Abstract
Young vine decline (YVD) is a grapevine trunk disease (GTD) which results in stunted and delayed growth, reduced yield, root necrosis and eventually death of young vines. Given losses associated with root trunk disease, and increasing limits on chemical fungicides, there is a need for sustainable approaches to combat disease; (1) Cover cropping is a commonly used practice in agricultural systems and has potential to reduce disease in vineyards but there is a risk that cover crop species may act as a host for grapevine pathogens, increasing the risk of infection; (2) We tested 25 plant species commonly used in cover crops to assess their potential to act as a host for a Ilyonectria liriodendri, which is a causal agent of young vine decline. We inoculated greenhouse pots with a pathogeninc strain of Ilyonectria and assayed the roots for the presence of the pathogen; (3) Of the 25 cover crops tested, many of the species showed increased root abundance of Ilyonectria, compared to background levels. In particular phacelia (Phacelia tanacetifolia) and buckwheat (Fagopyrum esculentum) showed very high levels of root colonization. (4) This is the first study to our knowledge that highlights the potential of cover crops to soil borne fungal pathogens.
1. Introduction
Pathogen spillover is a mechanism by which pathogen abundance is increased in a community, leading to disease outbreaks [1]. This occurs in natural and managed ecosystems when pathogens can live asymptomatically in some plants, allowing the abundance of the pathogen to increase to the tipping point of infection for susceptible plant species [2]. In agricultural systems that use cover crops as part of their management, the species included in the cover crop may act as reservoir plants–plants capable of associating with and proliferating a pathogen while remaining largely asymptomatic. However, the capacity for a cover crop species to act for a reservoir species is largely overlooked when growers are selecting cover crop mixes.
Although YVD is a disease complex, the main culprits are fungal pathogens including fungi belonging to the genera Ilyonectria, Dactylonectria, and Cylindrodendrum among others [3,4]. These organisms may be present in soils [5] or enter vineyards via infected nursery material [6]. Fungal spores are easily distributed via contaminated tools, irrigation equipment, and by air from fruiting bodies on decomposing/infected tissue [7]. Causal agents of YVD occur in all major growing areas of the world [8] and although it may start with a few infected vines, the rate of infection will increase as the vineyard ages [9]. Young vine decline continues to contribute to economic losses around the world [10] and currently, options to prevent infection and mitigate decline in vineyards are limited.
Options like fungicide treatment are limited in many countries and are not always effective [11]. Furthermore, most fungicides are designed to combat foliar diseases and not infections in the roots [12]. Since fungicides accumulate in the soil and are considered to be pollutants, legislation in major regions aims to minimize their use as much as possible [13]. This paired with the inclination of consumers to purchase sustainable wine has increased demand for organic wine production, pressuring growers to use low impact strategies to manage disease [14]. Such strategies include biological control in which organisms that inhibit pathogen growth are introduced into the crop system. A prime example is Trichoderma, a predatorial fungus capable of consuming GTD pathogens and has been studied extensively in the past decade [15,16,17]. Another approach is establishing groundcover systems or cover crops in the vineyard to suppress pathogens.
Traditionally, cover crops are plants that are grown during the main production season or during off seasons in order to maintain components of soil health which include erosion control, runoff, nutrient management, organic input and maintenance of soil macro and microorganisms [18]. Cover crops can help reduce pathogen pressure through a variety of mechanisms. Brassicaceous cover crops such as white mustard produce antifungal metabolites which can inhibit proliferation of fungi when introduced into the soil [19]. Cover crops also facilitate microbial diversity [20] which could lead to an increase in beneficial/antagonistic microbes such as plant growth promoting rhizobacteria (PGPR) [21] and Arbuscular mycorrhizal fungi [22] increased activity from antagonistic and beneficial microbes could help combat disease in vineyards.
Although cover crops confer many benefits to grapevines [23], they may be associated with increases in disease. Common cover crops like hairy vetch (Vicia villosa) may facilitate Ilyonectria pathogens if grown in vineyards [24]. Vukicevich et al. [25] found that grapevines grown in soil conditioned by native grasses and forbs were associated with increases in necrotic tissue compared to other groundcover treatments [25]. Likewise, Langenhoven et al. [26] isolated Dactylonectria spp. (black-Foot) and Pythium spp.(crown rot) pathogens from Triticale and ryegrass cover crops [26]. Furthermore, weeds from Spanish vineyards and nurseries tested positive for black-Foot as well as Petri disease pathogens [27]. These studies raise the concern that cover crops act as hosts or maintain an inoculum source in vineyards and nurseries.
Causal agents of YVD are often referred to as generalist, opportunistic [28] and/or weak pathogens [8]. Due to these strategies, YVD pathogens may benefit from root turnover and exudation [29] or even persist inside the living roots of cover crops. It is possible that YVD pathogens could have evolved alongside various plants to enter vascular tissue and survive as endophytes until tissue death, where they would be first in line to decompose the material [30]. This mechanism has not been investigated in a viticultural setting and the priority effects of YVD pathogens on the fungal community is not well studied [31].
If certain cover crops associate with or facilitate grapevine pathogens, they could be detrimental in the vineyard and this would greatly impact how we use cover crops to maintain soil health. In this study, we surveyed native plants as well as commercial cover crop species to determine if they associate with Ilyonectria liriodendri, a widely distributed grapevine trunk pathogen.
2. Materials and Methods
2.1. Plant Material and Soil
This experiment was designed to test the capacity of commonly used cover crops to host a common trunk pathogen. To achieve this goal, we grew only cover crop species in soil that was inoculated with the pathogen. We quantified the amount of inoculum added to each pot so that we could differentiate between positives in the soil that were due to inoculum alone (therefore no hosting capacity of the crop) versus inoculum that had been established in a host. Cover crops (Table 1) were grown in a greenhouse at the Summerland Research and Development Centre (SuRDC), British Columbia, Canada (49°33′57.8″ N 119°38′10.0″ W) from 25 October 2019 to 3 February 2020. The experiment was set up in a randomized complete block design with seven replicates, totaling 175 pots. This room was cooled by a fog system which kept temperatures below 28 °C during the summer months.

Table 1.
List of vineyard cover crops that were inoculated with Ilyonectria liriodendri.
Soil was collected at SuRDC in September 2019 from field 7, a viticulture research block. This soil is described as a Skaha loamy sand ((Brown Chernozemic soil) (Wittneben 1986; Soil Classification Working Group 1998)), with the following physio-chemical characteristics (0–20 cm depth): conductivity: 33 uS/cm; pH: 6.79; sulphur P-Extr 0.89 ppm; aluminium: 318 ppm; boron: 0.2 ppm; calcium: 768 ppm; copper: 1.68 ppm; iron: 105 ppm; potassium: 119 ppm; magnesium: 89.4 ppm; manganese: 120 ppm; sodium: 3.4 ppm; phosphorus: 30.7 ppm; sulfur: 2.7 ppm; zinc: 1.1 ppm; clay: 5.74%; silt: 10.19% and sand: 84.07%. We chose this soil because it came from a viticulture system, making it the most suitable soil for this study. It had been selected for previous studies largely due being pathogen free, allowing us to use it in manipulative studies with our isolate of Ilyonectria liriodendra. Three-litre nursery pots were filled, leaving a four-centimetre gap from the top to retain water, and placed in the SuRDC greenhouse.
2.2. Pathogen Inoculation and Plant Growing Conditions
Three isolates of Ilyonectria liriodendri (SuRDC 340, 60, 393) were introduced to each nursery pot via a 106 conidia spore suspension, close to the roots. Each isolate was incubated at 22 °C for one week on 5% potato dextrose agar (PDA) solution. To ensure plates were ready, sporulation was observed with a compound light microscope. Agar plates were flooded with a 1% tween solution which helped free the spores during agitation with a metal utensil. The resulting solution was filtered in a cheese cloth to remove large chunks of agar and hyphae. A hemocytometer was used to make the stock solution and the final concentration was made using the following formula:
where C1V1 = Concentration/amount (start) and Volume (start) C2V2 = Concentration/amount (final) and Volume (final). Then, each pot received 50 mL of inoculum 10 day after seeding.
c1v1 = c2v2
Each nursery pot was standardised with approximately 10 plants per pot for the duration of the experiment. During the first week, pots were watered by hand with an equal amount of water. Fertilizer supplement was applied once a week during the growing period. Each pot received 50 mL of 20–20–20 fertilizer (Miracle-Gro, Marisville, OH, USA) at the recommended concentration. During harvest, as much soil as possible was washed away from roots with reverse osmosis water. Roots and shoots were put into plastic bags and stored at 4 °C for 24 h until they were dried and weighed.
2.3. Accessing Colonization by I. liriodendra
To determine the extent of colonisation by I. liriodendri, we extracted DNA from each cover crop root system and soil. Root samples were collected from each cover crop after growing in soil inoculated with I. liriodendri for 3 months. Soil samples were collected from the pot after roots were removed. To quantify the abundance of I. liriodendri in each root sample, we used a digital droplet (dd) PCR assay.
At the end of the growing period, approximately five grams of fresh root samples were collected, sub sampled, pooled, and stored at −20 °C until DNA extraction.
Roots were submerged in 10% bleach for 5 min then rinsed with reverse osmosis water three times for one minute. After surface sterilization, roots were crushed with a mortar and pestle in liquid nitrogen. 0.25 g were taken from each sample and loaded into a lysing tube. Roots were lysed at 6.5 m/s and centrifuged for 10 min to facilitate separation of root tissues and nucleic acids.
Root DNA was isolated with the FastDNA Spin Kit for Soil (MPBio ©2018, Irvine, CA, USA) by following manufacturer’s instructions. DNA per sample was eluted in 100 µL and, DNA concentration as well as quality was assessed with a nanodrop device 1000c (Thermo Fisher Scientific, Wilmington, NC, USA). DNA was stored at −80 °C until digital PCR amplification.
We used a specific primer/probe assay to amplify the Ilyonectria isolates used in the inoculum. This assay targets the beta-tubulin region which is highly conserved region and single copy gene in fungi. The forward primer, 5′-CGAGGGACATACTTGTTTCCAGAG-3′ (Tm 61, GC 60%), reverse 5′-TCAACGAGGTACGCGAAATC-3′ -R (Tm 62, GC 50%), and probe TGTCAAACTCACACCACGTAGGCC (FAM) were designed and tested at the University of British Columbia laboratories [32].
For each 20 µL reaction, 10 µL Supermix (ddPCR Supermix for probes, Bio-Rad Inc., Hercules, CA, USA), 7 µL molecular grade water, 1 µL primers and probe (20× concentration), and 2 µL sample DNA, was used. Droplets were generated manually with the Bio-Rad QX100 Droplet Generator by adding 70 µL of Bio-Rad Droplet Generator Oil for Probes. PCR reactions were completed in the C1000 Thermal Cycler (Bio-Rad, Hercules, CA, USA) as per following conditions: initial heating at 95 °C for 10 min; denaturation at 94 °C for one minute; annealing at 59 °C for two minutes. Denaturation and annealing steps were repeated for 44 cycles, followed by enzyme inactivation at 98 °C for 10 min.
We measured droplet fluorescence with the QX 100 Droplet Reader (Bio-Rad, Quantalife software (version 1.7.4) and used FAM-HEX as the fluorescent dye. The threshold was set automatically via the Quantasoft algorithm (Bio-Rad-USA). Data (copy number) for each sample was back calculated to represent the number of copies per gram of soil and root using a formula described in Kokkoris et al. [33].
2.4. Data Analyses
All statistical analyses were performed in R (version 3.6.2) via Rstudio (version 1.2.5033) (R Core Team 2019). Digital PCR data (copies per gram of soil and root) were fitted to a generalized linear mixed-effects model with block as a random factor using the lme4 package (1.1-21). Soil and root data were analyzed separately using Type II analysis of variance in the car package (3.0-6). Tukey’s honest significance test in the emmeans package (1.4.3.01) was used for post hoc comparisons. All plots were created in ggplot2 (3.2.1).
3. Results
3.1. Abundance of Ilyonectria in Roots
After a brief growth period in the greenhouse, Ilyonectria liriodendri was isolated from the roots of various cover crops, in which copy number was significantly different among treatment groups (p = 2.2 × 10−16). Phacelia roots had the largest presence of Ilyonectria DNA, averaging 10,569 copies per gram of root followed by Buckwheat and common yarrow with 4817 and 1621 copies per gram, respectively, (Figure 1). The only cover crop that did not yield any pathogenic DNA was Crescendo ladino clover. This cover crop treatment was not significantly different from the others (see Appendix A for copy number summary statistics).
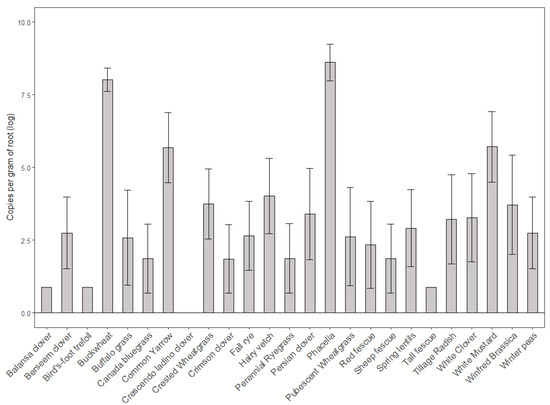
Figure 1.
Log concentration of Ilyonectria DNA (copy number per gram of root) isolated from surface sterilized cover crop roots grown for three months. Log-transformed data are displayed. Dotted line represents the amount of inoculum added to each pot, for comparison.
3.2. Abundance of Ilyonectria in Soil
Similar patterns were observed in DNA isolated from soil samples. Overall analysis of variance resulted in a significant difference in Ilyonectria copy number between cover crop treatments (p = 2.2 × 10−16). As expected, soil conditioned by phacelia contained the highest amount of pathogenic DNA with 3384 copies per gram. Surprisingly, Persian clover soil yielded the second highest concentration at 1564 copies per gram of soil followed by Buckwheat with 1466 copies per gram (Figure 2). Ilyonectria DNA was recovered from all soil samples (Appendix A).
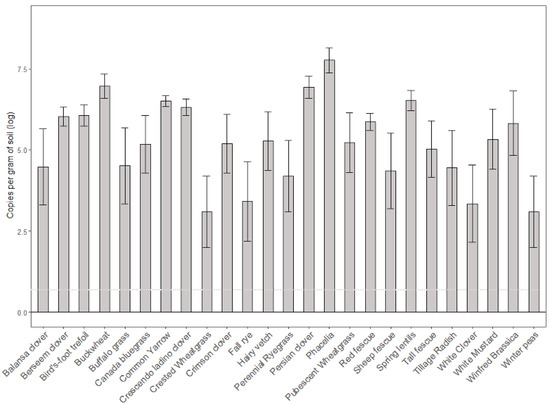
Figure 2.
Log concentration of Ilyonectria DNA (copy number per gram of soil) isolated from soil conditioned by each cover crop after a three-month growth period. Data is shown in the log transformation. Dotted line represents the amount of inoculum added to each pot, for comparison.
4. Discussion
This study shows that cover crops used in perennial agriculture can act as alternate hosts for a common grapevine pathogen. In our study, some cover crop speciessignificantly increased the abundance of Ilyonectria spp. in both roots and soil.
In itself this is not surprising; Ilyonectria liriodendri has a cosmopolitan distribution, having been isolated from soils in the Americas, Europe, and Oceania [34,35]. Moreover, this pathogen is present in multiple perennial cropping systems including apple [5], cherry [36], tea [37], and avocado [38] which highlights its generalist nature as a pathogen. Unlike previous studies, our work shows that the pathogen can infect non-crop species, across a wide taxonomic distribution. Given that the plants in our study are commonly used as cover crops in areas where Ilyonectria spp. are a significant pathogen, growers should consider the ability of cover crops to act as a reservoir for pathogens when selecting candidate species.
Two plants in particular have the potential to greatly amplify the abundance nof Ilyonectria spp. in soil. Phacelia is a genus native to the Americas belonging to Boraginaceae, which are classified as asterids [39]. Phacelia tanacetifolia is grown extensively arable crop rotations to condition soil structure, especially in sandy loam soils [40,41,42]. This is the first study to our knowledge that shows the ability of P. tanacetifolia to associate with Ilyonectria spp. Previous studies show that phacelia has the capacity to host other fungal pathogens (Sclerotinia minor [43], Rhizoctonia solani [44]). Ilyonectria robusta has been isolated from the roots of Taraxacum officinale which is also a perennial asterid [45].
Buckwheat Fagopyrum esculentum) also augmented the concentration of Ilyonectria spp. far greater than background levels. It is commonly used in cover crops due to its rapid establishment, weed suppression, pollinator species, and ability to extract phosphorus [18]. Unlike phacelia, buckwheat is a native to Southeast Asia [46] which makes plant provenance an unlikely explanation for why these two cover crops are the most likely to act as reservoir hosts for Ilyonectria spp. However, buckwheat does meet the criteria outlined in Cronin et al. [47] in which ideal reservoir hosts grow rapidly, have a short lifespan, and have high phosphorus concentrations in their tissues. Previous studies show that buckwheat is prone to damping off and root rot by fusarium spp. [48] and Rhizoctonia spp. [49]. More recently, Zini et al. isolated Fusarium incarnatum-equiset from germinated buckwheat seeds [50]. Considering these findings, it is not surprising that Ilyonectria spp were isolated from buckwheat roots and that this species could act as a reservoir host for grapevine pathogens.
Many other crop species in our study increased pathogen incidence in roots, but to a lesser degree. This was particularly true for many brassica species (Tillage radish, White Clover, White Mustard, Winfred Brassica and Persian Clover). The levels of Illyonectria spp in the roots of these crops are surprising since brassicas are well known for their fungicidal properties [51] and are used by growers specifically to reduce fungal pathogens in the soil [52]. Of these, only Persian Clover had elevated soil concentrations of Ilyonectria. Thus, cover crops may host pathogens asymptomatically in the growing season, but unless the plants are mulched into the soil, they may have limited biofumigant properties.
Most of our cover crops showed little to no ability to associate with Ilyonectria spp. We could not detect any Ilyonectria spp in Crescendo Ladino Clover roots, while Balansa clover, Birdsfoot Trefoil and Tall Fescue had levels that were not different from zero. In areas where Illyonectria spp. is a problem, these taxa may be good candidates to prevent outbreaks.
It is important to note that the behaviour of IIlyonectria spp. in our study may have been affected by resident soil microbes, as microbial communities can influence eachtother through a variety of different mechanisms including competition, facilitation. Thus, our results reflect a specific set of conditions and microbial community. To fully understand the risk of these cover crop species to act as pathogen reservoirs, future analyses must be conducted in under different soil and growing conditions. This study provides an excellent basis on which to develop future work.
5. Conclusions
While the benefits of cover crops are many, including improved soil nutrients, water relations and soil stability, they may not be universally beneficial. Here, we showed that commonly used cover crops may have the ability to increase the abundance of grapevine pathogens by acting as an alternate host. In areas where soil borne disease is a problem, the choice of cover crop may make the difference between pathogen suppression and outbreak. In this survey phacelia and buckwheat were found to act as reservoir hosts for Ilyonectria liriodendri. For a grower dealing with YVD, using phacelia and buckwheat in a cropping mixture may increase the abundance of the pathogen, leading to disease outbreak under the right conditions.
Author Contributions
Conceptualization D.R..; methodology, D.R., M.S. and M.M.H.; validation, M.M.H. and M.S.; formal analysis, D.R. investigation, D.R.; data curation, D.R.; writing—original draft preparation, M.M.H.; writing—review and editing, M.M.H.; supervision, M.S.; project administration, M.M.H. and M.S.; funding acquisition, M.M.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by AAFC Going Forward Wine Cluster (MMH) and partially by Organic Science Cluster 3 (M.S.), which was supported by the AgriScience Program under Agriculture and Agri-Food Canada’s Canadian Agricultural Partnership investment.
Acknowledgments
Summerland Research and Development greenhouse facility we used in this study. We would like to acknowledge the technical support by Bill Rabie, Lana Fukumoto and greenhouse staff.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Appendix A
| sample.ID | block | cover.crop | root.positives | soil.positives | root.per.gram | soil.per.gram |
| 1 | 1 | Balansa clover | 0 | 0 | 0 | 0 |
| 26 | 2 | Balansa clover | 0 | 2 | 0 | 370.37037 |
| 51 | 3 | Balansa clover | 0 | 1 | 0 | 172.413793 |
| 76 | 4 | Balansa clover | 0 | 6 | 0 | 1034.48276 |
| 101 | 5 | Balansa clover | 1 | 0 | 200 | 0 |
| 126 | 6 | Balansa clover | 0 | 6 | 0 | 1034.48276 |
| 151 | 7 | Balansa clover | 3 | 600 | ||
| 2 | 1 | Berseem clover | 0 | 4 | 0 | 714.285714 |
| 27 | 2 | Berseem clover | 0 | 1 | 0 | 172.413793 |
| 52 | 3 | Berseem clover | 2 | 1 | 400 | 178.571429 |
| 77 | 4 | Berseem clover | 1 | 6 | 192 | 1153.84615 |
| 102 | 5 | Berseem clover | 0 | 3 | 0 | 517.241379 |
| 127 | 6 | Berseem clover | 1 | 1 | 192 | 217.391304 |
| 152 | 7 | Berseem clover | 4 | 740.740741 | ||
| 3 | 1 | Bird's-foot trefoil | 1 | 1 | 200 | 178.571429 |
| 28 | 2 | Bird's-foot trefoil | 0 | 1 | 0 | 192.307692 |
| 53 | 3 | Bird's-foot trefoil | 0 | 11 | 0 | 1964.28571 |
| 78 | 4 | Bird's-foot trefoil | 0 | 3 | 0 | 576.923077 |
| 103 | 5 | Bird's-foot trefoil | 0 | 3 | 0 | 576.923077 |
| 128 | 6 | Bird's-foot trefoil | 0 | 1 | 0 | 192.307692 |
| 153 | 7 | Bird's-foot trefoil | 3 | 625 | ||
| 4 | 1 | Buckwheat | 29 | 8 | 6042 | 1428.57143 |
| 29 | 2 | Buckwheat | 82 | 14 | 15769 | 2800 |
| 54 | 3 | Buckwheat | 10 | 9 | 1923 | 1875 |
| 79 | 4 | Buckwheat | 13 | 4 | 2500 | 689.655172 |
| 104 | 5 | Buckwheat | 9 | 1 | 1667 | 178.571429 |
| 129 | 6 | Buckwheat | 5 | 3 | 1000 | 600 |
| 154 | 7 | Buckwheat | 14 | 2692.30769 | ||
| 5 | 1 | Buffalo grass | 1 | 6 | 1667 | 1111.11111 |
| 30 | 2 | Buffalo grass | 0 | 2 | 0 | 416.666667 |
| 55 | 3 | Buffalo grass | 2 | 0 | 3333 | 0 |
| 80 | 4 | Buffalo grass | 0 | 3 | 0 | 652.173913 |
| 105 | 5 | Buffalo grass | 0 | 2 | 0 | 434.782609 |
| 130 | 6 | Buffalo grass | 0 | 0 | 0 | 0 |
| 155 | 7 | Buffalo grass | 2 | 400 | ||
| 6 | 1 | Canada bluegrass | 0 | 2 | 0 | 416.666667 |
| 31 | 2 | Canada bluegrass | 0 | 1 | 0 | 227.272727 |
| 56 | 3 | Canada bluegrass | 0 | 0 | 0 | 0 |
| 81 | 4 | Canada bluegrass | 0 | 3 | 0 | 625 |
| 106 | 5 | Canada bluegrass | 2 | 4 | 400 | 714.285714 |
| 131 | 6 | Canada bluegrass | 1 | 1 | 185 | 185.185185 |
| 156 | 7 | Canada bluegrass | 4 | 714.285714 | ||
| 7 | 1 | Common yarrow | 0 | 4 | 0 | 714.285714 |
| 32 | 2 | Common yarrow | 2 | 6 | 400 | 1153.84615 |
| 57 | 3 | Common yarrow | 23 | 2 | 4600 | 370.37037 |
| 82 | 4 | Common yarrow | 5 | 2 | 1000 | 416.666667 |
| 107 | 5 | Common yarrow | 5 | 3 | 1000 | 625 |
| 132 | 6 | Common yarrow | 2 | 5 | 345 | 1086.95652 |
| 157 | 7 | Common yarrow | 4 | 689.655172 | ||
| 8 | 1 | Crescendo ladino | 0 | 1 | 0 | 178.571429 |
| 33 | 2 | Crescendo ladino | 0 | 2 | 0 | 384.615385 |
| 58 | 3 | Crescendo ladino | 0 | 3 | 0 | 576.923077 |
| 83 | 4 | Crescendo ladino | 0 | 3 | 0 | 500 |
| 108 | 5 | Crescendo ladino | 0 | 8 | 0 | 1379.31035 |
| 133 | 6 | Crescendo ladino | 0 | 5 | 0 | 1041.66667 |
| 158 | 7 | Crescendo ladino | 3 | 555.555556 | ||
| 9 | 1 | Crested wheatgrass | 1 | 1 | 208 | 200 |
| 34 | 2 | Crested wheatgrass | 1 | 1 | 714 | 172.413793 |
| 59 | 3 | Crested wheatgrass | 0 | 0 | 0 | 0 |
| 84 | 4 | Crested wheatgrass | 0 | 1 | 0 | 200 |
| 109 | 5 | Crested wheatgrass | 1 | 2 | 200 | 370.37037 |
| 134 | 6 | Crested wheatgrass | 1 | 0 | 200 | 0 |
| 159 | 7 | Crested wheatgrass | 0 | 0 | ||
| 10 | 1 | Crimson clover | 0 | 0 | 0 | 0 |
| 35 | 2 | Crimson clover | 0 | 3 | 0 | 600 |
| 60 | 3 | Crimson clover | 0 | 1 | 0 | 200 |
| 85 | 4 | Crimson clover | 2 | 7 | 400 | 1400 |
| 110 | 5 | Crimson clover | 0 | 3 | 0 | 517.241379 |
| 135 | 6 | Crimson clover | 1 | 2 | 172 | 384.615385 |
| 160 | 7 | Crimson clover | 1 | 185.185185 | ||
| 11 | 1 | Fall rye | 1 | 0 | 192 | 0 |
| 36 | 2 | Fall rye | 0 | 6 | 0 | 1071.42857 |
| 61 | 3 | Fall rye | 1 | 1 | 200 | 166.666667 |
| 86 | 4 | Fall rye | 0 | 0 | 0 | 0 |
| 111 | 5 | Fall rye | 1 | 2 | 208 | 370.37037 |
| 136 | 6 | Fall rye | 0 | 2 | 0 | 370.37037 |
| 161 | 7 | Fall rye | 0 | 0 | ||
| 12 | 1 | Hairy vetch | 2 | 4 | 357 | 689.655172 |
| 37 | 2 | Hairy vetch | 0 | 2 | 0 | 312.5 |
| 62 | 3 | Hairy vetch | 0 | 3 | 0 | 576.923077 |
| 87 | 4 | Hairy vetch | 2 | 1 | 400 | 217.391304 |
| 112 | 5 | Hairy vetch | 6 | 0 | 1034 | 0 |
| 137 | 6 | Hairy vetch | 1 | 2 | 200 | 416.666667 |
| 162 | 7 | Hairy vetch | 5 | 961.538462 | ||
| 13 | 1 | Perennial ryegrass | 1 | 1 | 185 | 192.307692 |
| 38 | 2 | Perennial ryegrass | 0 | 0 | 0 | 0 |
| 63 | 3 | Perennial ryegrass | 2 | 4 | 417 | 740.740741 |
| 88 | 4 | Perennial ryegrass | 0 | 0 | 0 | 0 |
| 113 | 5 | Perennial ryegrass | 0 | 2 | 0 | 434.782609 |
| 138 | 6 | Perennial ryegrass | 0 | 2 | 0 | 416.666667 |
| 163 | 7 | Perennial ryegrass | 1 | 217.391304 | ||
| 14 | 1 | Persian clover | 0 | 6 | 0 | 1200 |
| 39 | 2 | Persian clover | 0 | 2 | 0 | 384.615385 |
| 64 | 3 | Persian clover | 5 | 5 | 962 | 862.068966 |
| 89 | 4 | Persian clover | 1 | 10 | 192 | 1923.07692 |
| 114 | 5 | Persian clover | 0 | 33 | 0 | 5500 |
| 139 | 6 | Persian clover | 20 | 3 | 3846 | 576.923077 |
| 164 | 7 | Persian clover | 3 | 500 | ||
| 15 | 1 | Phacelia | 18 | 2 | 3750 | 370.37037 |
| 40 | 2 | Phacelia | 99 | 8 | 19038 | 1428.57143 |
| 65 | 3 | Phacelia | 2 | 14 | 385 | 2500 |
| 90 | 4 | Phacelia | 69 | 10 | 13269 | 2083.33333 |
| 115 | 5 | Phacelia | 15 | 26 | 3125 | 5200 |
| 140 | 6 | Phacelia | 124 | 18 | 23846 | 3103.44828 |
| 165 | 7 | Phacelia | 45 | 9000 | ||
| 16 | 1 | Pubescent wheatgrass | 0 | 0 | 0 | 0 |
| 41 | 2 | Pubescent wheatgrass | 0 | 1 | 0 | 185.185185 |
| 66 | 3 | Pubescent wheatgrass | 5 | 8800 | 833.333333 | |
| 91 | 4 | Pubescent wheatgrass | 0 | 1 | 0 | 200 |
| 116 | 5 | Pubescent wheatgrass | 0 | 2 | 0 | 333.333333 |
| 141 | 6 | Pubescent wheatgrass | 4 | 3 | 769 | 517.241379 |
| 166 | 7 | Pubescent wheatgrass | 9 | 1500 | ||
| 17 | 1 | Red fescue | 0 | 1 | 0 | 208.333333 |
| 42 | 2 | Red fescue | 3 | 3 | 600 | 555.555556 |
| 67 | 3 | Red fescue | 0 | 4 | 0 | 869.565217 |
| 92 | 4 | Red fescue | 0 | 1 | 0 | 156.25 |
| 117 | 5 | Red fescue | 0 | 2 | 0 | 370.37037 |
| 142 | 6 | Red fescue | 11 | 1 | 2115 | 178.571429 |
| 167 | 7 | Red fescue | 4 | 666.666667 | ||
| 18 | 1 | Sheep fescue | 0 | 3 | 0 | 600 |
| 43 | 2 | Sheep fescue | 1 | 1 | 185 | 185.185185 |
| 68 | 3 | Sheep fescue | 0 | 0 | 0 | 0 |
| 93 | 4 | Sheep fescue | 0 | 0 | 0 | 0 |
| 118 | 5 | Sheep fescue | 0 | 2 | 0 | 400 |
| 143 | 6 | Sheep fescue | 2 | 1 | 400 | 185.185185 |
| 168 | 7 | Sheep fescue | 11 | 2115.38462 | ||
| 19 | 1 | Spring lentils | 0 | 5 | 0 | 925.925926 |
| 44 | 2 | Spring lentils | 1 | 2 | 192 | 416.666667 |
| 69 | 3 | Spring lentils | 0 | 2 | 0 | 416.666667 |
| 94 | 4 | Spring lentils | 1 | 12 | 200 | 2222.22222 |
| 119 | 5 | Spring lentils | 0 | 3 | 0 | 652.173913 |
| 144 | 6 | Spring lentils | 5 | 7 | 1000 | 1521.73913 |
| 169 | 7 | Spring lentils | 1 | 192.307692 | ||
| 20 | 1 | Tall fescue | 0 | 4 | 0 | 869.565217 |
| 45 | 2 | Tall fescue | 0 | 1 | 0 | 200 |
| 70 | 3 | Tall fescue | 0 | 0 | 0 | 0 |
| 95 | 4 | Tall fescue | 0 | 1 | 0 | 208.333333 |
| 120 | 5 | Tall fescue | 0 | 2 | 0 | 416.666667 |
| 145 | 6 | Tall fescue | 1 | 3 | 192 | 750 |
| 170 | 7 | Tall fescue | 1 | 172.413793 | ||
| 21 | 1 | Tillage radish | 0 | 0 | 0 | 0 |
| 46 | 2 | Tillage radish | 1 | 0 | 192 | 0 |
| 71 | 3 | Tillage radish | 1 | 6 | 200 | 1034.48276 |
| 96 | 4 | Tillage radish | 0 | 3 | 0 | 500 |
| 121 | 5 | Tillage radish | 0 | 2 | 0 | 333.333333 |
| 146 | 6 | Tillage radish | 31 | 3 | 6200 | 535.714286 |
| 171 | 7 | Tillage radish | 2 | 357.142857 | ||
| 22 | 1 | White clover | 0 | 4 | 0 | 833.333333 |
| 47 | 2 | White clover | 3 | 0 | 600 | 0 |
| 72 | 3 | White clover | 0 | 2 | 0 | 370.37037 |
| 97 | 4 | White clover | 0 | 1 | 0 | 208.333333 |
| 122 | 5 | White clover | 1 | 1 | 179 | 227.272727 |
| 147 | 6 | White clover | 17 | 0 | 3269 | 0 |
| 172 | 7 | White clover | 0 | 0 | ||
| 23 | 1 | White mustard | 0 | 2 | 0 | 400 |
| 48 | 2 | White mustard | 4 | 1 | 741 | 217.391304 |
| 73 | 3 | White mustard | 7 | 10 | 1346 | 2000 |
| 98 | 4 | White mustard | 1 | 0 | 192 | 0 |
| 123 | 5 | White mustard | 5 | 2 | 1000 | 434.782609 |
| 148 | 6 | White mustard | 20 | 3 | 4000 | 535.714286 |
| 173 | 7 | White mustard | 2 | 400 | ||
| 24 | 1 | Winfred brassica | 0 | 2 | 0 | 384.615385 |
| 49 | 2 | Winfred brassica | 8 | 8 | 1667 | 1333.33333 |
| 74 | 3 | Winfred brassica | 0 | 3 | 0 | 652.173913 |
| 99 | 4 | Winfred brassica | 2 | 4 | 370 | 714.285714 |
| 124 | 5 | Winfred brassica | 0 | 11 | 0 | 2115.38462 |
| 149 | 6 | Winfred brassica | 40 | 6 | 7692 | 1000 |
| 174 | 7 | Winfred brassica | 0 | 0 | ||
| 25 | 1 | Winter peas | 2 | 1 | 400 | 200 |
| 50 | 2 | Winter peas | 1 | 0 | 192 | 0 |
| 75 | 3 | Winter peas | 0 | 0 | 0 | 0 |
| 100 | 4 | Winter peas | 0 | 0 | 0 | 0 |
| 125 | 5 | Winter peas | 1 | 1 | 185 | 227.272727 |
| 150 | 6 | Winter peas | 0 | 1 | 0 | 166.666667 |
| 175 | 7 | Winter peas | 2 | 322.580645 | ||
| 176 | 1 | Exp control background inoculant level | 55556 | 6.70804154 | ||
| 176 | 2 | Exp control background inoculant level | 55556 | 6.70804154 | ||
| 176 | 3 | Exp control background inoculant level | 55556 | 6.70804154 | ||
| 176 | 4 | Exp control background inoculant level | 55556 | 6.70804154 | ||
| 176 | 5 | Exp control background inoculant level | 55556 | 6.70804154 | ||
| 176 | 6 | Exp control background inoculant level | 55556 | 6.70804154 | ||
| 176 | 7 | Exp control background inoculant level | 55556 | 6.70804154 | ||
References
- Power, A.G.; Mitchell, C.E. Pathogen spillover in disease epidemics. Am. Nat. 2004, 164, S79–S89. [Google Scholar] [CrossRef] [PubMed]
- Beckstead, J.; Meyer, S.E.; Connolly, B.M.; Huck, M.B.; Street, L.E. Cheatgrass facilitates spillover of a seed bank pathogen onto native grass species. J. Ecol. 2010, 98, 168–177. [Google Scholar] [CrossRef]
- Akgül, D.S.; Ahioğlu, M. Fungal pathogens associated with young grapevine decline in the Southern Turkey vineyards. BIO Web Conf. 2019, 15, 01027. [Google Scholar] [CrossRef]
- Úrbez-Torres, J.R.; Haag, P.; Bowen, P.; O’Gorman, D.T. Grapevine Trunk Diseases in British Columbia: Incidence and Characterization of the Fungal Pathogens Associated with Black Foot Disease of Grapevine. Plant Dis. 2014, 98, 456–468. [Google Scholar] [CrossRef]
- Manici, L.M.; Kelderer, M.; Franke-Whittle, I.H.; Rühmer, T.; Baab, G.; Nicoletti, F.; Caputo, F.; Topp, A.; Insam, H.; Naef, A. Relationship between root-endophytic microbial communities and replant disease in specialized apple growing areas in Europe. Appl. Soil Ecol. 2013, 72, 207–214. [Google Scholar] [CrossRef]
- Álvarez-Pérez, J.M.; González-García, S.; Cobos, R.; Olego, M.Á.; Ibañez, A.; Díez-Galán, A.; Garzón-Jimeno, E.; Coque, J.J. Use of endophytic and rhizosphere actinobacteria from grapevine plants to reduce nursery fungal graft infections that lead to young grapevine decline. Appl. Environ. Microbiol. 2017, 83, e01564-17. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, M.; Diniz, I.; Cabral, A.; Rego, C.; Oliveira, H. Unveiling inoculum sources of black foot pathogens in a commercial grapevine nursery. Phytopathol. Mediterr. 2013, 52, 298–312. [Google Scholar]
- Hrycan, J.; Hart, M.; Bowen, P.; Forge, T. Grapevine trunk disease fungi: Their roles as latent pathogens and stress factors that favour disease development and symptom expression. Phytopathol. Mediterr. 2020, 59, 395–424. [Google Scholar]
- Baumgartner, K.; Hillis, V.; Lubell, M.; Norton, M.; Kaplan, J. Managing grapevine trunk diseases in California’s southern san joaquin valley. Am. J. Enol. Vitic. 2019, 70, 267–276. [Google Scholar] [CrossRef]
- Claverie, M.; Notaro, M.; Fontaine, F.; Wery, J. Current knowledge on Grapevine Trunk Diseases with complex etiology: A systemic approach. Phytopathol. Mediterr. 2020, 59, 29–53. [Google Scholar] [CrossRef]
- Adaskaveg, J.; Gubler, D.; Michailides, T. Fungicides, Bactericides, and Biologicals for Deciduous Tree Fruit, Nut, Strawberry, and Vine Crops 2017. UC Davis: Department of Plant Pathology. Available online: https://www.vineyardteam.org/files/resources/2_Excerpt%20from%20fungicide%20efficacy%20timing.pdf (accessed on 17 May 2022).
- British Columbia Ministry of Agriculture. Pest Control Products Recommended for use on Grapes in British Columbia. Pest Manag. Available online: http://www.agf.gov.bc.ca/cropprot/grapeipm/grape_pesticides.pdf (accessed on 17 May 2022).
- Vallejo, A.; Millán, L.; Abrego, Z.; Sampedro, M.C.; Sánchez-Ortega, A.; Unceta, N.; Gómez-Caballero, A.; Goicolea, M.A.; Diez-Navajas, A.M.; Barrio, R.J. Fungicide distribution in vitiviniculture ecosystems according to different application strategies to reduce environmental impact. Sci. Total Environ. 2019, 687, 319–329. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, N.; Perito, M.A. consumers’ willingness to consume sustainable and local wine in italy. Ital. J. Food Sci. 2020, 32, 222–233. [Google Scholar]
- Naher, L.; Yusuf, U.K.; Ismail, A.; Hossain, K. Trichoderma spp.: A biocontrol agent for sustainable management of plant diseases. Pak. J. Bot. 2014, 46, 1489–1493. [Google Scholar]
- Mounier, E.; Cortes, F.; Cadious, M.; Pajot, E. The benefits of Trichoderma atroviride I-1237 for the protection of grapevines against trunk diseases: From the nursery to the vineyard. Phytopathol. Mediterr. 2014, 53, 591–592. [Google Scholar]
- van Jaarsveld, W.J.; Halleen, F.; Bester, M.C.; Pierron, R.J.; Stempien, E.; Mostert, L. Investigation of Trichoderma species colonization of nursery grapevines for improved management of black foot disease. Pest Manag. Sci. 2020, 77, 397–405. [Google Scholar] [CrossRef]
- Clark, A. Managing Cover Crops Profitably, 3rd ed.; Sustainable Agriculture Research and Education: South Burlington, VT, USA, 2012; p. 3. [Google Scholar]
- Runno-Paurson, E.; Lääniste, P.; Eremeev, V.; Tähtjärv, T.; Kaurilind, E.; Tosens, T.; Niinemets, Ü.; Williams, I.H. Does winter oilseed rape as a winter cover crop influence potato late blight development in an organic crop rotation? Biol. Agric. Hortic. 2019, 36, 71–83. [Google Scholar] [CrossRef]
- Nivelle, E.; Verzeaux, J.; Habbib, H.; Kuzyakov, Y.; Decocq, G.; Roger, D.; Lacoux, J.; Duclercq, J.; Spicher, F.; Nava-Saucedo, J.E.; et al. Functional response of soil microbial communities to tillage, cover crops and nitrogen fertilization. Appl. Soil Ecol. 2016, 108, 147–155. [Google Scholar] [CrossRef]
- Eisenhauer, N. Aboveground-belowground interactions drive the relationship between plant diversity and ecosystem function. Res. Ideas Outcomes 2018, 4, e23688. [Google Scholar] [CrossRef]
- Dietrich, P.; Roscher, C.; Clark, A.T.; Eisenhauer, N.; Schmid, B.; Wagg, C. Diverse plant mixtures sustain a greater arbuscular mycorrhizal fungi spore viability than monocultures after 12 years. J. Plant Ecol. 2020, 13, 478–488. [Google Scholar] [CrossRef]
- Gattullo, C.E.; Mezzapesa, G.N.; Stellacci, A.M.; Ferrara, G.; Occhiogrosso, G.; Petrelli, G.; Castellini, M.; Spagnuolo, M. Cover crop for a sustainable viticulture: Effects on soil properties and table grape production. Agronomy 2020, 10, 1334. [Google Scholar] [CrossRef]
- Benitez, M.S.; Taheri, W.I.; Lehman, R.M. Selection of fungi by candidate cover crops. Appl. Soil Ecol. 2016, 103, 72–82. [Google Scholar] [CrossRef]
- Vukicevich, E.; Lowery, T.D.; Úrbez-Torres, J.R.; Bowen, P.; Hart, M. Groundcover management changes grapevine root fungal communities and plant-soil feedback. Plant Soil 2018, 424, 419–433. [Google Scholar] [CrossRef]
- Langenhoven, S.D.; Halleen, F.; Spies, C.F.J.; Stempien, E.; Mostert, L. Detection and quantification of black foot and crown and root rot pathogens in grapevine nursery soils in the Western Cape of South Africa. Phytopathol. Mediterr. 2018, 57, 519–537. [Google Scholar]
- Agustí-Brisach, C.; Gramaje, D.; León, M.; García-Jiménez, J.; Armengol, J. Evaluation of vineyard weeds as potential hosts of black-foot and petri disease pathogens. Plant Dis. 2011, 95, 803–810. [Google Scholar] [CrossRef]
- Van Niekerk, J.M.; Bester, W.; Halleen, F.; Crous, P.W.; Fourie, P.H. The distribution and symptomatology of grapevine trunk disease pathogens are influenced by climate. Phytopathol. Mediterr. 2011, 50, 98–111. [Google Scholar]
- Preece, C.; Peñuelas, J. Rhizodeposition under drought and consequences for soil communities and ecosystem resilience. Plant Soil 2016, 409, 1–17. [Google Scholar] [CrossRef]
- Hiscox, J.; Savoury, M.; Müller, C.T.; Lindahl, B.D.; Rogers, H.J.; Boddy, L. Priority effects during fungal community establishment in beech wood. ISME J. 2015, 9, 2246–2260. [Google Scholar] [CrossRef]
- Fukami, T.; Dickie, I.A.; Paula, W.J.; Paulus, B.C.; Park, D.; Roberts, A.; Buchanan, P.K.; Allen, R.B. Assembly history dictates ecosystem functioning: Evidence from wood decomposer communities. Ecol. Lett. 2010, 13, 675–684. [Google Scholar] [CrossRef]
- Holland, T.; Bowen, P.; Kokkoris, V.; Urbez-Torres, J.R.; Hart, M. Does inoculation with arbuscular mycorrhizal fungi reduce trunk disease in grapevine rootstocks? Horticulturae 2019, 5, 61. [Google Scholar] [CrossRef]
- Kokkoris, V.; Li, Y.; Hamel, C.; Hanson, K.; Hart, M. Site specificity in establishment of a commercial arbuscular mycorrhizal fungal inoculant. Sci. Total Environ. 2019, 660, 1135–1143. [Google Scholar] [CrossRef]
- Pathrose, B.; Jones, E.E.; Jaspers, M.V.; Ridgway, H.J. High genotypic and virulence diversity in Ilyonectria liriodendri isolates associated with black foot disease in New Zealand vineyards. Plant Pathol. 2014, 63, 613–624. [Google Scholar] [CrossRef]
- Martínez-Diz, M.P.; Andrés-Sodupe, M.; Berbegal, M.; Bujanda, R.; Díaz-Losada, E.; Gramaje, D. Droplet Digital PCR Technology for Detection of Ilyonectria liriodendri from Grapevine Environmental Samples. Plant Dis. 2020, 104, 1144–1150. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, D.P.; Nouri, M.T.; Trouillas, F.P. Taxonomy and multi-locus phylogeny of cylindrocarpon-like species associated with diseased roots of grapevine and other fruit and nut crops in California. Fungal Syst. Evol. 2018, 1, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Arafat, Y.; Tayyab, M.; Khan, M.U.; Chen, T.; Amjad, H.; Awais, S.; Lin, X.; Lin, W.; Lin, S. Long-term monoculture negatively regulates fungal community composition and abundance of tea orchards. Agronomy 2019, 9, 466. [Google Scholar] [CrossRef]
- Dann, E.K.; Cooke, A.W.; Forsberg, L.I.; Pegg, K.G.; Tan, Y.P.; Shivas, R.G. Pathogenicity studies in avocado with three nectriaceous fungi, Calonectria ilicicola, Gliocladiopsis sp. and Ilyonectria liriodendri. Plant Pathol. 2012, 61, 896–902. [Google Scholar] [CrossRef]
- Gilbert, C.; Dempcy, J.; Ganong, C.; Patterson, R.; Spicer, G.S. Phylogenetic relationships within Phacelia subgenus Phacelia (Hydrophyllaceae) inferred from nuclear rDNA ITS sequence data. Syst. Bot. 2005, 30, 627–634. [Google Scholar] [CrossRef]
- Tiryaki, I.; Keles, H. Reversal of the inhibitory effect of light and high temperature on germination of Phacelia tanacetifolia seeds by melatonin. J. Pineal Res. 2012, 52, 332–339. [Google Scholar] [CrossRef]
- Bacq-Labreuil, A.; Crawford, J.; Mooney, S.J.; Neal, A.L.; Ritz, K. Phacelia (Phacelia tanacetifolia Benth.) affects soil structure differently depending on soil texture. Plant Soil 2019, 441, 543–554. [Google Scholar] [CrossRef]
- Büchi, L.; Wendling, M.; Amossé, C.; Jeangros, B.; Charles, R. Cover crops to secure weed control strategies in a maize crop with reduced tillage. Field Crop. Res. 2020, 247. [Google Scholar] [CrossRef]
- Koike Salinas, S.T.; Smith, R.F.; Jackson, L.E.; Wyland, L.J.; Inman, J.I.; Chaney, W.E. Phacelia, Lana woollypod vetch, and Austrian winter pea: Three new cover crop hosts of Sclerotinia minor in California. Plant Dis. 1996, 80. [Google Scholar] [CrossRef]
- Kluth, C.; Buhre, C.; Varrelmann, M. Susceptibility of intercrops to infection with Rhizoctonia solani AG 2-2 IIIB and influence on subsequently cultivated sugar beet. Plant Pathol. 2010, 59, 1409–1412. [Google Scholar] [CrossRef]
- León, M.; Berbegal, M.; Abad-Campos, P.; Ramón-Albalat, A.; Caffi, T.; Rossi, V.; Hasanaliyeva, G.; Noceto, P.A.; Wipf, D.; Širca, S.; et al. Evaluation of sown cover crops and spontaneous weed flora as a potential reservoir of black-foot pathogens in organic viticulture. Biology 2021, 10, 498. [Google Scholar] [CrossRef] [PubMed]
- Ohnishi, O. Search for the wild ancestor of buckwheat III. The wild ancestor of cultivated common buckwheat, and of tatary buckwheat. Econ. Bot. 1998, 52, 123–133. [Google Scholar] [CrossRef]
- Morrall, R.A.A.; McKenzie, D.L. Diseases of specialty crops in Saskatchewan: 1. Notes on Buckwheatt and Sunflower 1972-73. Can. Plant Dis. Surv. 1975, 55, 69–72. Available online: https://phytopath.ca/wp-content/uploads/2014/10/cpds-archive/vol55/CPDS_Vol_55_No_2_(69-72)1975.pdf (accessed on 25 January 2022).
- Pathak, N.; Prajneshu, M.; Ahmad, S.; Kumar, L.; Bhaduri, A.; Dhandapani, A.; Sharma, O.P. Phytochemical Analysis and Antifungal Activity of Weed Extracts against Rhizoctonia Root Rot in Buckwheat ( Fagopyrum tataricum. Biopest. Int. 2021, 16, 125–131. [Google Scholar]
- Zini, P.B.; Poletto, T.; Fantinel, V.S.; Andrade, N.; Nunes, U.R.; Muniz, M.D.F.B.; Jacques, R.J.S. Buckwheat seed quality and pathogenicity of Fusarium spp. in plants. J. Seed Sci. 2022, 44. [Google Scholar] [CrossRef]
- Olivier, C.; Vaughn, S.F.; Mizubuti, E.S.G.; Loria, R. Variation in allyl isothiocyanate production within Brassica species and correlation with fungicidal activity. J. Chem. Ecol. 1999, 25, 2687–2701. [Google Scholar] [CrossRef]
- Wen, L.; Lee-Marzano, S.; Ortiz-Ribbing, L.M.; Gruver, J.; Hartman, G.L.; Eastburn, D.M. Suppression of soilborne diseases of soybean with cover crops. Plant Dis. 2017, 101, 1918–1928. [Google Scholar] [CrossRef]
- Fahey, J.W.; Zalcmann, A.T.; Talalay, P. The chemical diversity and distribution of glucosinolates and isothiocyanates among plants. Phytochemistry 2001, 56, 5–51. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).