Abstract
The presence of isolated metal cations, far from any other atom, is not uncommon in protein crystal structures. A systematic survey of the Protein Data Bank showed that nearly 8% of the metal cations are naked, more frequently if they can interact only electrostatically with their neighbors. Surprisingly, this seemed to be only weakly related to the crystallographic resolution.
1. Introduction
It has been repeatedly observed that metal cations are sometimes isolated in protein crystal structures, far from any other atom [1,2,3]. Some of these are physical and chemical nonsense that might be due to low-quality electron density maps or refinement protocols. A systematic analysis of all naked cations found in the crystal structures deposited with Protein Data Bank was described in the present communication.
2. Materials and Methods
Since this is not an analysis of the stereochemistry of the metal first coordination spheres, any type of contact between a metal ion and other atoms was considered, independently of the type of metal cation, its oxidation state, its spin state, its coordination number, and independently of the type of atom(s) surrounding the metal and of the type of interatomic interactions (covalent, ionic). This was justified by the aim of the work summarized here: we were not trying to understand the grounds for the metal isolation but to only describe this phenomenon.
The Protein Data Bank [4,5] (downloaded on August the 27th 2019; 150,916 files) was scanned, and only X-ray crystal structures containing metal cations and refined to at least 4 Å resolution were retained. Multi-model structures, Cα only structures, structures refined in non-standard space groups, and structures not containing proteins were discharged to ensure data homogeneity. This resulted in 53,669 PDB entries containing 237,105 metal cations, the most common of which are shown in Table 1. A total of 19,322 were isolated (no other atom was within 3 Å) in the asymmetric unit, and 18,373 were naked even when adjacent asymmetric units were considered (they were included in the computations as described in [6,7]). Table S1 (Supplementary Materials) lists all isolated metal cations.

Table 1.
The metal cations more frequently (at least 500 times) found in the Protein Data Bank. The following quantities were reported for each cation: The total number of occurrences in the Protein Data Bank (Total number), the occurrences in which the cation is naked (Naked), and the percentage of naked cations (% naked). The numbers in parentheses are the number occurrences, computed as the sum of the occupancy values deposited in the Protein Data Bank.
3. Results
Some metals were uncommonly naked. Iron, for example, which was observed more than 35,000 times, was naked only 63 times (nearly never). On the contrary, some metals were quite often naked. Magnesium, for example, which was observed very frequently—more than 60,000 times—was naked more than 10,000 times (in nearly 20% of the cases). One obvious explanation is that magnesium(II) salts are often used in buffers and crystallization cocktails, with the consequence that some electron density peaks might be wrongly interpreted as magnesium ions despite they are not—they might be water molecules that form hydrogen bonds with surrounding water molecules with intermolecular oxygen-oxygen distances longer than 3 Å.
For some metals, the high percentage of naked cations (strontium, for example, 58%) might be due to their large dimension: here a cation is considered naked if it lacks neighbors within 3 Å, and for the largest metals, this threshold might be too strict, especially at lower resolution, where the accuracy of interatomic distances might be limited. In this regard, it has been observed in a recent survey of the metal-oxygen distances in small molecule crystal structures, showing that even most of the strontium-oxygen distances are not longer than 3 Å [8]. Since strontium is not an essential metal ion, and there is no specific strontium-binding protein known, the observed binding of strontium in proteins is likely due to the preparation of heavy-atom derivatives.
In general, cations that interact electrostatically with their neighbors are observed to be naked more frequently than cations that interact covalently with their neighbors. On average, 11% of the alkali, 13% of the alkaline earth, and 11% of the lanthanide cations were found to be naked; on the contrary, less than 2% of the first-row transition element cations (from scandium to zinc) were observed to be naked.
Many toxic heavy metal cations, like mercury, strontium, tungsten, or platinum, are commonly used to derivatize the crystals for phase determination; their coordination geometry is often unknown because they can be detected at very low resolution when electron density for side chains remains unresolved, and because heavy atoms cause ripple of Fourier series termination to make their ligands invisible. In the case of strontium, a rare alkaline earth metal, it is most likely that their ligands are surface carboxylates.
A weak relationship between observation of a naked cation and diffraction data was observed. Figure 1 shows the distribution of the resolution values for the entire set of metal-containing protein crystal structures (broken line) and for the protein crystal structures that contain naked metals (continuous line). The second histogram was slightly displaced towards lower resolution, suggesting that naked cations were observed more frequently in lower resolution crystal structures. This was expected since it is at a very low resolution that electron density maps might be problematic. However, the two histograms of Figure 1 were largely superposed, suggesting that the observation of naked metals depends on local problems in the electron density maps, perhaps due to series termination errors, rather than on the global crystal diffraction quality.
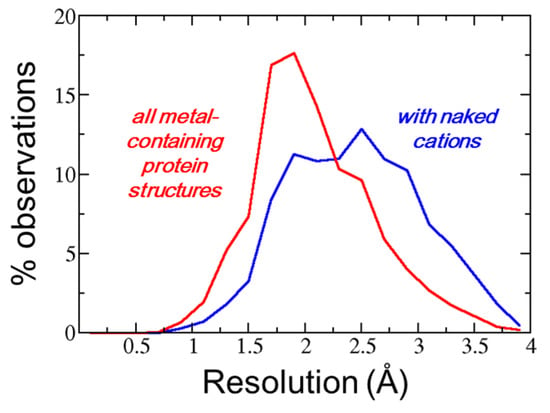
Figure 1.
Distribution of the resolution values for the entire set of metal-containing protein crystal structures (red line) and for the protein crystal structures that contain naked metals (blue line).
It was verified that the occurrence of naked cations in protein crystal structures is unrelated to the software used in the refinement (Buster, CNS, Phenix, Peofft, Prolsq, Refmac, Shelxl, TNT, X-Plor), as well as to the deposition year. This supports the hypothesis that it is really a local problem in the electron density map and, perhaps, it is related to different experimentalists’ preferences since most of the deposited structures do not have naked cations, which concentrate in relatively few structures: when there is a naked cation, on average, one third of the cations are naked.
4. Discussion
Crystallographic results, per se, cannot indicate whether a naked cation is a simple misinterpretation and a mistake, though they can, in some cases, suggest alternative interpretations like a water molecule instead of magnesium(II); even extensive refinement optimizations, e.g., with PDB-REDO [9] are relatively impotent in handling naked cations. Additional experiments may solve the problem by detecting the presence of specific chemical elements in protein crystals like X-ray fluoresces scans, use of soft X-rays in diffraction experiments [10]. Furthermore, inductively coupled plasma mass spectroscopy [11], and elucidation of the cation coordination sphere—by, e.g., X-ray absorption fine structure (EXAFS) or X-ray absorption near-edge structure (XANES) [12,13,14]—can be used to validate the presence of naked ions in macromolecular structures.
Despite excellent efforts and achievements in protein crystal structure validation [15,16,17], even focused on metal cations [18,19], the quality check of protein crystal structures may still improve. Highly doubtful structural details, which are likely a consequence of the “publish or perish” paranoia, should deserve better scrutiny before release in public databases.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4352/9/11/581/s1, Table S1 shows the following quantities for each metal cation: The total number of occurrences in the Protein Data Bank, the occurrences in which the cation is naked, and the percentage of naked cations.
Author Contributions
Conceptualization, K.D.-C.; O.C.; Methodology: O.C.; Writing-original draft, O.C.; Writing-review and editing, O.C.; K.D.-C.
Funding
Not applicable.
Acknowledgments
L. Cherubini is gratefully acknowledged for constant support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Djinovic-Carugo, K.; Carugo, O. Structural biology of the lanthanides-mining rare earths in the Protein Data Bank. J. Inorg. Biochem. 2015, 143, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Carugo, O. Silver and gold in the Protein Data Bank. J. Inorg. Biochem. 2017, 175, 244–247. [Google Scholar] [CrossRef] [PubMed]
- Carugo, O. Structural features of uranium-protein complexes. J. Inorg. Biochem. 2018, 189, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, F.C.; Koetzle, T.F.; Williams, G.J.B.; Meyer, E.F.J.; Brice, M.D.; Rodgers, J.R.; Kennard, O.; Shimanouchi, T.; Tasumi, M.J. The Protein Data Bank: A computer-based archival file for macromolecular structures. Mol. Biol. 1977, 112, 535–542. [Google Scholar] [CrossRef]
- Carugo, O.; Djinović-Carugo, K. How many packing contacts are observed in protein crystals? J. Struct. Biol. 2012, 180, 96–100. [Google Scholar] [CrossRef] [PubMed]
- Carugo, O.; Djinovic-Carugo, K. Packing bridges in protein crystal structures. J. Appl. Cryst. 2014, 47, 458–461. [Google Scholar] [CrossRef]
- Gagné, O.C.; Hawthorne, F.C. Comprehensive derivation of bond-valence parameters for ion pairs involving oxygen. Acta Cryst. 2016, B72, 601–625. [Google Scholar] [CrossRef] [PubMed]
- Joosten, R.P.; Long, F.; Murshudov, G.N.; Perrakis, A. The PDB_REDO server for macromolecular structure model optimization. IUCrJ 2014, 1, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Mueller-Dieckmann, C.; Panjikar, S.; Schmidt, A.; Mueller, S.; Kuper, J.; Geerlof, A.; Wilmanns, M.; Singh, R.K.; Tucker, P.A.; Weiss, M.S. On the routine use of soft X-rays in macromolecular crystallography. Part IV. Efficient determination of anomalous substructures in biomacromolecules using longer X-ray wavelengths. Acta Crystallogr. 2007, D63, 366–380. [Google Scholar] [CrossRef] [PubMed]
- van Heuveln, F.; Meijering, H.; Wieling, J. Inductively coupled plasma-MS in drug development: Bioanalytical aspects and applications. Bioanalysis 2012, 4, 1933–1965. [Google Scholar] [CrossRef] [PubMed]
- Arcovito, A.; della Longa, S. Local structure and dynamics of hemeproteins by X-ray absorption near edge structure spectroscopy. J. Inorg. Biochem. 2012, 112, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L. X-Ray Absorption Spectroscopy of Metalloproteins. Methods Mol. Biol. 2019, 1876, 179–195. [Google Scholar] [PubMed]
- Hummer, A.A.; Rompel, A. The use of X-ray absorption and synchrotron based micro-X-ray fluorescence spectroscopy to investigate anti-cancer metal compounds in vivo and in vitro. Metallomics 2013, 5, 597–614. [Google Scholar] [CrossRef] [PubMed]
- Read, R.J.; Adams, P.D.; Arendall, W.B.r.; Brunger, A.T.; Emsley, P.; Joosten, R.P.; Kleywegt, G.J.; Krissinel, E.B.; Lütteke, T.; Otwinowski, Z.; et al. A new generation of crystallographic validation tools for the protein data bank. Structure 2011, 19, 1395–1412. [Google Scholar] [CrossRef] [PubMed]
- Terwilliger, T.C.; Bricogne, G. Continuous mutual improvement of macromolecular structure models in the PDB and of X-ray crystallographic software: The dual role of deposited experimental data. Acta Cryst. 2014, D70, 2533–2543. [Google Scholar] [CrossRef] [PubMed]
- Domagalski, M.J.; Zheng, H.; Zimmerman, M.D.; Dauter, Z.; Wlodawer, A.; Minor, W. The quality and validation of structures from structural genomics. Methods Mol. Biol. 2014, 2091, 297–314. [Google Scholar]
- Zheng, H.; Chordia, M.D.; Cooper, D.R.; Chruszcz, M.; Müller, P.; Sheldrick, G.M.; Minor, W. Validation of metal-binding sites in macromolecular structures with the CheckMyMetal web server. Nat. Protoc. 2014, 9, 156–170. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Cooper, D.R.; Porebski, P.J.; Shabalin, I.G.; Handing, K.B.; Minor, W. CheckMyMetal: A macromolecular metal-binding validation tool. Acta Cryst. 2017, D73, 223–233. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).