Simple Summary
DNMT3A mutation has been associated with adverse outcomes. In this study, we aimed to investigate the impact of DNMT3A status on NPM1 MRD predictive value for survival in a retrospective cohort of acute myeloid leukemia (AML) patients aged over 60 years old treated intensively. A total of 138 patients treated for NPM1-mutated AML in two French institutions were analyzed retrospectively. A 4log reduction of NPM1 MRD was associated with a better outcome. DNMT3A negative patients who achieved a 4log reduction had a superior outcome to those who did not. However, postinduction NPM1 MRD1 reduction was not predictive of OS and LFS in DNMT3Amut patients. These results confirm that post-induction NPM1 MRD1 is a reliable tool to assess disease outcome in elderly AML patients. However, the presence of DNMT3A also identify a subgroup of patients at high risk of relapse.
Abstract
Minimal residual disease (MRD) is now a powerful surrogate marker to assess the response to chemotherapy in acute myeloid leukemia (AML). DNMT3A mutation has been associated with adverse outcomes. In this study, we aimed to investigate the impact of DNMT3A status on NPM1 MRD predictive value for survival in a retrospective cohort of AML patients aged over 60 years old treated intensively. A total of 138 patients treated for NPM1-mutated AML in two French institutions were analyzed retrospectively. DNMT3A status did not influence the probability of having a ≥ 4log MRD1 reduction after induction. Only 20.4% of FLT3-ITD patients reached ≥ 4log MRD1 reduction compared to 47.5% in FLT3wt cases. A 4log reduction of NPM1 MRD was associated with a better outcome, even in FLT3-ITD mutated patients, independent of the allelic ratio. DNMT3A negative patients who reached a 4log reduction had a superior outcome to those who did not (HR = 0.23; p < 0.001). However, postinduction NPM1 MRD1 reduction was not predictive of OS and LFS in DNMT3Amut patients. These results confirm that post-induction NPM1 MRD1 is a reliable tool to assess disease outcome in elderly AML patients. However, the presence of DNMT3A also identifies a subgroup of patients at high risk of relapse.
1. Introduction
Although the outcome in younger adults with acute myeloid leukemia (AML) has improved these last decades, the treatment of elderly patients remains challenging [1]. Indeed, recent data suggest that the European LeukemiaNet (ELN) 2017 risk stratification per se was not accurate to stratify properly patients older than 60 years old treated with intensive chemotherapy [2]. AML patients over 60 years carried adverse cytogenetics more frequently than younger adults but also a specific gene-expression and molecular alterations that support a molecular basis for poor outcomes [3]. Moreover, approximately 70% of patients over 60 years carry unfavorable cytogenetic or molecular markers whereas only 30 to 35% of them are effectively transplanted [4,5]. This might be explained by the fact that very few patients over the age of 70 years old reach transplantation due to excess toxicity during induction, comorbidities, or a less complete remission rate. In the absence of hematopoietic stem cell transplantation (HSCT), it is still unclear if these patients may benefit from intensive chemotherapy, even in the ELN 2017 favorable risk group, over alternative strategies, especially in the era of venetoclax-based combination [6].
Minimal residual disease is now a powerful surrogate marker to assess the response to chemotherapy. Several techniques are used routinely to assess MRD such as RT-qPCR, flow cytometry, and next generation sequencing (NGS). However, only NPM1 and CBF MRD by RT-qPCR are considered reliable tools to re-allocate patients into different risk groups. In younger adults, NPM1 MRD has recently been demonstrated as a favorable predictive marker for event-free survival (EFS) and overall survival (OS) independent of fms-like tyrosine kinase-3 internal tandem duplications (FLT3-ITD) status. Balsat et al. [7] prospectively showed in a cohort of 229 patients aged below 60 that a 4-log reduction of NPM1 MRD after induction conferred a better survival, independent of FLT3-ITD status. These results were in line with those previously published by Krönke et al. [8]. However, there are very few data regarding the predictive value of NPM1 MRD in elderly AML patients (aged over 60 years old). In elderly patients, it is also rather unclear whether good molecular responders or at least those with undetectable MRD by flow-cytometry may benefit from allogenic HSCT, especially in the intermediate subgroup [9]. Numerous studies have suggested the negative impact of DNMT3A mutation in NPM1-mutated AML patients, especially in those with concurrent FLT3-ITD mutation [2,10,11].
In this study, we aimed to investigate the impact of DNMT3A status on NPM1 MRD predictive value for survival in a retrospective cohort of AML patients aged over 60 years old treated intensively.
2. Patients and Methods
2.1. Patients
A total of 138 patients (aged 60 years or more) treated in Lyon and Hauts de France centers (CHU Lille, CHU Amiens, Hôpital St Vincent, CH Valenciennes, CH Arras, CH Boulogne-sur-mer, CH Roubaix, CH Lens, CH Dunkerque), France, with newly diagnosed NPM1-mutated AML (de novo or secondary) for which post induction bone marrow (BM) minimal residual disease (MRD) were available, were included in this study. At diagnosis, blood and BM samples were examined for cytogenetic abnormalities and molecular markers (NPM1, FLT3-ITD, DNMT3A) according to local procedures. Patients were assigned to risk groups according to the 2017 ELN classification, regarding the FLT3-ITD ratio [12]. Karyotype was not considered in the risk stratification. As the TP53, ASXL1, and RUNX1 mutational status were not available, these variables were not included in the risk stratification.
2.2. Treatments
All patients were treated by an anthracycline- and cytarabine-based induction chemotherapy regimen. One hundred and fifteen of them were treated in or according to the observational Acute Leukemia French Association (ALFA)-1200 study, with or without midostaurin (depending on the presence of FLT3-ITD and drug availability). The 23 other patients were included in other protocols (eight in ALFA-0701, 10 in MyloFrance 4 (NCT02473146), three in BIG1 protocol (NCT02416388)], one in ALFA-0702, one in BRIGHT protocol (NCT03416179) [13,14]. Patients achieving composite complete remission (CRc) after one or two courses of induction were given consolidation chemotherapy according to the protocol in which they were included. Nineteen patients underwent HSCT in their first CRc after reduced intensity conditioning (RIC).
2.3. Clinical and Molecular Markers
Cytogenetic analysis was performed according to the International System for Human Cytogenetic Nomenclature guidelines [15]. NPM1 and FLT3-ITD mutations were detected and quantified as previously described [16,17]. MRD for NPM1 was assessed after at least the first cycle of induction (MRD1). Quantification of the different NPM1 mutation types was performed by allele-specific oligonucleotide real-time quantitative polymerase chain reaction (ASO-RQ-PCR) using a common forward primer, a mutation-specific reverse primer and a common hydrolysis probe, as previously described [18,19,20,21,22]. The NPM1m copy number value was then normalized on the number of ABL1 transcripts used as an endogenous reference gene [23,24,25]. All patients with NPM1, FLT3-ITD status, and at least post-induction NPM1 MRD on BM were included in this retrospective study. Patients with atypical NPM1 transcripts were also include into the descriptive analysis. Only patients with monitorable transcripts were included in the survival analysis.
2.4. Outcome Parameters
CRc (CR + Cri + CRp) status was defined on BM aspirates with less than 5% of blasts recovery and classical hematological recovery characteristics [12]. Overall survival (OS) was calculated from treatment assignment to death from any cause. Leukemia-free survival (LFS) was determined for responders from CRc until disease relapse or death of any cause. Living patients were censored for OS at the last follow-up date, and patients in CR were censored for LFS at the last disease assessment.
2.5. Statistical Analyses
Comparative descriptive statistics were used to characterize patients and their disease in their entirety. Continuous variables were reported as the median ± standard deviation (SD) followed by a t test if the distribution was normal in both groups; if not, the median, range (min-max), and inter-quartile range (IQR) were assessed with a Mann–Whitney test to compare groups. The discrete and qualitative variables were reported as count and percentage. The probabilities of LFS and OS were estimated using the Kaplan–Meier method, and the log-rank rest evaluated the differences between survival distributions. Univariate and multivariate analyses including the baseline demographic, and clinical and molecular features were studied thanks to Cox regressions. The statistical results were two-sided with a p-value < 0.05 considered statistically significant (XLSTAT©).
3. Results
3.1. Initial Patient Characteristics
The median age of the entire cohort was 66.1 years old (range: 60 to 78.2). Table 1 shows the distribution of patient’s characteristics by DNMT3A status. Of the 138 NPM1mut patients, DNMT3A status was available in 98 of them. Overall, 10 out of 138 patients were considered to have a secondary AML (previous history of myelodysplastic syndrome or therapy-related AML). The most frequent DNMT3A alteration was R882 missense mutation (51.9%). DNMT3A mutations were usually part of the main clone with a median variant allelic frequency of 42% (Table 1). FLT3-ITD was evidenced in 52 of 138 patients (37.6%) with a median FLT3-ITD allele ratio (AR) of 0.53 (range: 0.05 to 3). FLT3-ITD mutations co-occurred more frequently in DNMT3Amut patients (48.1% vs. 21.4%, p < 0.001). In this cohort, 40 patients were classified in the unfavorable ELN subgroup because of the presence of FLT3-ITD. Higher median NPM1 baseline levels were detected in cases with DNMT3Amut and FLT3-ITD compared to double negative cases (1384% vs. 1685%; p = 0.045) (Supplementary Figure S1). No significant correlation was found between the DNMT3A mutational status and age, FLT3-ITD allelic ratio (AR), or karyotype.

Table 1.
Patient characteristics.
3.2. General Outcome According to DNMT3A and FLT3-ITD Status
With a median follow-up of 20 months (0.07 to 128.4), the overall CRc rate was 89.9% with no influence of DNMT3A or FLT3 mutational status on the probability of CR. MRD1 response was available in 126/138 patients at the end of induction. The median OS of the entire cohort was 30.6 months with an OS rate of 69.1% at 1 year and 47.5% at 3 years. LFS was 59.1% at 1 year and 33.5% at 3 years (median PFS: 15.4 months) (Supplementary Figure S2). The presence of FLT3-ITD mutation was associated with a worse OS (median OS: 10.9 months) compared to NPM1mut FLT3wt patients (median OS: 46.3 months, p < 0.001) (Figure 1A). There was no significant difference in OS and LFS between NPM1mut FLT3wt and NPM1mut FLT3-ITDlow. Even if there was no difference between NPM1mut FLT3-ITDhigh (median OS = 10.6 months) and NPM1mut FLT3-ITDlow (median OS = 22.6 months), patients with high FLT3-ITD AR have a worse outcome compared to wild type ones (Figure 1B, Supplementary Figure S3). Regarding the prognostic impact of DNMT3A mutation, the presence of the mutation was associated to a worse outcome in terms of OS (DNMT3Amut vs. DNMT3Awt median OS: 18.5 months vs. not reached (NR), p = 0.002) and LFS (DNMT3Amut vs. DNMT3Awt median LFS: 9.3 months vs. 54.7 months, p < 0.001) (Figure 1C, Supplementary Figure S5). Among the 52 patients having DNMT3A and FLT3-ITD status available, the presence of DNMT3Amut and FLT3-ITD significantly worsen the OS and LFS compared to isolated NPM1-mutated patients. In NPM1mut DNMT3Amut cases, FLT3-ITD impaired significantly OS and LFS (Figure 1D, Supplementary Figure S6).
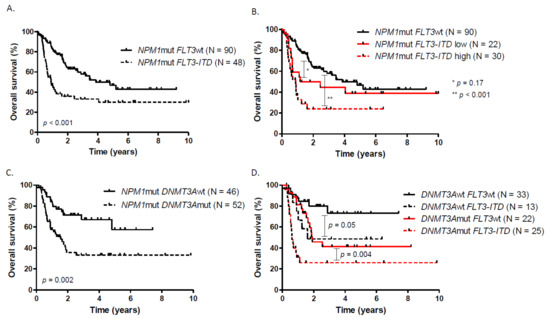
Figure 1.
Overall survival according to (A) FLT3-ITD status, (B) FLT3-ITD allelic ratio, (C) DNMT3A status, (D) DNMT3A and FLT3-ITD status.
3.3. Prognostic Impact of Postinduction NPM1 BM-MRD Log Reduction
Postinduction NPM1 MRD1 on BM was available in 127 patients (92%). In this elderly cohort of NPM1mut patients, a 4log reduction of NPM1 MRD was associated with a better outcome in terms of OS (median OS: NR vs. 13.4 months, HR = 0.35, p < 0.01) and LFS (Figure 2A, Supplementary Figure S7). Similarly, a 5log reduction identifies a subgroup of patients with a very favorable outcome when compared to a 4log reduction (Figure 2B). Overall, DNMT3A status did not influence the probability of having a ≥4log MRD1 reduction after induction. However, only nine out of 44 (20.4%) FLT3-ITD patients reached ≥4log MRD1 reduction whereas 38 out of 80 FLT3wt (47.5%) were good molecular responders (p < 0.001). A high FLT3-ITD allelic ratio or DNMT3A co-occurrence have no significant impact on the probability of reaching a ≥ 4log MRD1 reduction. FLT3-ITD-mutated patients who achieved a 4log reduction had a superior outcome to those who did not (HR = 0.34; 95% CI, 0.16 to 0.70; p < 0.001). Similarly, NPM1mut FLT3wt patients with a 4log reduction in NPM1 BM-MRD had a longer OS (three-year OS, 68.1%; 95% CI, 48.8 to 82.9) than those without good molecular response (three-year OS, 46.5%; 95% CI, 30.2 to 61.7) (Figure 2C). DNMT3A negative patients who achieved a 4log reduction had a superior outcome to those who did not reached at least a 4log reduction (HR = 0.23; 95% CI, 0.07 to 0.72; p < 0.001). However, postinduction NPM1 MRD1 reduction was not predictive of OS in DNMT3A positive patients (Figure 2D). DNMT3Amut patients had a very poor LFS which was even worse in poor NPM1 MRD1 responders compared to those who reached at least a 4log reduction (Supplementary Figure S8).
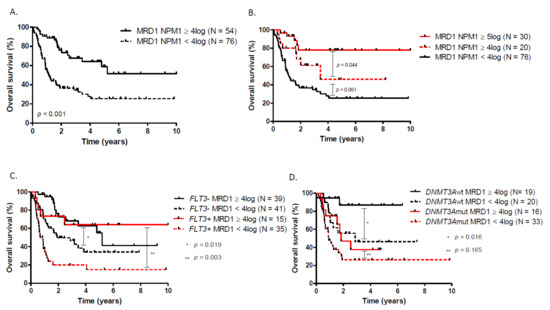
Figure 2.
Overall survival according to (A) NPM1 MRD1, (B) NPM1 MRD1 with a 5log reduction cut-off, (C) NPM1 MRD1 in patients with or without FLT3-ITD, (D) NPM1 MRD1 in patients with or without DNMT3A.
3.4. Multivariate Analysis and Prognosis Model
In the univariate analysis for OS, the variables associated with a poorer survival were the presence of FLT3-ITD mutation, DNMT3A mutation, absence of MRD1 4log reduction and unfavorable ELN risk group. Age, HSCT in CR1 and karyotype were not of prognostic value (Supplementary Table S1). In multivariate analysis, only the DNMT3A mutational status and a 4-log reduction in NPM1 BM-MRD were significantly associated with survival (Table 2).

Table 2.
Multivariate analysis.
Based on these results, we identified among NPM1 positive patients three groups with distinct prognosis, based on FLT3-ITD, DNMT3A status, and the NPM1 BM-MRD post-induction response (Figure 3A). Within the ELN 2017 favorable risk group (n = 98), the NPM1 scoring system (NPM1 SS) reallocated 45.9 and 25.5% of patients into intermediate and unfavorable risk groups, respectively (Figure 3B). The median OS of the favorable, intermediate, and unfavorable NPM1 SS risk groups were NR, 30.6, and 13.2 months, respectively (Figure 3C). The median LFS of the favorable, intermediate, and unfavorable NPM1 SS risk group were NR, 17.2, and 7.7 months, respectively (Figure 3D, Supplementary Table S2). We then used ROC curve comparison to assess if NPM1 SS could be more accurate in OS prediction than ELN classification. When compared to ELN (AUC = 0.695), the NPM1 scoring system was more accurate for OS prediction in patients within the intermediate (AUC = 0.833) and unfavorable (AUC = 0.863) NPM1 SS risk group. However, there was no significant difference in AUC between the NPM1 SS favorable risk group and ELN favorable risk group (Figure 3E).
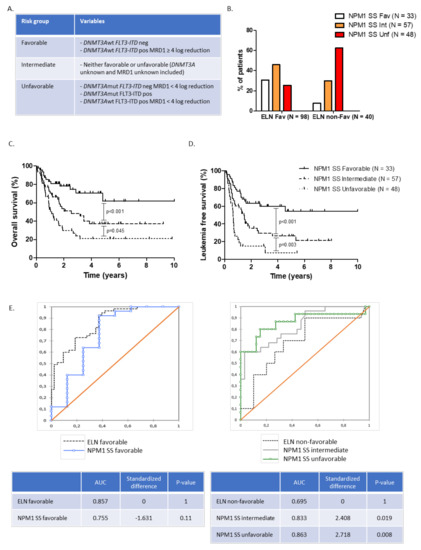
Figure 3.
(A) NPM1 scoring system (SS) risk groups, (B) ELN 2017 risk groups reallocation according to NPM1 SS, (C) overall survival, and (D) leukemia-free survival in NPM1 SS favorable, intermediate, and unfavorable risk groups. (E) ROC curve analysis assessing overall survival prediction according to ELN and NPM1 SS.
4. Discussion
The management of AML in patients older than 60 years remains a major challenge as it is still rather unclear which patients will benefit or not from intensive chemotherapy compared to low-intensity regimens according to clinical, molecular, or cytogenetics markers, especially in the absence of HSCT [9]. Several factors may be involved in the poor prognosis of older AML patients. Silva et al. reported that elderly AML is a specific and distinct entity that harbors genetic and epigenetic patterns not shared by younger adults [19]. Beside a specific methylation signature, these AMLs are enriched in genetic alterations in spliceosome machinery, epigenetic regulators, and in DNA repair factors known to be associated with global treatment resistance. Aging has also been related to an increased frequency of unfavorable karyotype incidence, probably due to the increasing prevalence of secondary AML [20]. Nevertheless, after 60 years old, Nagel et al. showed that there were no major modifications among the ELN risk group distribution under and beyond 70 years old, which was the same regarding the NPM1 and FLT3-ITD mutations distribution, suggesting that other cofounding factors such as secondary mutations may impact on survival [3]. Indeed, ELN 2017 classification shows some limitation into stratifying patients, especially those within favorable or intermediate subgroups that are not referred systematically to HSCT. Recently, Gardin et al. [13] showed that integrating secondary AML-like gene mutations (ASXL1, SRSF2, STAG2, BCOR, U2AF1, EZH2, SF3B1, and ZRSR2) identifies a specific subset of high-risk patients and may thus improve the definition of high-risk older patients with AML [13].
FLT3-ITD mutations and its high allelic ratio is well known to be associated with a high risk of relapse and dismal outcomes in younger adults, whereas its impact seems to be reduced or absent in elderly patients [23,24,26]. In our study, patients with low FLT3-ITD AR have a similar outcome than those without FLT3 mutation, suggesting that other confounding factors might significantly impact survival. We also showed that the sole presence of FLT3-ITD was not an independent prognostic factor among NPM1-mutated patients when integrating other molecular markers such as DNMT3A and MRD monitoring. Regarding DNMT3A, its mutation has been associated with adverse outcomes among patients with an intermediate-risk cytogenetic profile or FLT3 mutations, regardless of age [11,27,28]. In AML, the exact pathogenic mechanism by which DNMT3A mutations act on and negatively affect the outcome is rather unclear. Despite the apparent biological consequences of DNMT3A loss in normal hematopoietic and leukemic stem cells (LSC), the correlation between the differentially methylated genes and gene expression have never been demonstrated, suggesting that other mechanisms may be involved [29,30]. Moreover, significant differences in DNA methylation signatures between AML with DNMT3A-R882 and non-R882 mutations have been reported, suggesting that mutations in different DNMT3A domains lead to different neomorphic functions [31]. Additionally, Guryanova et al. [10,32] showed recently that DNMT3A cooperates with FLT3-ITD and NPM1 to induce AML in vivo, and promotes resistance to anthracycline chemotherapy through impaired nucleosome eviction and chromatin remodeling in response to anthracyclines. As with Papaemmanuil et al. [10], our results suggest also that the co-occurrence of FLT3-ITD and DNMT3A mutations might be detrimental regarding the outcome.
NPM1 gene mutations are important as molecular markers for the diagnosis, prognosis, and monitoring of MRD in AML patients. Several groups have reported the prognostic impact of the NPM1 mutation MRD response, especially when assessed early [7,33,34,35,36]. Ivey et al. prospectively showed that NPM1 MRD performed after the first cycle of consolidation was highly predictive of relapse-free survival (RFS) and OS, independent of the FLT3-ITD and DNMT3A mutational status [36]. These results were recently confirmed with postinduction MRD, which showed that patients harboring at least a 4-log reduction did not benefit from HSCT, independent of the FLT3-ITD status [7]. This is, to our knowledge, the largest study assessing the impact of NPM1 BM-MRD1 on survival in elderly AML patients treated with intensive chemotherapy. Although multiple studies evaluating NPM1mut MRD also included some patients aged > 60 years, none of them specifically focused on the application and clinical significance of MRD monitoring in elderly patients with NPM1mut AML [36,37,38]. As in younger adults, we showed that a 4log reduction of NPM1 BM-MRD1 after induction was of strong prognostic significance, especially in patients with FLT3-ITD mutation. More stringent MRD responses such as 5log reduction may be also more accurate to identify patients with a more favorable outcome. Our results suggest that the presence of DNMT3A may abrogate the predictive value of NPM1 BM-MRD1 on survival, at least in the elderly. However, these results should be validated prospectively, especially in younger AML patients.
More recently, Herold et al. [2] showed that ELN 2017 classification could be refined within each risk group by integrating the DNMT3A mutational status that systematically identified a subgroup of patients with significantly inferior OS compared with DNMT3A wild-type patients. Interestingly, the incidence of DNMT3A mutation seems to increase after the age of 40 years old, suggesting that this mutation is critical in leukemogenesis as it is involved in clonal hematopoiesis [39]. By integrating DNMT3A, NMP1 MRD and FLT3-ITD status, we identified among NPM1-mutated elderly patients three subgroups with different outcomes. NPM1 SS identifies among ELN a favorable subgroup—elderly patients with dismal prognoses that might benefit from HSCT in their first CR. However, the number of transplanted patients were too small to validate this hypothesis. We should also be cautious about the interpretation of results in this elderly patient population. Although the sample size is fairly large in this specific NPM1-mutated cohort of patients, we still cannot be completely confident about the impact of other confounding factors. Limitations in our study mainly concern its retrospective profile, MRD assessment on bone marrow over peripheral blood, and the absence of other molecular parameters such as TP53 or secondary mutations in the prognostic model.
In conclusion, our work confirms that post-induction NPM1 MRD1 is a reliable tool to assess the disease outcome in elderly AML patients. Even if these results have some limitations, the presence of DNMT3A seems to abrogate the predictive value of NPM1 good molecular responses, but deserves to be addressed and confirmed in larger studies. Future efforts should focus on identifying older patients who clearly derive a survival benefit from allogeneic transplantation in their first remission. Nevertheless, new therapeutic agents may drastically change the prognosis of AML in elderly patients, especially FLT3 inhibitors (i.e., gilteritinib) and venetoclax [6,40].
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/cancers13092156/s1, Figure S1: NPM1 RT-qPCR absolute value according to FLT3-ITD and DNMT3A mutational status; Figure S2: A. Overall survival and B. Leukemia free survival of the entire cohort (N = 138); Figure S3: Leukemia free survival according to NPM1 and FLT3-ITD status; Figure S4: Leukemia free survival according to FLT3-ITD allelic ratio; Figure S5: Leukemia free survival according to DNMT3A status; Figure S6: Leukemia free survival according to DNMT3A and FLT3-ITD status; Figure S7: Leukemia free survival according NPM1 BM-MRD1; Figure S8: Leukemia free survival according NPM1 BM-MRD1 and DNMT3A status; Table S1: univariate analysis; Table S2: Overall survival and leukemia free survival according to NPM1 SS subgroups.
Author Contributions
M.H. treated patients, gathered biological and clinical data, and wrote the paper. N.D., S.H. (Sarah Huet), S.H. (Sandrine Hayette), and P.S. realized the molecular analysis. A.M., D.L., L.G., L.S., I.P., N.C., S.D.-L., M.B., G.F., H.L.-W. and F.B. treated patients. X.T. and C.P. treated patients and reviewed the paper. H.G. and C.P. gave final approval. All authors have read and agreed to the published version of the manuscript.
Funding
This work was partially funded by “Programme de Recherche Translationnelle en Cancérologie”: TRANSLA10-060, INCA-DGOS_9967 and PHRC 2007/1911.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the French Ministry of Health Ethic Committee (“Comité consultatif sur le traitement de l’information en matière de recherche dans le domaine de la santé”) (HEMILY Database, file n°08-40, 23/04/2014).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors have no conflict of interest to declare.
References
- Appelbaum, F.R.; Gundacker, H.; Head, D.R.; Slovak, M.L.; Willman, C.L.; Godwin, J.E.; Anderson, J.E.; Petersdorf, S.H. Age and acute myeloid leukemia. Blood 2006, 107, 3481–3485. [Google Scholar] [CrossRef]
- Herold, T.; Rothenberg-Thurley, M.; Grunwald, V.V.; Janke, H.; Goerlich, D.; Sauerland, M.C.; Konstandin, N.P.; Dufour, A.; Schneider, S.; Neusser, M.; et al. Validation and refinement of the revised 2017 European LeukemiaNet genetic risk stratification of acute myeloid leukemia. Leukemia 2020, 34, 3161–3172. [Google Scholar] [CrossRef] [PubMed]
- Nagel, G.; Weber, D.; Fromm, E.; Erhardt, S.; Lübbert, M.; Fiedler, W.; Kindler, T.; Krauter, J.; Brossart, P.; Kündgen, A.; et al. Epidemiological, genetic, and clinical characterization by age of newly diagnosed acute myeloid leukemia based on an academic population-based registry study (AMLSG BiO). Ann. Hematol. 2017, 96, 1993–2003. [Google Scholar] [CrossRef]
- Devillier, R.; Forcade, E.; Garnier, A.; Thepot, S.; Guillerm, G.; Hicheri, Y.; Bulabois, C.-E.; Lioure, B.; Roth Guepin, G.; Dumas, P.-Y.; et al. Allogeneic Hematopoietic Stem Cell Transplantation Improves Outcome of Elderly Patients with Acute Myeloid Leukemia in First Complete Remission: A Time-Dependent and Multistate Analysis from the French Innovative Leukemia Organization. Blood 2018, 132, 209. [Google Scholar] [CrossRef]
- Von dem Borne, P.A.; de Wreede, L.C.; Halkes, C.J.M.; Marijt, W.A.F.; Falkenburg, J.H.F.; Veelken, H. Effectivity of a strategy in elderly AML patients to reach allogeneic stem cell transplantation using intensive chemotherapy: Long-term survival is dependent on complete remission after first induction therapy. Leuk. Res. 2016, 46, 45–50. [Google Scholar] [CrossRef]
- DiNardo, C.D.; Jonas, B.A.; Pullarkat, V.; Thirman, M.J.; Garcia, J.S.; Wei, A.H.; Konopleva, M.; Döhner, H.; Letai, A.; Fenaux, P.; et al. Azacitidine and Venetoclax in Previously Untreated Acute Myeloid Leukemia. N. Engl. J. Med. 2020, 383, 617–629. [Google Scholar] [CrossRef]
- Balsat, M.; Renneville, A.; Thomas, X.; De Botton, S.; Caillot, D.; Marceau, A.; Lemasle, E.; Marolleau, J.P.; Nibourel, O.; Berthon, C.; et al. Postinduction minimal residual disease predicts outcome and benefit from allogeneic stem cell transplantation in acute myeloid leukemia with NPM1 mutation: A study by the acute leukemia French association group. J. Clin. Oncol. 2017, 35, 185–193. [Google Scholar] [CrossRef]
- Krönke, J.; Schlenk, R.F.; Jensen, K.O.; Tschürtz, F.; Corbacioglu, A.; Gaidzik, V.I.; Paschka, P.; Onken, S.; Eiwen, K.; Habdank, M.; et al. Monitoring of minimal residual disease in NPM1-mutated acute myeloid leukemia: A study from the German-Austrian acute myeloid leukemia study group. Proc. J. Clin. Oncol. 2011, 29, 2709–2716. [Google Scholar] [CrossRef] [PubMed]
- Heiblig, M.; Labussière-Wallet, H.; Nicolini, F.E.; Michallet, M.; Hayette, S.; Sujobert, P.; Plesa, A.; Balsat, M.; Paubelle, E.; Barraco, F.; et al. Prognostic value of genetic alterations in elderly patients with acute myeloid leukemia: A single institution experience. Cancers 2019, 11, 570. [Google Scholar] [CrossRef]
- Papaemmanuil, E.; Gerstung, M.; Bullinger, L.; Gaidzik, V.I.; Paschka, P.; Roberts, N.D.; Potter, N.E.; Heuser, M.; Thol, F.; Bolli, N.; et al. Genomic classification and prognosis in acute myeloid leukemia. N. Engl. J. Med. 2016, 374, 2209–2221. [Google Scholar]
- Renneville, A.; Boissel, N.; Nibourel, O.; Berthon, C.; Helevaut, N.; Gardin, C.; Cayuela, J.M.; Hayette, S.; Reman, O.; Contentin, N.; et al. Prognostic significance of DNA methyltransferase 3A mutations in cytogenetically normal acute myeloid leukemia: A study by the Acute Leukemia French Association. Leukemia 2012, 26, 1247–1254. [Google Scholar] [CrossRef]
- Döhner, H.; Estey, E.; Grimwade, D.; Amadori, S.; Appelbaum, F.R.; Büchner, T.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Larson, R.A.; et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 2017, 129, 424–447. [Google Scholar] [CrossRef]
- Gardin, C.; Pautas, C.; Fournier, E.; Itzykson, R.; Lemasle, E.; Bourhis, J.H.; Adès, L.; Marolleau, J.P.; Malfuson, J.V.; Gastaud, L.; et al. Added prognostic value of secondary AML-like gene mutations in ELN intermediate-risk older AML: ALFA-1200 study results. Blood Adv. 2020, 4, 1942–1949. [Google Scholar] [CrossRef]
- Lambert, J.; Pautas, C.; Terré, C.; Raffoux, E.; Turlure, P.; Caillot, D.; Legrand, O.; Thomas, X.; Gardin, C.; Gogat-Marchant, K.; et al. Gemtuzumab ozogamicin for de novo acute myeloid leukemia: Final efficacy and safety updates from the open-label, phase III ALFA-0701 trial. Haematologica 2019, 104, 113–119. [Google Scholar] [CrossRef]
- Fenwarth, L.; Duployez, N.; Thomas, X.; Boissel, N.; Geffroy, S.; Marceau-Renaut, A.; Caillot, D.; Raffoux, E.; Lemasle, E.; Marolleau, J.P.; et al. Clofarabine improves relapse-free survival of acute myeloid leukemia in younger adults with micro-complex karyotype. Cancers 2020, 12, 88. [Google Scholar] [CrossRef]
- Simons, A.; Shaffer, L.G.; Hastings, R.J. Cytogenetic nomenclature: Changes in the ISCN 2013 compared to the 2009 edition. Cytogenet. Genome Res. 2013, 141, 1–6. [Google Scholar] [CrossRef]
- Huet, S.; Jallades, L.; Charlot, C.; Chabane, K.; Nicolini, F.E.; Michallet, M.; Magaud, J.-P.; Hayette, S. New Quantitative Method to Identify NPM1 Mutations in Acute Myeloid Leukaemia. Leuk. Res. Treatment 2013, 2013, 756703. [Google Scholar] [CrossRef]
- Meshinchi, S.; Woods, W.G.; Stirewalt, D.L.; Sweetser, D.A.; Buckley, J.D.; Tjoa, T.K.; Bernstein, I.D.; Radich, J.P. Prevalence and prognostic significance of Flt3 internal tandem duplication in pediatric acute myeloid leukemia. Blood 2001, 97, 89–94. [Google Scholar] [CrossRef]
- Silva, P.; Neumann, M.; Schroeder, M.P.; Vosberg, S.; Schlee, C.; Isaakidis, K.; Ortiz-Tanchez, J.; Fransecky, L.R.; Hartung, T.; Türkmen, S.; et al. Acute myeloid leukemia in the elderly is characterized by a distinct genetic and epigenetic landscape. Leukemia 2017, 31, 1640–1644. [Google Scholar] [CrossRef]
- Rao, A.V.; Valk, P.J.M.; Metzeler, K.H.; Acharya, C.R.; Tuchman, S.A.; Stevenson, M.M.; Rizzieri, D.A.; Delwel, R.; Buske, C.; Bohlander, S.K.; et al. Age-specific differences in oncogenic pathway dysregulation and anthracycline sensitivity in patients with acute myeloid leukemia. J. Clin. Oncol. 2009, 27, 5580–5586. [Google Scholar] [CrossRef]
- Gorello, P.; Cazzaniga, G.; Alberti, F.; Dell’Oro, M.G.; Gottardi, E.; Specchia, G.; Roti, G.; Rosati, R.; Martelli, M.F.; Diverio, D.; et al. Quantitative assessment of minimal residual disease in acute myeloid leukemia carrying nucleophosmin (NPM1) gene mutations. Leukemia 2006, 20, 1103–1108. [Google Scholar] [CrossRef]
- Schnittger, S.; Schoch, C.; Kern, W.; Mecucci, C.; Tschulik, C.; Martelli, M.F.; Haferlach, T.; Hiddemann, W.; Falini, B. Nucleophosmin gene mutations are predictors of favorable prognosis in acute myelogenous leukemia with a normal karyotype. Blood 2005, 106, 3733–3739. [Google Scholar] [CrossRef] [PubMed]
- Whitman, S.P.; Maharry, K.; Radmacher, M.D.; Becker, H.; Mrózek, K.; Margeson, D.; Holland, K.B.; Wu, Y.Z.; Schwind, S.; Metzeler, K.H.; et al. FLT3 internal tandem duplication associates with adverse outcome and gene- and microRNA-expression signatures in patients 60 years of age or older with primary cytogenetically normal acute myeloid leukemia: A cancer and leukemia group B study. Blood 2010, 116, 3622–3626. [Google Scholar] [CrossRef]
- Straube, J.; Ling, V.Y.; Hill, G.R.; Lane, S.W. The impact of age, NPM1 mut, and FLT3 ITD allelic ratio in patients with acute myeloid leukemia. Blood 2018, 131, 1148–1153. [Google Scholar] [CrossRef] [PubMed]
- Gabert, J.; Beillard, E.; van der Velden, V.H.J.; Bi, W.; Grimwade, D.; Pallisgaard, N.; Barbany, G.; Cazzaniga, G.; Cayuela, J.M.; Cave, H.; et al. Standardization and quality control studies of ‘real-time’ quantitative reverse transcriptase polymerase chain reaction of fusion gene transcripts for residual disease detection in leukemia—A Europe Against Cancer program. Leukemia 2003, 17, 2318–2357. [Google Scholar] [CrossRef]
- Foran, J.M.; Sun, Z.; Paietta, E.; Racevskis, J.; Claxton, D.F.; Lazarus, H.M.; Arber, D.A.; Rowe, J.M.; Godwin, J.; Zhang, Y.; et al. FLT3-ITD Mutations Are Prevalent and Significantly Impact Outcome after Intensive Therapy in Elderly Adults with Acute Myeloid Leukemia (AML): Analysis of the North American Intergroup E2906 Phase III Trial in Patients Age ≥60 Years. Blood 2018, 132, 3995. [Google Scholar] [CrossRef]
- Cappelli, L.V.; Meggendorfer, M.; Dicker, F.; Jeromin, S.; Hutter, S.; Kern, W.; Haferlach, T.; Haferlach, C.; Höllein, A. DNMT3A mutations are over-represented in young adults with NPM1 mutated AML and prompt a distinct co-mutational pattern. Leukemia 2019, 33, 2741–2746. [Google Scholar] [CrossRef] [PubMed]
- Ley, T.J.; Ding, L.; Walter, M.J.; McLellan, M.D.; Lamprecht, T.; Larson, D.E.; Kandoth, C.; Payton, J.E.; Baty, J.; Welch, J.; et al. DNMT3A mutations in acute myeloid leukemia. N. Engl. J. Med. 2010, 363, 2424–2433. [Google Scholar] [CrossRef]
- Hájková, H.; Marková, J.; Haškovec, C.; Šárová, I.; Fuchs, O.; Kostečka, A.; Cetkovský, P.; Michalová, K.; Schwarz, J. Decreased DNA methylation in acute myeloid leukemia patients with DNMT3A mutations and prognostic implications of DNA methylation. Leuk. Res. 2012, 36, 1128–1133. [Google Scholar] [CrossRef]
- Emperle, M.; Adam, S.; Kunert, S.; Dukatz, M.; Baude, A.; Plass, C.; Rathert, P.; Bashtrykov, P.; Jeltsch, A. Mutations of R882 change flanking sequence preferences of the DNA methyltransferase DNMT3A and cellular methylation patterns. Nucleic Acids Res. 2019, 47, 11355–11367. [Google Scholar] [CrossRef]
- Russler-Germain, D.A.; Spencer, D.H.; Young, M.A.; Lamprecht, T.L.; Miller, C.A.; Fulton, R.; Meyer, M.R.; Erdmann-Gilmore, P.; Townsend, R.R.; Wilson, R.K.; et al. The R882H DNMT3A Mutation Associated with AML Dominantly Inhibits Wild-Type DNMT3A by Blocking Its Ability to Form Active Tetramers. Cancer Cell 2014, 25, 442–454. [Google Scholar] [CrossRef]
- Guryanova, O.A.; Shank, K.; Spitzer, B.; Luciani, L.; Koche, R.P.; Garrett-Bakelman, F.E.; Ganzel, C.; Durham, B.H.; Mohanty, A.; Hoermann, G.; et al. DNMT3A mutations promote anthracycline resistance in acute myeloid leukemia via impaired nucleosome remodeling. Nat. Med. 2016, 22, 1488–1495. [Google Scholar] [CrossRef]
- Hubmann, M.; Köhnke, T.; Hoster, E.; Schneider, S.; Dufour, A.; Zellmeier, E.; Fiegl, M.; Braess, J.; Bohlander, S.K.; Subklewe, M.; et al. Molecular response assessment by quantitative real-time polymerase chain reaction after induction therapy in NPM1-mutated patients identifies those at high risk of relapse. Haematologica 2014, 99, 1317–1325. [Google Scholar] [CrossRef] [PubMed]
- Schnittger, S.; Kern, W.; Tschulik, C.; Weiss, T.; Dicker, F.; Falini, B.; Haferlach, C.; Haferlach, T. Minimal residual disease levels assessed by NPM1 mutation-specific RQ-PCR provide important prognostic information in AML. Blood 2009, 114, 2220–2231. [Google Scholar] [CrossRef]
- Shayegi, N.; Kramer, M.; Bornhäuser, M.; Schaich, M.; Schetelig, J.; Platzbecker, U.; Röllig, C.; Heiderich, C.; Landt, O.; Ehninger, G.; et al. The level of residual disease based on mutant NPM1 is an independent prognostic factor for relapse and survival in AML. Blood 2013, 122, 83–92. [Google Scholar] [CrossRef]
- Ivey, A.; Hills, R.K.; Simpson, M.A.; Jovanovic, J.V.; Gilkes, A.; Grech, A.; Patel, Y.; Bhudia, N.; Farah, H.; Mason, J.; et al. Assessment of minimal residual disease in standard-risk AML. N. Engl. J. Med. 2016, 374, 422–433. [Google Scholar] [CrossRef]
- Kristensen, T.; Møller, M.B.; Friis, L.; Bergmann, O.J.; Preiss, B. NPM1 mutation is a stable marker for minimal residual disease monitoring in acute myeloid leukaemia patients with increased sensitivity compared to WT1 expression. Eur. J. Haematol. 2011, 87, 400–408. [Google Scholar] [CrossRef]
- Ommen, H.B.; Schnittger, S.; Jovanovic, J.V.; Ommen, I.B.; Hasle, H.; Østergaard, M.; Grimwade, D.; Hokland, P. Strikingly different molecular relapse kinetics in NPM1c, PML-RARA, RUNX1-RUNX1T1, and CBFB-MYH11 acute myeloid leukemias. Blood 2010, 115, 198–205. [Google Scholar] [CrossRef] [PubMed]
- Bezerra, M.F.; Lima, A.S.; Piqué-Borràs, M.R.; Silveira, D.R.; Coelho-Silva, J.L.; Pereira-Martins, D.A.; Weinhäuser, I.; Franca-Neto, P.L.; Quek, L.; Corby, A.; et al. Co-occurrence of DNMT3A, NPM1, FLT3 mutations identifies a subset of acute myeloid leukemia with adverse prognosis. Blood 2020, 135, 870–875. [Google Scholar] [CrossRef] [PubMed]
- Perl, A.E.; Martinelli, G.; Cortes, J.E.; Neubauer, A.; Berman, E.; Paolini, S.; Montesinos, P.; Baer, M.R.; Larson, R.A.; Ustun, C.; et al. Gilteritinib or chemotherapy for relapsed or refractory FLT3-mutated AML. N. Engl. J. Med. 2019, 381, 1728–1740. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).