852 (accessed on 31 October 2023).
Acknowledgments
Authors wish to thank Murad Awad for the support with microbiome sequencing and the staff of Soil Science Lab. at HAO-DEMETER for soil chemical analysis.
Conflicts of Interest
The authors declare no conflict of interest.
References
- FAO. The Global Economy of Pulses; FAO: Rome, Italy, 2019; ISBN 978-92-5-109730-4. [Google Scholar]
- Tsanakas, G.F.; Mylona, P.V.; Koura, K.; Gleridou, A.; Polidoros, A.N. Genetic Diversity Analysis of the Greek Lentil (Lens culinaris) Landrace ‘Eglouvis’ Using Morphological and Molecular Markers. Plant Genet. Resour. 2018, 16, 469–477. [Google Scholar] [CrossRef]
- Polidoros, A.N.; Avdikos, I.D.; Gleridou, A.; Kostoula, S.D.; Koura, E.; Sakellariou, M.A.; Stavridou, E.; Gerasopoulos, D.; Lagopodi, A.; Mavromatis, A.; et al. Lentil Gene Pool for Breeding. In Cash Crops; Priyadarshan, P.M., Jain, S.M., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 407–475. ISBN 978-3-030-74925-5. [Google Scholar]
- Sakellariou, M.; Psiloglou, B.E.; Giannakopoulos, C.; Mylona, P.V. Integration of Abandoned Lands in Sustainable Agriculture: The Case of Terraced Landscape Re-Cultivation in Mediterranean Island Conditions. Land 2021, 10, 457. [Google Scholar] [CrossRef]
- Cregger, M.A.; Veach, A.M.; Yang, Z.K.; Crouch, M.J.; Vilgalys, R.; Tuskan, G.A.; Schadt, C.W. The Populus Holobiont: Dissecting the Effects of Plant Niches and Genotype on the Microbiome. Microbiome 2018, 6, 31. [Google Scholar] [CrossRef]
- Lund, M.; Agerbo Rasmussen, J.; Ramos-Madrigal, J.; Sawers, R.; Gilbert, M.T.P.; Barnes, C.J. Rhizosphere Bacterial Communities Differ Among Traditional Maize Landraces. Environ. DNA 2022, 4, 1241–1249. [Google Scholar] [CrossRef]
- Awad, M.; Giannopoulos, G.; Mylona, P.V.; Polidoros, A.N. Comparative Analysis of Grapevine Epiphytic Microbiomes among Different Varieties, Tissues, and Developmental Stages in the Same Terroir. Appl. Sci. 2022, 13, 102. [Google Scholar] [CrossRef]
- Morales Moreira, Z.P.; Helgason, B.L.; Germida, J.J. Assembly and Potential Transmission of the Lens culinaris Seed Microbiome. FEMS Microbiol. Ecol. 2022, 97, fiab166. [Google Scholar] [CrossRef]
- Liu, K.; Bandara, M.; Hamel, C.; Knight, J.D.; Gan, Y. Intensifying Crop Rotations with Pulse Crops Enhances System Productivity and Soil Organic Carbon in Semi-Arid Environments. Field Crops Res. 2020, 248, 107657. [Google Scholar] [CrossRef]
- Pramanik, K.; Das, A.; Banerjee, J.; Das, A.; Chatterjee, S.; Sharma, R.; Kumar, S.; Gupta, S. Metagenomic Insights into Rhizospheric Microbiome Profiling in Lentil Cultivars Unveils Differential Microbial Nitrogen and Phosphorus Metabolism under Rice-Fallow Ecology. Int. J. Mol. Sci. 2020, 21, 8895. [Google Scholar] [CrossRef]
- Gruet, C.; Muller, D.; Moënne-Loccoz, Y. Significance of the Diversification of Wheat Species for the Assembly and Functioning of the Root-Associated Microbiome. Front. Microbiol. 2022, 12, 782135. [Google Scholar] [CrossRef]
- Brescia, F.; Sillo, F.; Franchi, E.; Pietrini, I.; Montesano, V.; Marino, G.; Haworth, M.; Zampieri, E.; Fusini, D.; Schillaci, M.; et al. The ‘Microbiome Counterattack’: Insights on the Soil and Root-associated Microbiome in Diverse Chickpea and Lentil Genotypes after an Erratic Rainfall Event. Environ. Microbiol. Rep. 2023, 15, 459–483. [Google Scholar] [CrossRef]
- Kuppe, C.W.; Schnepf, A.; Von Lieres, E.; Watt, M.; Postma, J.A. Rhizosphere Models: Their Concepts and Application to Plant-Soil Ecosystems. Plant Soil. 2022, 474, 17–55. [Google Scholar] [CrossRef]
- Berendsen, R.L.; Pieterse, C.M.J.; Bakker, P.A.H.M. The Rhizosphere Microbiome and Plant Health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef]
- Bazghaleh, N.; Prashar, P.; Woo, S.; Vandenberg, A. Effects of Lentil Genotype on the Colonization of Beneficial Trichoderma Species and Biocontrol of Aphanomyces Root Rot. Microorganisms 2020, 8, 1290. [Google Scholar] [CrossRef]
- Schaedel, M.; Hidrobo, G.; Grossman, J. From Microns to Meters: Exploring Advances in Legume Microbiome Diversity for Agroecosystem Benefits. Front. Sustain. Food Syst. 2021, 5, 668195. [Google Scholar] [CrossRef]
- Quiza, L.; Tremblay, J.; Pagé, A.P.; Greer, C.W.; Pozniak, C.J.; Li, R.; Haug, B.; Hemmingsen, S.M.; St-Arnaud, M.; Yergeau, E. The Effect of Wheat Genotype on the Microbiome Is More Evident in Roots and Varies through Time. ISME Commun. 2023, 3, 32. [Google Scholar] [CrossRef]
- Cordero, J.; De Freitas, J.R.; Germida, J.J. Bacterial Microbiome Associated with the Rhizosphere and Root Interior of Crops in Saskatchewan, Canada. Can. J. Microbiol. 2020, 66, 71–85. [Google Scholar] [CrossRef]
- Vierheilig, H.; Schweiger, P.; Brundrett, M. An Overview of Methods for the Detection and Observation of Arbuscular Mycorrhizal Fungi in Roots †. Physiol. Plant. 2005, 125, 393–404. [Google Scholar] [CrossRef]
- R Core Team. A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022. [Google Scholar]
- Chen, H.; Boutros, P.C. VennDiagram: A Package for the Generation of Highly-Customizable Venn and Euler Diagrams in R. BMC Bioinform. 2011, 12, 35. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H. Vegan: Community Ecology Package, R Package Version 2.2-0.; R Foundation for Statistical Computing: Vienna, Austria, 2014. [Google Scholar]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Use R! 2nd ed.; Springer International Publishing: Cham, Switzerland, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- Ling, N.; Wang, T.; Kuzyakov, Y. Rhizosphere Bacteriome Structure and Functions. Nat. Commun. 2022, 13, 836. [Google Scholar] [CrossRef]
- Csárdi, G.; Nepusz, T.; Müller, K.; Horvát, S.; Traag, V.; Zanini, F.; Noom, D. Igraph for R: R Interface of the Igraph Library for Graph Theory and Network Analysis. 2023. Available online: https://CRAN.R-project.org/package=igraph (accessed on 31 October 2023).
- Oksanen, A.J.; Blanchet, F.G.; Kindt, R.; Legen-, P.; Minchin, P.R.; Hara, R.B.O.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H. Community Ecology Package. Ecol. Package 2012, 3, 263. [Google Scholar]
- Casañas, F.; Simó, J.; Casals, J.; Prohens, J. Toward an Evolved Concept of Landrace. Front. Plant Sci. 2017, 8, 145. [Google Scholar] [CrossRef]
- Lombardi, M.; Materne, M.; Cogan, N.O.I.; Rodda, M.; Daetwyler, H.D.; Slater, A.T.; Forster, J.W.; Kaur, S. Assessment of Genetic Variation within a Global Collection of Lentil (Lens culinaris Medik.) Cultivars and Landraces Using SNP Markers. BMC Genet. 2014, 15, 150. [Google Scholar] [CrossRef]
- Khazaei, H.; Caron, C.T.; Fedoruk, M.; Diapari, M.; Vandenberg, A.; Coyne, C.J.; McGee, R.; Bett, K.E. Genetic Diversity of Cultivated Lentil (Lens culinaris Medik.) and Its Relation to the World’s Agro-Ecological Zones. Front. Plant Sci. 2016, 7, 1093. [Google Scholar] [CrossRef]
- Idrissi, O.; Piergiovanni, A.R.; Toklu, F.; Houasli, C.; Udupa, S.M.; De Keyser, E.; Van Damme, P.; De Riek, J. Molecular Variance and Population Structure of Lentil (Lens culinaris Medik.) Landraces from Mediterranean Countries as Revealed by Simple Sequence Repeat DNA Markers: Implications for Conservation and Use. Plant Genet. Resour. 2018, 16, 249–259. [Google Scholar] [CrossRef]
- Brisson, V.L.; Schmidt, J.E.; Northen, T.R.; Vogel, J.P.; Gaudin, A.C.M. Impacts of Maize Domestication and Breeding on Rhizosphere Microbial Community Recruitment from a Nutrient Depleted Agricultural Soil. Sci. Rep. 2019, 9, 15611. [Google Scholar] [CrossRef]
- Brown, S.P.; Grillo, M.A.; Podowski, J.C.; Heath, K.D. Soil Origin and Plant Genotype Structure Distinct Microbiome Compartments in the Model Legume Medicago truncatula. Microbiome 2020, 8, 139. [Google Scholar] [CrossRef]
- Yang, T.; Liu, K.; Poppy, L.; Mulenga, A.; Gampe, C. Minimizing Lentil Harvest Loss through Improved Agronomic Practices in Sustainable Agro-Systems. Sustainability 2021, 13, 1896. [Google Scholar] [CrossRef]
- Hartman, K.; Van Der Heijden, M.G.; Roussely-Provent, V.; Walser, J.-C.; Schlaeppi, K. Deciphering Composition and Function of the Root Microbiome of a Legume Plant. Microbiome 2017, 5, 2. [Google Scholar] [CrossRef]
- Priya, S.; Bansal, R.; Kumar, G.; Dikshit, H.K.; Kumari, J.; Pandey, R.; Singh, A.K.; Tripathi, K.; Singh, N.; Kumari, N.K.P.; et al. Root Trait Variation in Lentil (Lens culinaris Medikus) Germplasm under Drought Stress. Plants 2021, 10, 2410. [Google Scholar] [CrossRef]
- Pérez-Jaramillo, J.E.; Carrión, V.J.; Bosse, M.; Ferrão, L.F.V.; De Hollander, M.; Garcia, A.A.F.; Ramírez, C.A.; Mendes, R.; Raaijmakers, J.M. Linking Rhizosphere Microbiome Composition of Wild and Domesticated Phaseolus Vulgaris to Genotypic and Root Phenotypic Traits. ISME J. 2017, 11, 2244–2257. [Google Scholar] [CrossRef]
- Saleem, M.; Law, A.D.; Sahib, M.R.; Pervaiz, Z.H.; Zhang, Q. Impact of Root System Architecture on Rhizosphere and Root Microbiome. Rhizosphere 2018, 6, 47–51. [Google Scholar] [CrossRef]
- Kavamura, V.N.; Robinson, R.J.; Hughes, D.; Clark, I.; Rossmann, M.; Melo, I.S.D.; Hirsch, P.R.; Mendes, R.; Mauchline, T.H. Wheat Dwarfing Influences Selection of the Rhizosphere Microbiome. Sci. Rep. 2020, 10, 1452. [Google Scholar] [CrossRef]
- Lei, S.; Xu, X.; Cheng, Z.; Xiong, J.; Ma, R.; Zhang, L.; Yang, X.; Zhu, Y.; Zhang, B.; Tian, B. Analysis of the Community Composition and Bacterial Diversity of the Rhizosphere Microbiome across Different Plant Taxa. MicrobiologyOpen 2019, 8, e00762. [Google Scholar] [CrossRef]
- Ellouze, W.; Hamel, C.; Vujanovic, V.; Gan, Y.; Bouzid, S.; St-Arnaud, M. Chickpea Genotypes Shape the Soil Microbiome and Affect the Establishment of the Subsequent Durum Wheat Crop in the Semiarid North American Great Plains. Soil. Biol. Biochem. 2013, 63, 129–141. [Google Scholar] [CrossRef]
- Hamel, C.; Gan, Y.; Sokolski, S.; Bainard, L.D. High Frequency Cropping of Pulses Modifies Soil Nitrogen Level and the Rhizosphere Bacterial Microbiome in 4-Year Rotation Systems of the Semiarid Prairie. Appl. Soil. Ecol. 2018, 126, 47–56. [Google Scholar] [CrossRef]
- Wattenburger, C.J.; Gutknecht, J.; Zhang, Q.; Brutnell, T.; Hofmockel, K.; Halverson, L. The Rhizosphere and Cropping System, but Not Arbuscular Mycorrhizae, Affect Ammonia Oxidizing Archaea and Bacteria Abundances in Two Agricultural Soils. Appl. Soil. Ecol. 2020, 151, 103540. [Google Scholar] [CrossRef]
- Morales Moreira, Z.P.; Helgason, B.L.; Germida, J.J. Environment Has a Stronger Effect than Host Plant Genotype in Shaping Spring Brassica Napus Seed Microbiomes. Phytobiomes J. 2021, 5, 220–230. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Schlaeppi, K.; Spaepen, S.; Van Themaat, E.V.L.; Schulze-Lefert, P. Structure and Functions of the Bacterial Microbiota of Plants. Annu. Rev. Plant Biol. 2013, 64, 807–838. [Google Scholar] [CrossRef]
- Serna-Chavez, H.M.; Fierer, N.; Van Bodegom, P.M. Global Drivers and Patterns of Microbial Abundance in Soil. Glob. Ecol. Biogeogr. 2013, 22, 1162–1172. [Google Scholar] [CrossRef]
- Gqozo, M.P.; Bill, M.; Siyoum, N.; Labuschagne, N.; Korsten, L. Fungal Diversity and Community Composition of Wheat Rhizosphere and Non-Rhizosphere Soils from Three Different Agricultural Production Regions of South Africa. Appl. Soil. Ecol. 2020, 151, 103543. [Google Scholar] [CrossRef]
- Lu, J.; Yang, F.; Wang, S.; Ma, H.; Liang, J.; Chen, Y. Co-Existence of Rhizobia and Diverse Non-Rhizobial Bacteria in the Rhizosphere and Nodules of Dalbergia odorifera Seedlings Inoculated with Bradyrhizobium Elkanii, Rhizobium Multihospitium–Like and Burkholderia Pyrrocinia–Like Strains. Front. Microbiol. 2017, 8, 2255. [Google Scholar] [CrossRef]
- Martínez-Hidalgo, P.; Hirsch, A.M. The Nodule Microbiome: N2-Fixing Rhizobia Do Not Live Alone. Phytobiomes J. 2017, 1, 70–82. [Google Scholar] [CrossRef]
- Alexander, A.; Singh, V.K.; Mishra, A.; Jha, B. Plant Growth Promoting Rhizobacterium Stenotrophomonas maltophilia BJ01 Augments Endurance against N2 Starvation by Modulating Physiology and Biochemical Activities of Arachis hypogea. PLoS ONE 2019, 14, e0222405. [Google Scholar] [CrossRef]
- Singh, R.P.; Jha, P.N. The PGPR Stenotrophomonas maltophilia SBP-9 Augments Resistance against Biotic and Abiotic Stress in Wheat Plants. Front. Microbiol. 2017, 8, 1945. [Google Scholar] [CrossRef]
- Sun, S.-L.; Yang, W.-L.; Fang, W.-W.; Zhao, Y.-X.; Guo, L.; Dai, Y.-J. The Plant Growth-Promoting Rhizobacterium Variovorax boronicumulans CGMCC 4969 Regulates the Level of Indole-3-Acetic Acid Synthesized from Indole-3-Acetonitrile. Appl. Environ. Microbiol. 2018, 84, e00298-18. [Google Scholar] [CrossRef]
- Sumbul, A.; Ansari, R.A.; Rizvi, R.; Mahmood, I. Azotobacter: A Potential Bio-Fertilizer for Soil and Plant Health Management. Saudi J. Biol. Sci. 2020, 27, 3634–3640. [Google Scholar] [CrossRef]
- Mylona, P.V.; Premakumar, R.; Pau, R.N.; Bishop, P.E. Characteristics of Orf1 and Orf2 in the anfHDGK Genomic Region Encoding Nitrogenase 3 of Azotobacter vinelandii. J. Bacteriol. 1996, 178, 204–208. [Google Scholar] [CrossRef][Green Version]
- Taffner, J.; Erlacher, A.; Bragina, A.; Berg, C.; Moissl-Eichinger, C.; Berg, G. What Is the Role of Archaea in Plants? New Insights from the Vegetation of Alpine Bogs. mSphere 2018, 3, e00122-18. [Google Scholar] [CrossRef]
- Akinola, S.A.; Babalola, O.O. The Fungal and Archaeal Community within Plant Rhizosphere: A Review on Their Contribution to Crop Safety. J. Plant Nutr. 2021, 44, 600–618. [Google Scholar] [CrossRef]
- Mittelstrass, J.; Sperone, F.G.; Horton, M.W. Using Transects to Disentangle the Environmental Drivers of Plant-microbiome Assembly. Plant Cell Environ. 2021, 44, 3745–3755. [Google Scholar] [CrossRef]
- Peiffer, J.A.; Spor, A.; Koren, O.; Jin, Z.; Tringe, S.G.; Dangl, J.L.; Buckler, E.S.; Ley, R.E. Diversity and Heritability of the Maize Rhizosphere Microbiome under Field Conditions. Proc. Natl. Acad. Sci. USA 2013, 110, 6548–6553. [Google Scholar] [CrossRef]
- Majdura, J.; Jankiewicz, U.; Gałązka, A.; Orzechowski, S. The Role of Quorum Sensing Molecules in Bacterial–Plant Interactions. Metabolites 2023, 13, 114. [Google Scholar] [CrossRef]
- Zafeiriou, I.; Ntoanidou, S.; Baira, E.; Kasiotis, K.M.; Barmpouni, T.; Machera, K.; Mylona, P.V. Ingenious Characterization and Assessment of Lentil Germplasm Collection to Aphid Acyrthosiphon pisum Stress Unveils Distinct Responses. Front. Plant Sci. 2022, 13, 1011026. [Google Scholar] [CrossRef]
- Berlanga-Clavero, M.V.; Molina-Santiago, C.; De Vicente, A.; Romero, D. More than Words: The Chemistry behind the Interactions in the Plant Holobiont. Environ. Microbiol. 2020, 22, 4532–4544. [Google Scholar] [CrossRef]
- Panth, M.; Hassler, S.C.; Baysal-Gurel, F. Methods for Management of Soilborne Diseases in Crop Production. Agriculture 2020, 10, 16. [Google Scholar] [CrossRef]
- Liu, F.; Hewezi, T.; Lebeis, S.L.; Pantalone, V.; Grewal, P.S.; Staton, M.E. Soil Indigenous Microbiome and Plant Genotypes Cooperatively Modify Soybean Rhizosphere Microbiome Assembly. BMC Microbiol. 2019, 19, 201. [Google Scholar] [CrossRef]
- Barret, M.; Guimbaud, J.; Darrasse, A.; Jacques, M. Plant Microbiota Affects Seed Transmission of Phytopathogenic Microorganisms. Mol. Plant Pathol. 2016, 17, 791–795. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).