Abstract
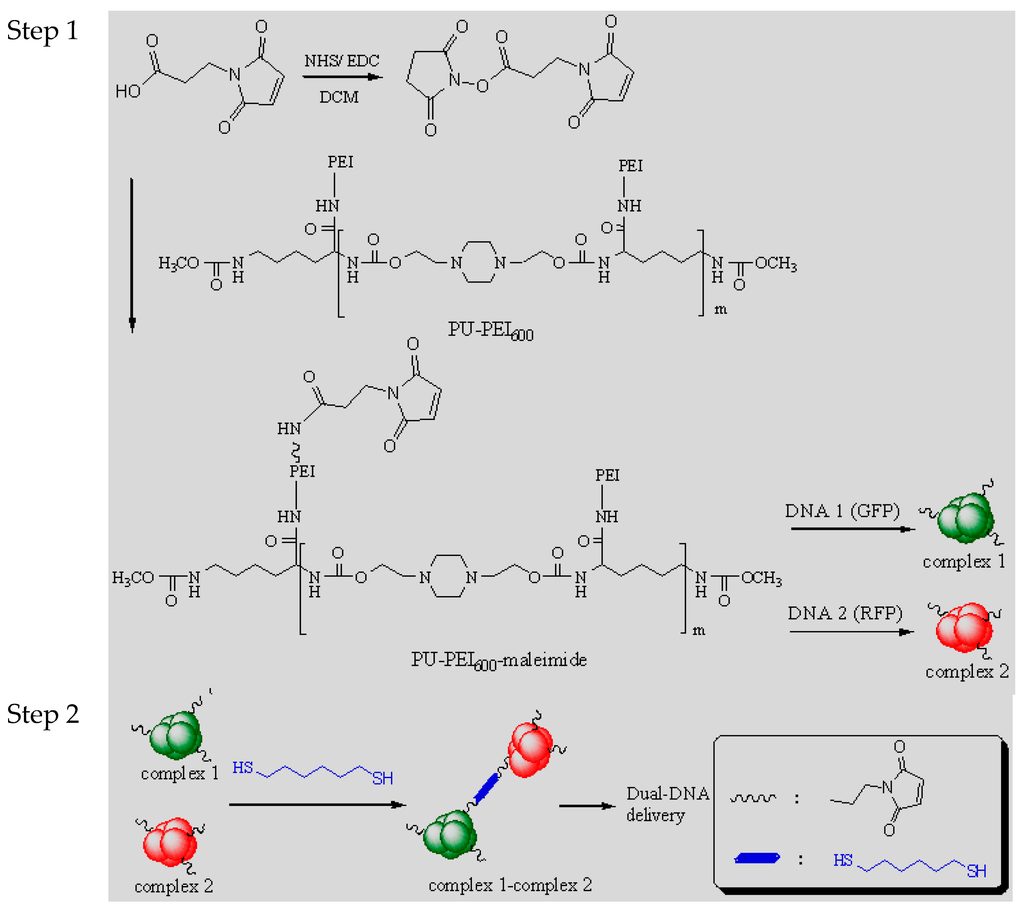
A polyurethane (PU) grafted with small molecular weight polyethylenimine (PEI600) was synthesized. This PU-PEI600 can assemble DNA via electrostatic interactions into nano-sized polymer/DNA complexes. The complexes exhibited great transfection efficiency in delivering DNA along with a reduced cell toxicity comparing to commercial PEI25k (Mw ~25,000). In order to establish a system for concurrently delivering two different DNA or RNA molecules for cell reprogramming (e.g., induced pluripotent stem cells) or the necessity of multi-expression (e.g., double knock down), the PU-PEI600 was further functionalized with maleimide molecules. The novel PU-PEI600-maleimide would still effectively interact with assigned DNA and different functions of PU-PEI600-maleimide/DNA complexes were self-conjugated in presence of a dithiol molecule (1,6-hexanedithiol). In this study, two reporter genes (pEGFP-C2 and pLanRFP-N) were used and evidence of green/red fluorescence co-expression in cells was observed. This article brings a new concept and a practical method for a plurality of different DNA molecules that are more efficient to be concurrently delivered and co-expressed. This method is very helpful in studying cellular multi-regulation or in the treatment of disease with multiple gene defects in vivo.
1. Introduction
Non-viral gene delivery, mostly referred to as polymer-based delivery systems, has been considered potentially safer (low toxicity and immunogenicity) as well as lower restriction of cell specificity in comparison to viral-mediated delivery [1]. The optimum of designing synthetic gene delivery vehicles is to build DNA/vehicle assemblies that are biocompatible and efficient enough to be used in human therapy. In our previous reports, cationic polyurethane exhibited high transfection efficiency with relatively low cytotoxicity in vitro and in vivo [2,3]. A high molecular weight polyethylenimine (PEI25k, Mw ~25,000) revealed its highly effective potential among vectors based on cationic polymers because of its ability of prominent endolysosomal escape and its character of structural protonation [4]. However, these advantages are compromised with its high toxicity that limits its applications to gene therapy. In this study, a cationic polyurethane (PU-PEI600) bearing small molecular weight PEI (Mw = 600) in PU’s side chains was designed and showed superior capability of transfection efficiency and less cytotoxicity than PEI25k. So far, nearly all research is dealing with administration of one kind DNA or RNA at a time (“mono transfection”). However, in work such as cell reprogramming (e.g., induced pluripotent stem cells) or the need of multi-expression (e.g., double knock down), requirements of sending two different (or more) DNA or RNA (“dual transfection”) are in necessity [5,6,7,8,9]. The conventional manipulation is to dispense a mixture of distinct complexes to cells. Here, we proposed a new concept in that cells were treated with covalent conjugates of separately-prepared complexes to increase the success rate of co-expression. In order to achieve this concept, the PU-PEI600 was further functionalized with maleimide molecules. The novel PU-PEI600-maleimide is as effective as PEI25k at interacting individually with two reporter DNA (pEGFP-C2 and pLanRFP-N) and distinct PU-PEI600-maleimide/DNA complexes were covalently-conjugated in presence of a dithiol molecule(s) (1,6-hexanedithiol). These conjugates were found to be small enough and each contained on average two to three PU-PEI600-maleimide/DNA complexes and showed still effectively transferred into the studied cells. Then, co-expression of both green and red fluorescence in cells was evaluated and the feasibility of dual transfection (co-delivering two different DNA) was discussed.
2. Experimental Section
2.1. Materials
l-Lysine methyl ester diisocyanate (LDI, Kyowa Hakko Kyoko, Tokyo, Japan), N,N-dimethyl formamide (DMF, Fluka, Taiwan), N,N′-bis-(2-hydroxyethyl)-piperazine (PPA, Aldrich, Taiwan), Short branch polyethylenimine (Mw = 600, PEI600, Sigma, Taiwan), N-hydroxysuccinimide (NHS, Alfa Aesar, Ward Hill, MA, USA), 1-ethyl-3-(3-dimethylaminopropyl)-carboiimide (EDC, TCI, Tokyo, Japan), 3-(2,5-dioxo-2,5-dihydro-1H-pyrrol-1-yl)propanoic acid (Sigma, Taiwan), 1,6-hexanedithiol (Alfa Aesar, Ward Hill, MA, USA), acetonitrile (ACN) and triethylamine (J.T.Baker, Taiwan) were used. Chemicals of agarose, HEPES, Tris-EDTA, and Luria Broth purchased from Sigma (Taiwan) were used.
2.2. Polymer Characterization
The structures of synthesized polymers were characterized by nuclear magnetic resonance (NMR, Bruker AMX-400 spectrometer, Berlin, Germany) and Fourier transformed infrared spectroscopy (FT-IR, Mattson Galaxy Series 5000, Middleton, WI, USA). All of the chemical shifts in 1H NMR spectra were reported in parts per million (ppm). A 99.8% pure dimethylsulfoxide (DMSO-d6) and deuteroxide (D2O) were used as the solvent to characterize the polymers. IR spectra were obtained using KBr pellets. The molecular weights of the polymers were determined by gel permeation chromatography analysis (GPC, Waters model LC-2410, Milford, MA, USA) in tetrahydrofuran (THF) after calibration with polystyrene standards. The sample concentration for GPC in THF was 8 mg/mL and the flow rate was 1 mL/min.
2.3. Synthesis of Polyurethane (PU) and PU-PEI600
The PU was synthesized via a two-step process as shown in Scheme 1. One hundred forty-five milligrams of l-lysine-diisocyanate (LDI) and 102 mg of N,N′-bis-(2-hydroxyethyl)-piperazine (PPA) at a NCO/OH molar ratio of 1.2/1 were mixed in 1 mL anhydrous DMF solvent in a three-neck reaction flask under a dry nitrogen purge. The reaction was carried out at 60 °C for 12 h using a 0.5 wt % dibutyltin dilaurate catalyst. Then, methanol (4 mL) was slowly added into the reaction mixture until no unreacted isocyanate was detected. The PU was precipitated and purified in an excess of ethyl ether and dried at 40 °C under vacuum. The characteristics of the polymer were confirmed by FTIR and 1H NMR [2]. Briefly, 1H NMR (400 MHz, DMSO-d6, ppm) δ: 1.21–1.81 (6H,–CH(COOCH3)CH2CH2CH2CH2–), 2.50–2.71 (–N2(CH2CH2)2), 2.90 (–CH2CH2NH–), 2.99, 3.9 (–NCH2CH2O–), 3.12 (–NHCH(COOCH3)CH2–), 3.4 (–COOCH3), 3.51 (–CH2NHCOOCH3), 3.67 (–NHCOOCH3), 8.01 (–NHCH(COOCH3)CH2–).

Scheme 1.
Synthesis of polyurethane grafted polyethylenimine (PU-PEI600).
Also shown in Scheme 1, PU-PEI600 was synthesized using the aminolysis reaction of PU and small molecular weight polyethylenimine (Mw = 600, PEI600). First, 100 mg PU was dissolved in 1 mL anhydrous DMF and an excess amount of PEI600 (600 mg) dissolving in 1 mL MeOH was added slowly to react at 45 °C for at least 48 h using 1 mL triethylamine (Et3N) as a catalyst. The product was precipitated in anhydrous ethyl ether and vacuum-dried at 40 °C. The polymer was further purified with dissolving in MeOH and re-precipitated in ethyl ether before characterization by FTIR and 1H NMR. Distinct signals of 1H NMR (400 MHz, D2O, ppm) from the grafting of PEI600 were observed. δ: 0.95 (–NHCH2CH2NH–), 1.21–1.81 (6H, –CH(COOCH3)CH2CH2CH2CH2–), 2.0 (–CONHCH2CH2–), 2.48, 3.14 (–NHCH2CH2NH–), 2.50–2.71 (–N2(CH2CH2)2), 2.90 (–CH2CH2NH–), 2.99, 3.9 (–NCH2CH2O–), 3.12 (–NHCH(COOCH3)CH2–), 3.51 (–CH2NHCOOCH3), 3.67 (–NHCOOCH3), 3.8 (–CONHCH2CH2–), 8.01 (–NHCH(COOCH3)CH2–).
2.4. Synthesis PU-PEI600-Maleimide
In Scheme 2, 5 mg of 3-(2,5-dioxo-2,5-dihydro-1H-pyrrol-1-yl)propanoic acid was firstly dissolved in 1 mL anhydrous DCM. The solution was added with NHS (4 mg) and EDC (4 mg) and allowed to react at 35 °C for at least 30 min to have (maleimide-NHS). Further, 200 mg PU-PEI600 dissolving in 1 mL MeOH was reacted with maleimide-NHS at molar ratio of 1/1 for 12 h at RT. The product was vacuum-dried at 40 °C to remove DCM and purified with rinses of ACN. The product, PU-PEI600-maleimide, was also characterized by FTIR and 1H NMR. 1H NMR (400 MHz, D2O, ppm) δ: 0.95 (–NHCH2CH2NH–), 1,21–1.81 (6H, –CH(COOCH3)CH2CH2CH2CH2–), 2.0 (–CONHCH2CH2–), 2.48, 3.14 (–NHCH2CH2NH–), 2.50–2.71 (–N2(CH2CH2)2), 2.90 (–CH2CH2NH–), 2.94, 4.02 (–COCH2CH2N(CO–)2), 2.99, 3.9 (–NCH2CH2O–), 3.12 (–NHCH(COOCH3)CH2–), 3.51 (–CH2NHCOOCH3), 3.67 (–NHCOOCH3), 3.8 (–CONHCH2CH2–), 5.05 (–CH2CH2NHCOCH2CH2–), 8.01 (–NHCH(COOCH3)CH2–), 8.22 (–COCH=CHCO–).
Scheme 2.
Synthesis of PU-PEI600-maleimide and process of dual-DNA transfection.
2.5. Preparation of Plasmid DNA and Cell Culture
Reporter DNA pCMVLacZ, pEEGFP-C2, and pLanRFP-N encode the genes of β-galactosidase, green fluorescent protein, and red fluorescent protein, respectively, were amplified in Escherichia coli (DH5α strain) and purified by column chromatography (Qiagen Plasmid Mega kit, Hilden, Germany) [10]. The purified plasmid DNA was dissolved in Tris-EDTA buffer (10 mM Tris and 1 mM EDTA, pH 8.0) and the purity of each DNA was determined by the ratio of UV absorbance at 260/280 nm. The cell line COS-7 (SV 40 virus transformed African green monkey cell line, ATCC CRL-1651) was cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco-BRL Co., Ltd., Taiwan) supplemented with 10% fetal bovine serum, penicillin (final 100 IU/mL), streptomycin (final 100 μg/mL) and amphotericin B (final 0.25 μg/mL) at 37 °C in a humidified atmosphere of 5% CO2.
2.6. Formation of PU-PEI600/DNA Complexes and PU-PEI600-Maleimide/DNA Complexes
Ten milligrams per milliliter of the polymers were dissolved in 20 mM HEPES buffer (pH 7.4) and serial dilutions were prepared and added to react with DNA (final concentration = 5 μg/mL) for various polymers/DNA ratios (w/w). The polymer/DNA complexes were self-assembled in a HEPES buffer at room temperature for at least 30 min.
2.7. Electrophoretic Mobility Assay of Polymer/DNA Complexes
The electrophoretic mobility of the polymer/DNA complexes was measured in a 0.7% agarose gel prepared in Tris-acetate-EDTA (TAE) buffer containing ethidium bromide (0.6 μg/mL). After electrophoresis at 100 V for 30 min, DNA bands were visualized by UV irradiation and photographed.
2.8. Sizes of Polymer/DNA Complexes by Dynamic Light Scattering
The hydrodynamic sizes of polymer/DNA complexes and conjugates of the PU-PEI600-maleimide/DNA complexes were determined by dynamic light scattering (DLS) (Nicomp 380 system, San Diego, CA, USA) at 25 °C with a 5 mW He–Ne laser (λ = 633 nm) as the incident beam.
2.9. XTT Cell Viability Assay for Cell Toxicity
The influence of polymer/DNA complexes and PU-PEI600-maleimide/DNA conjugates on cell viability was evaluated in cell culture using the XTT assay [10]. COS-7 cells were cultured in complete DMEM with a seeding density of 1 × 104 cells/well in a 96-well plate. The cells were incubated in an incubator with 5% CO2 at 37 °C for 24 h. Subsequently, the cells were treated with 200 μL FBS-free DMEM containing polymer/DNA complexes or PU-PEI600-maleimide/DNA conjugates. After incubation for 1 h with the polymer/DNA complexes or the conjugates, the cells were washed with 200 μL PBS and replaced by complete DMEM for further 48 h incubation. Then, 50 μL of XTT reagent was added to each well and incubated at 37 °C for 1 h. The results were expressed as the relative cell viability (%) with respect to control wells containing cells that were only treated with DMEM.
2.10. Transfection Protocol for Cells Delivered with pCMV-LacZ DNA by ONPG Assay
COS-7 cells were used to evaluate the transfection efficiency of PU-PEI600/DNA complexes and PU-PEI600-maleimide/DNA complexes. The cells in complete DMEM were seeded in a 96-well plate (1 × 104 cells per well) and incubated for 24 h before transfection. The DNA concentration was kept constant at 5 μg/mL (1 μg/well) and the amounts of polymers were varied for designated polymer/DNA ratios. Two-hundred-microliter solutions of the complexes were taken and incubated with cells for 1 h at 37 °C. For the diluted experiments of dual transfection, 4-fold dilution of these complexes was made in DMEM before cell incubation. The medium was replaced afterwards with complete DMEM and the cells were incubated for further 48 h. For evaluating transfection efficiency, the cells were washed with 0.3 mL PBS and then permeabilized with 20 μL cell lysis buffer at 4 °C for 20 min. An ONPG solution (180 μL/well) was added after lysis treatment and the cells were incubated at 37 °C for another 1 h. The expression of pCMV-LacZ gene was measured spectrometrically using an Enzyme-linked immunosorbent assay (ELISA) reader at a wavelength of 405 nm.
2.11. Dual DNA Transfection via Thiol-Conjugates of PU-PEI600-Maleimide/DNA Complexes
For dual DNA transfection, two distinct PU-PEI600-maleimide/pEEGFP-C2 complexes and PU-PEI600-maleimide/pLanRFP-N complexes were prepared as the method stated in experimental section. To obtain conjugates of PU-PEI600-maleimide/DNA complexes, these two different PU-PEI600-maleimide/DNA complexes were reacted with 1.6-hexanedithiol dissolved in HEPES buffer via a reaction between maleimide and SH [11,12]. The thiol-conjugated reaction was conducted for 30 min at room temperature and the condition (as an optimal molar ratio of PU-PEI600-maleimide/dithiol) for preparation of the conjugates was studied.
The cells were seeded in a 24-well plate (6 × 104 cells per well) in complete DMEM as a culture medium and incubated for 24 h before transfection. An amount of 1200 μL cell medium containing the conjugates of two distinct PU-PEI600-maleimide/DNA complexes was taken and incubated with cells for 1 h at 37 °C. The medium was replaced afterwards with complete DMEM and the cells were incubated for further 48 h. For evaluation of dual DNA transfection efficiency, the co-expression of pEEGFP-C2 (green fluorescence protein, GFP) and pLanRFP-N (red fluorescence protein, RFP) was observed using fluorescence (Nikon ECLIPSE TE2000-U, Tokyo, Japan) microscope under two magnifications of 100× and 400×.
3. Results and Discussion
3.1. Synthesis and Characterization of PU-PEI600 and PU-PEI600-Maleimide
PU-PEI600 and PU-PEI600-maleimide were synthesized as shown in Scheme 1 and Scheme 2. The chemical structures of these polymers were confirmed by FT-IR and 1H NMR spectroscopy. The peaks in the FT-IR spectra at 1720 cm−1 (C=O stretching, urethane), 1651 cm−1 (C=O stretching, amide), 1553 cm−1 (N–H bending, amide), and 3320 cm−1 (N–H stretching, urethane) represent the absorption of urethane links in PU-PEI600 and PU-PEI600-maleimide. There was no obvious difference in FT-IR spectra between PU-PEI600 and PU-PEI600-maleimide because only a few maleimide groups were grafted in PU-PEI600 (see below). The chemical shifts of characterized protons of PU-PEI600 and PU-PEI600-maleimide are reported in the experimental section on polymer synthesis. PU-PEI600 was formed as evidenced by disappearance of the characteristic peak of CH(COO)– (3.4 ppm) from PU and appearance of the peak of PEI, –CH–CH– (2.48–3.14 ppm), in PU-PEI600. Furthermore, the synthesized PU-PEI600-maleimide was identified by a particular peak of maleimide of –C=C– (8.22 ppm). The GPC data showed that the weight averaged molecular weights of PU and PU-PEI600 were 15,428 and 36,413, respectively with a polydispersity range from 1 to 1.15. The increase in molecular weight indicates that about 30 PEI600 molecules have been successfully connected to the side chains of PU. On the other hand, small difference between PU-PEI600 and PU-PEI600-maleimide (Mw ~37,306) suggested that only a few maleimide groups were grafted on PU-PEI600 (calculated as 4–6 maleimide molecules grafted on a PU-PEI600 structure).
3.2. The Gel Retardation and Size Analysis of PU-PEI600/DNA Complexes
The ability of cationic polymers interacting electrostatically with DNA is positively related to a reduction in DNA electrophoretic mobility. Therefore, the relative ability of polymers to do DNA condensation into complexes can be assessed by comparing the polymer/DNA ratios where free DNA band is still observed. It can be seen in Figure 1A that at 1/1 ratio (w/w) of PU-PEI600/DNA, the disappearance of DNA bands starting from lane 2 suggests that amine groups of PU-PEI600 with positive charges can effectively interact with DNA to form complexes. It is worth noting that commercial PEI25k also acts to retain DNA in their complexes at 1/1 weight ratio [13,14]. Since a very similar structure is shared in both outer shell of PEI25k and the side chains of PU-PEI600, respectively, the difference in the “interior” polymer’s structures having little contribution to DNA condensation is also suggested. However, the unlikeness in polymer’s structure (e.g., backbone) may change the profile of buffering capacity due to different amount of total amines that react in acid titration (see Supplementary Materials).
The size of polymer/DNA complexes is influenced by a polymer capable of doing DNA condensation. Figure 1B shows the size of PU-PEI600/DNA complexes at ratios ranging from 1/1 to 100/1 (w/w). The average diameters of these complexes were within 100–200 nm at ratios starting from 5/1 (w/w) and were small enough for cellular endocytosis (<0.2 μm) [15]. The discrepancy in the results at weight ratios between DNA condensation (at 1/1) and size measurement (at 5/1) says that noticeable DNA condensation starts early before smallest polymer/DNA complexes were formed. Since the size of polymer/DNA complexes would be affected by polymer’s structural arrangement (including backbone structure of polymers) [16,17] and density distribution of positive charges [18], the size difference in the complexes using PEI600 in comparison with PU-PEI600 was observed (Figure 1B). Therefore, the size of polymer/DNA complexes is more informative for describing the extent of polymer/DNA interactions than DNA electrophoretic mobility.

Figure 1.
(A) DNA gel retardation of PU-PEI600/DNA complexes at various ratios (w/w); lanes 1, free DNA; lane 2, PU-PEI600/DNA = 1/1; lane 3, PU-PEI600/DNA = 2/1; lane 4, PU-PEI600/DNA = 5/1; lane 5, PU-PEI600/DNA = 10/1; lane 6, PU-PEI600/DNA = 20/1; lane 7, PU-PEI600/DNA = 100/1; (B) Particle sizes of polymer/DNA complexes prepared at various ratios (w/w). Data are presented as mean ± SD (n = 3).
3.3. The Cytotoxicity and Transfection Assay of PU-PEI600/DNA Complexes
The cytotoxicity is an important factor for a success of gene delivery. To determine the cytotoxicity of PU-PEI600/DNA in comparison with that of PEI600/DNA, cells were incubated with increasing amounts of PU-PEI600/DNA or PEI600/DNA complexes at fixed DNA concentration (5 μg/mL) and relative viability of these complexes was measured. As shown in Figure 2A, PU-PEI600/DNA complexes exhibited no toxicity on COS-7 cells till a ratio of 100/1 (w/w) was reached. The PEI600/DNA complexes also exhibited no toxicity as PU-PEI600/DNA complexes. This phenomenon is attributed to the use of low molecular weight PEI [19] since a higher molecular weight polymer with dense positive charges (e.g., PEI25k) would result in a prominent cytotoxicity due to a great extent of disturbance on cell membrane.
The transfection efficiency of complexes in delivering pCMV-LacZ DNA was evaluated by the expression of β-galactosidase (equivalent to ONPG absorbance). The effect of ratios of PU-PEI600/DNA and PEI600/DNA complexes on their transfection efficiency in COS-7 cells are shown in Figure 2B. PU-PEI600 with a polymer structure composed of PEI600 (Mw ~36000) bears a great transfection potential. In contrast, the use of PEI600 (Mw = 600) barely resulted in any DNA expression even though a capability of condensation with DNA into small complexes by PEI600 was noted (Figure 1B). This is because of early dissociation between small cationic molecules (PEI600) and DNA in cellular compartments and limited stability of the formed complexes [20]. The best transfection efficiency of PU-PEI600/DNA complexes was reached at a broad range of ratios from 5/1 to 20/1 (w/w), which is in discrepancy to PEI25k/DNA complexes in which a sharp change occurred before/after its optimal ratio at 1/1 (data not shown). This observation demonstrates that grafting of short PEI600 into the side chain of PU (as PU-PEI600) has created a great improvement in the transfection profile in comparison with PEI25k or PEI600 (see Figure 2C).
Figure 2.
(A) Cytotoxicity of PU-PEI600/DNA and PEI600/DNA complexes in COS-7 cells; (B) Transfection efficiency of the polymers/DNA complexes. Data are presented as mean ± SD (n ≥ 3); (C) Transfection efficiency of PU-PEI600/DNA complexes expressing pEEGFP-C2 DNA under a fluorescent microscope (100×).
3.4. The Gel Retardation, Size Analysis and Transfection Efficiency of PU-PEI600-Maleimide/DNA Complexes
PU-PEI600 was further modified to carry maleimide groups via a reaction between the amine group of PU-PEI600 and maleimide-NHS. A similarity in the profiles of DNA condensation and sizes between PU-PEI600-maleimide (Figure 3A,B) and PU-PEI600 (Figure 1A,B) is shown. These results are in agreement with FT-IR, NMR and GPC data that only a few maleimide structures were grafted on PU-PEI600 and therefore, had minor influence on DNA condensation and the complex sizes. However, as shown in Figure 3C the optimal ratio of PU-PEI600-maleimide/DNA complexes for the best transfection was slightly shifted to 10/1 (from PU-PEI600/DNA at 5/1; w/w). The rational explanation is that less amine groups available for protonation after maleimide modification and the introduction of hydrophobic property of maleimide groups into the polymer structure.

Figure 3.
(A) DNA gel retardation of PU-PEI600-maleimide/DNA complexes at various ratios (w/w); PU-PEI600-maleimide/DNA: 1/1 (lane 1), 5/1 (lane 2), 10/1 (lane 3), 15/1 (lane 4), 20/1 (lane 5), 25/1 (lane 6), 30/1 (lane 7); (B) Particle sizes of PU-PEI600-maleimide/DNA complexes prepared at various ratios (w/w). Data are presented as mean ± SD (n = 3); (C) Transfection efficiency of PU-PEI600-maleimide/DNA complexes. Data are presented as mean ± SD (n ≥ 3).
3.5. The Effect of Thiol-Conjugation on the Size and Cytotoxicity of PU-PEI600-Maleimide/DNA Complexes
In order to achieve dual DNA transfection into a cell, conjugation of two distinct polymer/DNA complexes was designed. PU-PEI600-maleimide/DNA1 and PU-PEI600-maleimide/DNA2 complexes were individually prepared at PU-PEI600-maleimide/DNA ratio of 10/1 (see Section 3.4. and Figure 3C). The maleimide groups of these two different complexes can be conjugated via a presence of 1,6-hexanedithiol as a thioether linker. Figure 4A shows the effect of 1,6-hexanedithiol on the size of the conjugates of PU-PEI600-maleimide/DNA complexes. An increase in the size of the conjugates was detected as an increased amount of the dithiol was added. Our result also shows the average size of the conjugates of PU-PEI600-maleimide/DNA complexes was about 200 nm when a molar ratio at 1.25/1 of dithiol/PU-PEI600-maleimide was applied. Since the average size of PU-PEI600-maleimide/DNA complexes (at w/w ratio of 10/1) was below 100 nm (Figure 3B), it can be estimated that a conjugate (at 1.25/1 molar ratio of dithiol/PU-PEI600-maleimide-DNA) could consist of 2–3 PU-PEI600-maleimide/DNA complexes and the probability to have two different kinds of complexes in a conjugate is estimated as 50% and 75%, respectively.
The cytotoxicity of 1,6-hexanedithiol and the conjugates of PU-PEI600-maleimide/DNA complexes containing the dithiol was determined by XTT assay. Figure 4B shows that the cytotoxicity of these conjugates is dependent to the increased dithiol added for thiol-conjugation. It is clear that there was more than 90% cell viability when dithiol/PU-PEI600-maleimide-DNA molar ratios were below 5/1.
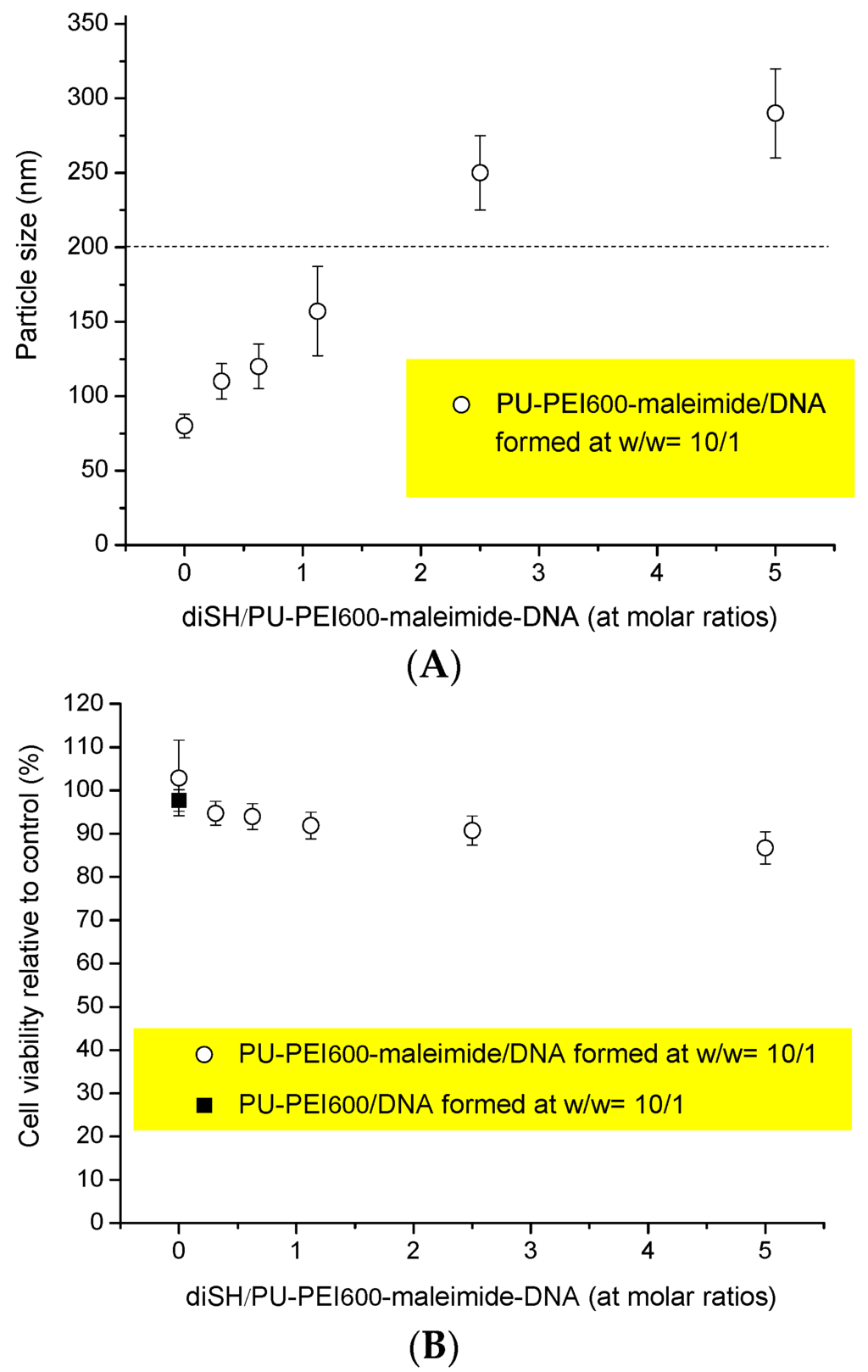
Figure 4.
(A): Particle sizes of the conjugates of PU-PEI600-maleimide/DNA complexes at various diSH/PU-PEI600-maleimide-DNA molar ratios; (B) Cytotoxicity of the conjugates of PU-PEI600-maleimide/DNA complexes in COS-7 cells. Data are presented as mean ± SD (n = 3).
3.6. Cellular Delivery of pCMV-lacZ DNA via PU-PEI600-Maleimide after Thiol-Conjugation
In order to find out an optimal condition for thiol-conjugation, the PU-PEI600-maleimide/DNA (pCMV-lacZ) complexes were conjugated with various concentration of the dithiol and transfection efficiency of the conjugates was evaluated by ONPG absorbance. As can be seen in Figure 5A, the transfection efficiency of PU-PEI600-maleimide/DNA complexes without the dithiol was only marginally higher than the conjugates of PU-PEI600-maleimide/DNA with a molar ratio of dithiol/PU-PEI600-maleimide-DNA at 1.25/1. The reason for this slight reduction of transfection efficiency is due to a slight increase in particle size of the conjugates (~0.2 μm) (Figure 4A) and less favorable for cellular endocytosis.


Figure 5.
(A) Transfection efficiency of the conjugates of PU-PEI600-maleimide/DNA complexes at various diSH/PU-PEI600-maleimide-DNA molar ratios. Results are presented as mean ± SD (n = 3); (B,C) The dual DNA transfection of the conjugates of two different PU-PEI600-maleimid/DNA complexes after thio conjugation and 100× and 400× magnification, respectively; (D) Transfection of a mix of two different PU-PEI600-maleimide/DNA complexes without dithiol conjugaion (100×); (E) The dual DNA transfection of the four-fold diluted conjugates of two different PU-PEI600-maleimid/DNA complexes; (F) Transfection of a four-fold diluted mix of two different PU-PEI600-maleimide/DNA complexes without dithiol conjugaion; (G) Transfection of PU-PEI600-maleimide in complexation with a mix of two DNA after four-fold dilution without dithiol conjugaion.
3.7. Dual Transfection of pEGFP-C2 and pLanRFP-N DNA via the Conjugates of PU-PEI600-Maleimide/DNA Complexes
In this study, two varieties of DNA (pEGFP-C2 and pLanRFP-N) encoding green and red fluorescence protein (GFP and RFP, respectively) were used to study the hypothesis of co-expression of different PU-PEI600-maleimide/DNA complexes via the thiol-conjugation. After PU-PEI600-maleimide/pEGFP-C2 complexes and PU-PEI600-maleimide/pLanRFP-N complexes (both prepared at an optimal weight ratio of 10/1, Figure 3C) were prepared, the conjugates, at PU-PEI600-maleimide-DNA/dithiol molar ratio of 1/1.25 can be formed to have ideally small particles (Figure 5A). The conjugates mainly contain two to three PU-PEI600-maleimide/DNA complexes with 50% to 75% chance of enclosing two distinct DNA, respectively. The efficiency of dual DNA transfection of the conjugates is expressed by showing concurrent GFP and RFP expression in a COS-7 cell under fluorescence observation.
Figure 5B,C shows the fluorescent images of co-expression of GFP (left) and RFP (right) in many cells transfected by the conjugates of PU-PEI600-maleimide/DNA complexes. Remarkably, a substantial percentage of cells showed both expression of GFP and RFP (Figure 5B). Traditionally, for the intention of dual DNA transfection, cells can be treated by a mix of these different PU-PEI600-maleimide/DNA complexes without the dithiol [21]. We knew that about 20% of cells can be transfected and express either GFP or RFP by “mono transfection” of PU-PEI600-maleimide/pEGFP-C2 complexes or PU-PEI600-maleimide/pLanRFP-N complexes, respectively. Theoretically, however, the probability of having simultaneous GFP and RFP co-expression in a cell is far less around 4% (=20% × 20%), as shown in Figure 5D. Indeed, the level of co-expression would be enhanced using a mix of two concentrated polymer/DNA complexes but, on the other hand, this treatment would result in an increase in cytotoxicity [22]. Another clear demonstration is shown (Figure 5E,F) when cells were treated with the four-fold diluted conjugates (Figure 5E) comparing to a mix of two PU-PEI600-maleimide/DNA complexes after four-fold dilution (Figure 5F). The latter method showed that, without conjugation, the co-expression of both PU-PEI600-maleimide/DNA complexes was very low (Figure 5F). Since several DNA molecules could be incorporated into a particle of formed complexes [23], another comparison was also included in which PU-PEI600-maleimide was in complexation with a mix of two different DNA without diSH before adding into cells. Again, the result showed an unbalanced DNA transfection and the co-expression of both GFP and RFP was very low (Figure 5G). Taken together, the use of the conjugates of two different PU-PEI600-maleimide/DNA complexes would greatly increase the success rate of dual DNA transfection into a cell with good biocompatibility.
4. Conclusions
A new polymer, PU-PEI600-maleimide, was synthesized. The transfection efficiency of PU-PEI600-maleimide/DNA complexes in COS-7 cells was much better at delivering a pCMV-lacZ DNA than short chain PEI600. For dual DNA transfection, the PU-PEI600-maleimide/DNA complexes containing two different kinds of DNA were thiol-conjugated. This practical method shows that a plurality of different DNA molecules is more efficient to be concurrently delivered and co-expressed. The cell compatibility accompanied by the dual DNA transfection was also remarkable. This method, therefore, is very helpful in studying cellular multi-regulation or in the treatment of disease with multiple gene defects in vivo.
Supplementary Materials
The supplementary materials is available online at: www.mdpi.com/2073-4360/7/10/1503/s1. Figure S1, the representative GPC curve of PU (methods were listed in the experimental section: polymer characterization); Figure S2, the representative GPC curve of PU-PEI600; Figure S3, the representative GPC curve of PU-PEI600-maleimide; Figure S4, the buffering capacity profile of PU, PU-PEI600-maleimide and PEI25k; Figure S5, FT-IR of PU-PEI600; Figure S6, NMR of PU-PEI600; Figure S7, FT-IR of PU-PEI600-maleimide; Figure S8, NMR of PU-PEI600-maleimide.
Acknowledgments
We are very grateful to Ministry of Science and Technology of Taiwan for providing the research funding (NSC101-2113-M-194-002 and MOST 103-2113-M-194-004).
Author Contributions
Wei-Chih Hung performed experiments and analyzed the data. Jong-Yuh Cherng was responsible for validation of the methods, the progress of this project and managed the final manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Boulaiz, H.; Marchal, J.A.; Prados, J.; Melguizo, C.; Aránega, A. Non-viral and viral vectors for gene therapy. Cell. Mol. Biol. 2005, 51, 3–22. [Google Scholar] [PubMed]
- Cherng, J.Y.; Hung, W.C.; Kao, H.C. Blending of polyethylenimine with a cationic polyurethane greatly enhances both DNA delivery efficacy and reduces the overall cytotoxicity. Curr. Pharm. Biotechnol. 2011, 12, 839–846. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.P.; Chien, Y.; Chiou, G.Y.; Cherng, J.Y.; Wang, M.L.; Lo, W.L.; Chang, Y.L.; Huang, P.I.; Chen, Y.W.; Shih, Y.H.; et al. Inhibition of cancer stem cell-like properties and reduced chemoradioresistance of glioblastoma using microRNA145 with cationic polyurethane-short branch PEI. Biomaterials 2012, 33, 1462–1476. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Xu, Y.; Wang, B.; Qiao, W.; Liu, D.; Li, Z. Cationic compounds used in lipoplexes and polyplexes for gene delivery. J. Control. Release 2004, 100, 165–180. [Google Scholar] [PubMed]
- Peng, C.H.; Cherng, J.Y.; Chiou, G.Y.; Chen, Y.C.; Chien, C.H.; Kao, C.L.; Chang, Y.L.; Chien, Y.; Chen, L.K.; Liu, J.H.; et al. Delivery of Oct4 and SirT1 with cationic polyurethanes-short branch PEI to aged retinal pigment epithelium. Biomaterials 2011, 32, 9077–9088. [Google Scholar] [PubMed]
- Jokinen, E.; Koivunen, J.P. Bcl-xl and Mcl-1 are the major determinants of the apoptotic response to dual PI3K and MEK blockage. Int. J. Oncol. 2015, 17, 1103–1110. [Google Scholar]
- Zhao, M.; He, H.W.; Sun, H.X.; Ren, K.H.; Shao, R.G. Dual knockdown of N-ras and epiregulin synergistically suppressed the growth of human hepatoma cells. Biochem. Biophys. Res. Commun. 2009, 387, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Nguyen, P.; Jia, F.; Hu, S.; Gong, Y.; de Almeida, P.E.; Wang, L.; Nag, D.; Kay, M.A.; Giaccia, A.J.; et al. Double knockdown of prolyl hydroxylase and factor-inhibiting hypoxia-inducible factor with nonviral minicircle gene therapy enhances stem cell mobilization and angiogenesis after myocardial infarction. Circulation 2011, 124, S46–S54. [Google Scholar] [CrossRef] [PubMed]
- Blancafort, A.; Giró-Perafita, A.; Oliveras, G.; Palomeras, S.; Turrado, C.; Campuzano, Ò.; Carrión-Salip, D.; Massaguer, A.; Brugada, R.; Palafox, M.; et al. Dual fatty acid synthase and HER2 signaling blockade shows marked antitumor activity against breast cancer models resistant to anti-HER2 drugs. PLoS ONE 2015. [Google Scholar] [CrossRef] [PubMed]
- Hung, W.C.; Cherng, J.Y. Self-assembly of PEG-oligonucleotide-based matrices and lipoplexes as DNase-responsive delivery systems. Polymer 2015, 67, 148–156. [Google Scholar] [CrossRef]
- Park, E.J.; Gevrek, T.N.; Sanyal, R.; Sanyal, A. Indispensable platforms for bioimmobilization: maleimide-based thiol reactive hydrogels. Bioconjug. Chem. 2014, 25, 2004–2011. [Google Scholar] [CrossRef] [PubMed]
- Gevrek, T.N.; Bilgic, T.; Klok, H.A.; Sanyal, A. Maleimide-functionalized thiol reactive copolymer brushes: Fabrication and post-polymerization modification. Macromolecules 2014, 47, 7842–7851. [Google Scholar] [CrossRef]
- Zhao, D.; Gong, T.; Zhu, D.; Zhang, Z.; Sun, X. Comprehensive comparison of two new biodegradable gene carriers. Int. J. Pharm. 2011, 413, 260–270. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.H.; Gu, Z.P.; Zhang, X.; Fan, M.M. pH-Sensitive ternary nanoparticles for nonviral gene delivery. RSC Adv. 2015, 5, 44291–44298. [Google Scholar] [CrossRef]
- Cherng, J.Y.; van de Wetering, P.; Talsma, H.; Crommelin, D.J.A.; Hennink, W.E. Effect of size and serum proteins on transfection efficiency of poly((2-dimethylamino)ethyl methacrylate)-plasmid nanoparticles. Pharm. Res. 1996, 13, 1038–1042. [Google Scholar] [CrossRef] [PubMed]
- Van de Wetering, P.; Cherng, J.Y.; Talsma, H.; Crommelin, D.J.A.; Hennink, W.E. 2-(dimethylamino)ethyl methacrylate based (co)polymers as gene transfer agents. J. Control. Release 1998, 53, 145–153. [Google Scholar] [CrossRef]
- Liu, X.Y.; Ho, W.Y.; Hung, W.J.; Shau, M.D. The characteristics and transfection efficiency of cationic poly (ester-co-urethane)–short chain PEI conjugates self-assembled with DNA. Biomaterials 2009, 30, 6665–6673. [Google Scholar] [CrossRef] [PubMed]
- Hung, W.C.; Shau, M.D.; Kao, H.C.; Shih, M.F.; Cherng, J.Y. The synthesis of cationic polyurethanes to study the effect of amines and structures on their DNA transfection potential. J. Control. Release 2009, 133, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Jia, L.; Li, Z.; Zhang, D.; Zhang, Q.; Shen, J.; Guo, H.; Tian, X.; Liu, G.; Zheng, D.; Qi, L. Redox-responsive catiomer based on PEG-ss-chitosan oligosaccharide-ss-polyethylenimine copolymer for effective gene delivery. Polym. Chem. 2013, 4, 156–165. [Google Scholar] [CrossRef]
- Lavertu, M.; Méthot, S.; Tran-Khanh, N.; Buschmann, M.D. High efficiency gene transfer using chitosan/DNA nanoparticles with specific combinations of molecular weight and degree of deacetylation. Biomaterials 2006, 27, 4815–4824. [Google Scholar] [CrossRef] [PubMed]
- Needham, C.J.; Shah, S.R.; Dahlin, R.L.; Kinard, L.A.; Lam, J.; Watson, B.M.; Lu, S.; Kasper, F.K.; Mikos, A.G. Osteochondral tissue regeneration through polymeric delivery of DNA encoding for the SOX trio and RUNX2. Acta Biomater. 2014, 10, 4103–4112. [Google Scholar] [CrossRef] [PubMed]
- Cherng, J.Y.; Talsma, H.; Verrijk, R.; Crommelin, D.J.A.; Hennink, W.E. The effect of formulation parameters on the size of poly-((2-dimethylamino) ethyl methacrylate)-plasmid complexes. Eur. J. Pharm. Biopharm. 1999, 47, 215–224. [Google Scholar] [CrossRef]
- Li, G.Y.; Guan, R.L.; Ji, L.N.; Chao, H. DNA condensation induced by metal complexes. Coord. Chem. Rev. 2014, 281, 100–113. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).