Artificial Intelligence for Risk–Benefit Assessment in Hepatopancreatobiliary Oncologic Surgery: A Systematic Review of Current Applications and Future Directions on Behalf of TROGSS—The Robotic Global Surgical Society
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Search Strategy
2.2. Eligibility Criteria
2.3. Data Extraction
- Study characteristics: titles, authors, year of publication, study design, and country of origin.
- Population characteristics: sample size (total number of participants), surgical population.
- AI technologies: data source, AI model types, best-performing model, prediction target.
- Outcomes: effect sizes (OR, RR, HR) and corresponding 95% confidence intervals (CI) for AUROC validation, sensitivity, specificity, key predictors, external validation, and clinical use.
2.4. Quality Assessment and Synthesis Technique
3. Results
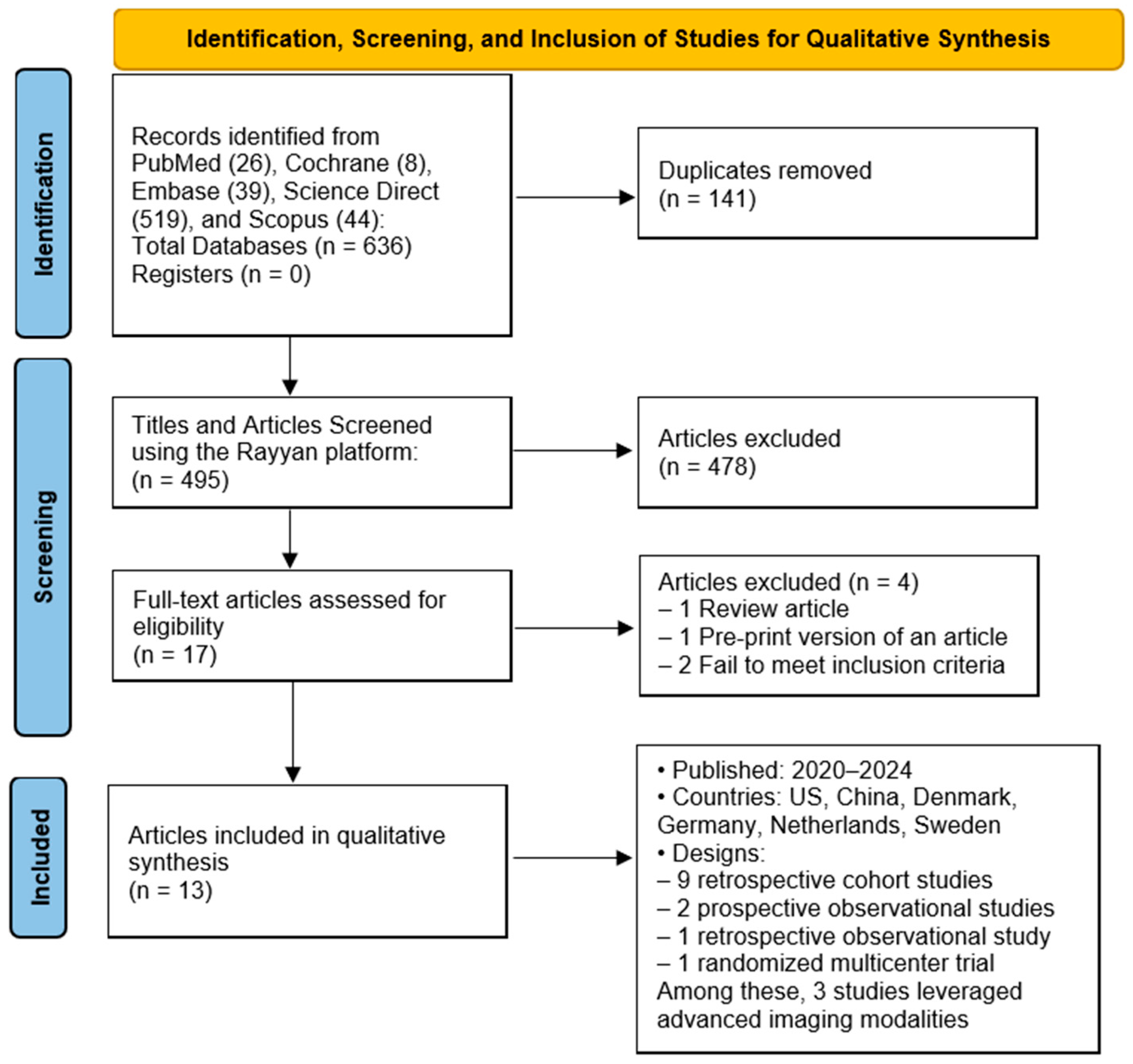
3.1. Study Selection
3.2. Study Characteristics
3.3. Study Objectives and Data Sources
3.4. Artificial Intelligence (AI) Models Applied
3.5. Model Performance Across Clinical Use Cases
3.5.1. Cancer Risk Prediction
3.5.2. Postoperative Complication Prediction
3.5.3. Survival Prognostication
3.6. Validation and Clinical Integration
4. Discussion
4.1. Observations in Producing an Effective Model
4.2. Limitations and Risk of Bias
4.3. Future Directions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ehnstrom, S.R.; Siu, A.M.; Maldini, G. Hepatopancreaticobiliary Surgical Outcomes at a Community Hospital. Hawaii J. Health Soc. Welf. 2022, 81, 309–315. [Google Scholar] [PubMed]
- Papis, D.; Vagliasindi, A.; Maida, P. Hepatobiliary and Pancreatic Surgery in the Elderly: Current Status. Ann. Hepato-Biliary-Pancreat. Surg. 2020, 24, 1–5. [Google Scholar] [CrossRef]
- Hendrix, J.M.; Garmon, E.H. American Society of Anesthesiologists Physical Status Classification System. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2025. [Google Scholar]
- Charlson Comorbidity Index Score (CCI Score). Available online: https://reference.medscape.com/calculator/879/charlson-comorbidity-index-score-cci-score (accessed on 28 July 2025).
- Amin, A.; Cardoso, S.A.; Suyambu, J.; Abdus Saboor, H.; Cardoso, R.P.; Husnain, A.; Isaac, N.V.; Backing, H.; Mehmood, D.; Mehmood, M.; et al. Future of Artificial Intelligence in Surgery: A Narrative Review. Cureus 2024, 16, e51631. [Google Scholar] [CrossRef]
- Rashidi, H.H.; Pantanowitz, J.; Hanna, M.G.; Tafti, A.P.; Sanghani, P.; Buchinsky, A.; Fennell, B.; Deebajah, M.; Wheeler, S.; Pearce, T.; et al. Introduction to Artificial Intelligence and Machine Learning in Pathology and Medicine: Generative and Nongenerative Artificial Intelligence Basics. Mod. Pathol. 2025, 38, 100688. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Ouzzani, M.; Hammady, H.; Fedorowicz, Z.; Elmagarmid, A. Rayyan—A Web and Mobile App for Systematic Reviews. Syst. Rev. 2016, 5, 210. [Google Scholar] [CrossRef]
- Wells, G.; Shea, B.; O’Connell, D.; Peterson, J.; Welch, V.; Losos, M.; Tugwell, P. The Newcastle–Ottawa Scale (NOS) for Assessing the Quality of Non-Randomized Studies in Meta-Analysis. Eur. J. Epidemiol. 2000, 25, 603–605. [Google Scholar]
- Whiting, P.F.; Rutjes, A.W.S.; Westwood, M.E.; Mallett, S.; Deeks, J.J.; Reitsma, J.B.; Leeflang, M.M.G.; Sterne, J.A.C.; Bossuyt, P.M.M.; QUADAS-2 Group. QUADAS-2: A Revised Tool for the Quality Assessment of Diagnostic Accuracy Studies. Ann. Intern. Med. 2011, 155, 529–536. [Google Scholar] [CrossRef] [PubMed]
- Haddaway, N.R.; Page, M.J.; Pritchard, C.C.; McGuinness, L.A. PRISMA2020: An R Package and Shiny App for Producing PRISMA 2020-compliant Flow Diagrams, with Interactivity for Optimised Digital Transparency and Open Synthesis. Campbell Syst. Rev. 2022, 18, e1230. [Google Scholar] [CrossRef]
- Leupold, M.; Chen, W.; Esnakula, A.K.; Frankel, W.L.; Culp, S.; Hart, P.A.; Abdelbaki, A.; Shah, Z.K.; Park, E.; Lee, P.; et al. Interobserver Agreement in Dysplasia Grading of Intraductal Papillary Mucinous Neoplasms: Performance of Kyoto Guidelines and Optimization of Endomicroscopy Biomarkers through Pathology Reclassification. Gastrointest. Endosc. 2025, 101, 1155–1165.e6. [Google Scholar] [CrossRef]
- Màlyi, A.; Bronsert, P.; Schilling, O.; Honselmann, K.C.; Bolm, L.; Szanyi, S.; Benyó, Z.; Werner, M.; Keck, T.; Wellner, U.F.; et al. Postoperative Pancreatic Fistula Risk Assessment Using Digital Pathology Based Analyses at the Parenchymal Resection Margin of the Pancreas—Results from the Randomized Multicenter RECOPANC Trial. HPB 2025, 27, 393–401. [Google Scholar] [CrossRef] [PubMed]
- Machicado, J.D.; Chao, W.-L.; Carlyn, D.E.; Pan, T.-Y.; Poland, S.; Alexander, V.L.; Maloof, T.G.; Dubay, K.; Ueltschi, O.; Middendorf, D.M.; et al. High Performance in Risk Stratification of Intraductal Papillary Mucinous Neoplasms by Confocal Laser Endomicroscopy Image Analysis with Convolutional Neural Networks (with Video). Gastrointest. Endosc. 2021, 94, 78–87.e2. [Google Scholar] [CrossRef]
- Khan, S.; Bhushan, B. Machine Learning Predicts Patients With New-Onset Diabetes at Risk of Pancreatic Cancer. J. Clin. Gastroenterol. 2024, 58, 681–691. [Google Scholar] [CrossRef]
- Wang, H.; Shen, B.; Jia, P.; Li, H.; Bai, X.; Li, Y.; Xu, K.; Hu, P.; Ding, L.; Xu, N.; et al. Guiding Post-Pancreaticoduodenectomy Interventions for Pancreatic Cancer Patients Utilizing Decision Tree Models. Front. Oncol. 2024, 14, 1399297. [Google Scholar] [CrossRef]
- Hu, K.; Bian, C.; Yu, J.; Jiang, D.; Chen, Z.; Zhao, F.; Li, H. Construction of a Combined Prognostic Model for Pancreatic Ductal Adenocarcinoma Based on Deep Learning and Digital Pathology Images. BMC Gastroenterol. 2024, 24, 387. [Google Scholar] [CrossRef]
- Cichosz, S.L.; Jensen, M.H.; Hejlesen, O.; Henriksen, S.D.; Drewes, A.M.; Olesen, S.S. Prediction of Pancreatic Cancer Risk in Patients with New-Onset Diabetes Using a Machine Learning Approach Based on Routine Biochemical Parameters. Comput. Methods Programs Biomed. 2024, 244, 107965. [Google Scholar] [CrossRef]
- Chen, W.; Zhou, B.; Jeon, C.Y.; Xie, F.; Lin, Y.-C.; Butler, R.K.; Zhou, Y.; Luong, T.Q.; Lustigova, E.; Pisegna, J.R.; et al. Machine Learning versus Regression for Prediction of Sporadic Pancreatic Cancer. Pancreatology 2023, 23, 396–402. [Google Scholar] [CrossRef]
- Ingwersen, E.W.; Stam, W.T.; Meijs, B.J.V.; Roor, J.; Besselink, M.G.; Groot Koerkamp, B.; De Hingh, I.H.J.T.; Van Santvoort, H.C.; Stommel, M.W.J.; Daams, F. Machine Learning versus Logistic Regression for the Prediction of Complications after Pancreatoduodenectomy. Surgery 2023, 174, 435–440. [Google Scholar] [CrossRef] [PubMed]
- Placido, D.; Yuan, B.; Hjaltelin, J.X.; Zheng, C.; Haue, A.D.; Chmura, P.J.; Yuan, C.; Kim, J.; Umeton, R.; Antell, G.; et al. A Deep Learning Algorithm to Predict Risk of Pancreatic Cancer from Disease Trajectories. Nat. Med. 2023, 29, 1113–1122. [Google Scholar] [CrossRef]
- Li, Q.; Bai, L.; Xing, J.; Liu, X.; Liu, D.; Hu, X. Risk Assessment of Liver Metastasis in Pancreatic Cancer Patients Using Multiple Models Based on Machine Learning: A Large Population-Based Study. Dis. Markers 2022, 2022, 1586074. [Google Scholar] [CrossRef] [PubMed]
- Aronsson, L.; Andersson, R.; Ansari, D. Artificial Neural Networks versus LASSO Regression for the Prediction of Long-Term Survival after Surgery for Invasive IPMN of the Pancreas. PLoS ONE 2021, 16, e0249206. [Google Scholar] [CrossRef] [PubMed]
- Merath, K.; Hyer, J.M.; Mehta, R.; Farooq, A.; Bagante, F.; Sahara, K.; Tsilimigras, D.I.; Beal, E.; Paredes, A.Z.; Wu, L.; et al. Use of Machine Learning for Prediction of Patient Risk of Postoperative Complications After Liver, Pancreatic, and Colorectal Surgery. J. Gastrointest. Surg. 2020, 24, 1843–1851. [Google Scholar] [CrossRef] [PubMed]
- Ben Hmido, S.; Abder Rahim, H.; Ploem, C.; Haitjema, S.; Damman, O.; Kazemier, G.; Daams, F. Patient Perspectives on AI-Based Decision Support in Surgery. BMJ Surg. Interv. Health Technol. 2025, 7, e000365. [Google Scholar] [CrossRef]
- Khanna, N.N.; Maindarkar, M.A.; Viswanathan, V.; Fernandes, J.F.E.; Paul, S.; Bhagawati, M.; Ahluwalia, P.; Ruzsa, Z.; Sharma, A.; Kolluri, R.; et al. Economics of Artificial Intelligence in Healthcare: Diagnosis vs. Treatment. Healthcare 2022, 10, 2493. [Google Scholar] [CrossRef]
- Collins, G.S.; Reitsma, J.B.; Altman, D.G.; Moons, K.G.M. Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis (TRIPOD): The TRIPOD Statement. BMJ 2015, 350, g7594. [Google Scholar] [CrossRef] [PubMed]
| Authors | Journal | Publication Year | Country | Study Type | Sample Size |
|---|---|---|---|---|---|
| Leupold M, et al. [12] | Gastrointestinal Endoscopy | 2024 | USA | Observational | 64 |
| Khan S, et al. [15] | Journal of Clinical Gastroenterology | 2024 | USA | Retrospective Cohort | 81,213 |
| Wang H, et al. [16] | Frontiers in Oncology | 2024 | China | Retrospective Cohort | 749 |
| Hu K, et al. [17] | BMC (BioMed Central) Gastroenterology | 2024 | China | Retrospective Cohort | 142 |
| Màlyi A, et al. [13] | HPB (Hepato-Pancreato-Biliary) | 2024 | Germany | Randomized Multicenter Trial | 320 |
| Cichosz SL, et al. [18] | Computer Methods and Programs in Biomedicine | 2024 | Denmark | Retrospective Cohort | 1432 |
| Chen W, et al. [19] | Pancreatology | 2023 | USA | Retrospective Cohort | 4,500,000 |
| Ingwersen EW, et al. [20] | Surgery | 2023 | Netherlands | Retrospective Observational | 4912 |
| Placido D, et al. [21] | Nature Medicine | 2023 | Denmark | Retrospective Cohort | 9,200,000 |
| Li Q, et al. [22] | Disease Markers | 2022 | China | Retrospective Cohort | 47,919 |
| Machicado JD, et al. [14] | Gastrointestinal Endoscopy | 2021 | USA | Prospective Single-Center | 35 |
| Aronsson L, et al. [23] | PLOS One | 2021 | Sweden | Retrospective Cohort | 440 |
| Merath K, et al. [24] | Journal of Gastroenterology Surgery | 2020 | USA | Retrospective Cohort | 15,657 |
| Authors | Publication Year | Country | Sample Size | AI Model Used | Best Model * |
|---|---|---|---|---|---|
| Leupold M, et al. [12] | 2024 | USA | 64 | EUS-nCLE | Combined model (EUS-nCLE + 2024 Kyoto High-Risk Stigmata) |
| Khan S, et al. [15] | 2024 | USA | 81,213 | XGBoost, END-PAC Boursi | XGBoost |
| Wang H, et al. [16] | 2024 | China | 749 | Decision Tree | Decision Tree |
| Hu K, et al. [17] | 2024 | China | 142 | DenseNet121, ResNet18 MobileNet_v3_small | Combined Model (Pathological Risk Signature + Clinical Risk Signature) |
| Màlyi A, et al. [13] | 2024 | Germany | 320 | QuPath AI Generalized Linear Model (GLM) | Generalized Linear Model (GLM) |
| Cichosz SL, et al. [18] | 2024 | Denmark | 1432 | Random Forest (RF) | Random Forest (RF) |
| Chen W, et al. [19] | 2023 | USA | 4,500,000 | Random Survival Forest (RSF) eXtreme Gradient Boosting (XGB) Cox Proportional Hazards (COX) | eXtreme Gradient Boosting (XGB) |
| Ingwersen EW, et al. [20] | 2023 | Netherlands | 4912 | Random Forest Neural Network Support Vector Machine Gradient Boosting | Gradient Boosting |
| Placido D, et al. [21] | 2023 | Denmark | 9,200,000 | Bag-of-Words Multilayer Perceptron (MLP) Gated Recurrent Unit (GRU) Transformer | Transformer |
| Li Q, et al. [22] | 2022 | China | 47,919 | Random Forest (RF) XGBoost SVM Deep Neural Network (DNN) Logistic Regression (LR) | Random Forest (RF) |
| Machicado JD, et al. [14] | 2021 | USA | 35 | Segmentation-Based Model (SBM) Holistic-Based Model (HBM) | Holistic-Based Model (HBM) |
| Aronsson L, et al. [23] | 2021 | Sweden | 440 | Artificial Neural Network (ANN) LASSO Logistic Regression | Artificial Neural Networks (ANNs) |
| Merath K, et al. [24] | 2020 | USA | 15,657 | Decision Tree | Decision Tree |
| Authors | Publication Year | Country | Sample Size | AUROC (DK) | AUROC (US Cross-Validation) | AUROC (US Independent Training) | AUROC (Internal) |
|---|---|---|---|---|---|---|---|
| Cichosz SL, et al. [18] | 2024 | Denmark | 1432 | 0.78 (CI 0.75–0.83) | N/A | N/A | N/A |
| Khan S, et al. [15] | 2024 | USA | 81,213 | N/A | 0.81 (XGBoost); 0.66 (END-PAC); 0.71 (Boursi) | N/A | 0.80 (XGBoost); 0.63 (END-PAC); 0.68 (Boursi) |
| Placido D, et al. [21] | 2023 | Denmark | 9,200,000 | 0.879 | 0.710 (Danish-trained model) | 0.775 (US-VA model) | N/A |
| Authors | Publication Year | Country | Sample Size | AUROC (Internal) | Sensitivity | Specificity |
|---|---|---|---|---|---|---|
| Wang H, et al. [16] | 2024 | China | 749 | 0.79 | 77% | N/A |
| Màlyi A, et al. [13] | 2024 | Germany | 320 | 0.73 | 80% | 62% |
| Ingwersen EW, et al. [20] | 2023 | Netherlands | 4912 | CR-POPF Gradient Boosting (ML model) = 0.74 DGE-Both models (ML and logistic regression) = 0.59 | N/A | N/A |
| Merath K, et al. [24] | 2020 | USA | 15,657 | 0.74 | N/A | N/A |
| Authors | Publication Year | Country | Sample Size | AUROC (US Cross Validation) | AUROC (Internal) | Sensitivity | Specificity |
|---|---|---|---|---|---|---|---|
| Wang H, et al. [16] | 2024 | China | 749 | N/A | 0.88 | 67% | N/A |
| Aronsson L, et al. [23] | 2021 | Sweden | 440 | ANN1 = 0.82 F1 0.89 | N/A | 95% | 42% |
| Authors | Publication Year | Country | Sample Size | External Validation | Clinical Use Case |
|---|---|---|---|---|---|
| Khan S, et al. [15] | 2024 | USA | 81,213 | N/A | Stratify patients with new-onset diabetes to identify high PDAC risk for early imaging (e.g., MRI, EUS). |
| Hu K, et al. [17] | 2024 | China | 142 | N/A | Prognosis of PDAC based on integrated histopathological and clinical data. |
| Cichosz SL, et al. [18] | 2024 | Denmark | 1432 | N/A | Future implication, not yet in clinical use Stratify patients ≥50 years old with new-onset diabetes (NOD) into high- vs. low-risk for pancreatic cancer (PCRD). |
| Chen W, et al. [19] | 2023 | USA | 4,500,000 | Externally validated using an independent cohort from the U.S. Veterans Affairs (VA) Health System | Risk stratification for PDAC screening using EHR data. |
| Placido D, et al. [21] | 2023 | Denmark | 9,200,000 | Cross-application of Danish-trained model to US-VA data | Identifying high-risk patients (e.g., top 0.1%) for cost-effective surveillance and early detection of pancreatic cancer. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Goyal, A.; Koutentakis, M.; Park, J.; Macias, C.A.; Ballard, I.; Law, S.H.; Babu, A.; Lau, E.C.A.; Mendoza, M.; Acosta, S.V.J.; et al. Artificial Intelligence for Risk–Benefit Assessment in Hepatopancreatobiliary Oncologic Surgery: A Systematic Review of Current Applications and Future Directions on Behalf of TROGSS—The Robotic Global Surgical Society. Cancers 2025, 17, 3292. https://doi.org/10.3390/cancers17203292
Goyal A, Koutentakis M, Park J, Macias CA, Ballard I, Law SH, Babu A, Lau ECA, Mendoza M, Acosta SVJ, et al. Artificial Intelligence for Risk–Benefit Assessment in Hepatopancreatobiliary Oncologic Surgery: A Systematic Review of Current Applications and Future Directions on Behalf of TROGSS—The Robotic Global Surgical Society. Cancers. 2025; 17(20):3292. https://doi.org/10.3390/cancers17203292
Chicago/Turabian StyleGoyal, Aman, Michail Koutentakis, Jason Park, Christian A. Macias, Isaac Ballard, Shen Hong Law, Abhirami Babu, Ehlena Chien Ai Lau, Mathew Mendoza, Susana V. J. Acosta, and et al. 2025. "Artificial Intelligence for Risk–Benefit Assessment in Hepatopancreatobiliary Oncologic Surgery: A Systematic Review of Current Applications and Future Directions on Behalf of TROGSS—The Robotic Global Surgical Society" Cancers 17, no. 20: 3292. https://doi.org/10.3390/cancers17203292
APA StyleGoyal, A., Koutentakis, M., Park, J., Macias, C. A., Ballard, I., Law, S. H., Babu, A., Lau, E. C. A., Mendoza, M., Acosta, S. V. J., Abou-Mrad, A., Marano, L., & Oviedo, R. J. (2025). Artificial Intelligence for Risk–Benefit Assessment in Hepatopancreatobiliary Oncologic Surgery: A Systematic Review of Current Applications and Future Directions on Behalf of TROGSS—The Robotic Global Surgical Society. Cancers, 17(20), 3292. https://doi.org/10.3390/cancers17203292








