Simple Summary
The aim of the present meta-analysis was to analyze all available studies reporting clinical characteristics of breast cancer gene 1 (BRCA1) gene hypermethylated breast cancer in women, and to pool the results in order to provide a unique clinical profile of this cancer setting population. Identifying the clinical profile of breast cancer in women harboring BRCA1 gene hypermethylation may help oncologists select a subgroup of patients who may be candidates for BRCA1 methylation assessment, thus, possibly enlarging the cancer population who may benefit from new target-therapy agents. Results showed that BRCA1 gene hypermethylation should be suspected in all breast cancer patients with advanced disease stages, positive lymph nodes, and premenopausal age at diagnosis. Multidisciplinary groups treating women with breast cancer should take into account the possibility of addressing patients with these characteristics with a BRCA1 gene methylation status analysis.
Abstract
Background: DNA aberrant hypermethylation is the major cause of transcriptional silencing of the breast cancer gene 1 (BRCA1) gene in sporadic breast cancer patients. The aim of the present meta-analysis was to analyze all available studies reporting clinical characteristics of BRCA1 gene hypermethylated breast cancer in women, and to pool the results to provide a unique clinical profile of this cancer population. Methods: On September 2020, a systematic literature search was performed. Data were retrieved from PubMed, MEDLINE, and Scopus by searching the terms: “BRCA*” AND “methyl*” AND “breast”. All studies evaluating the association between BRCA1 methylation status and breast cancer patients’ clinicopathological features were considered for inclusion. Results: 465 studies were retrieved. Thirty (6.4%) studies including 3985 patients met all selection criteria. The pooled analysis data revealed a significant correlation between BRCA1 gene hypermethylation and advanced breast cancer disease stage (OR = 0.75: 95% CI: 0.58–0.97; p = 0.03, fixed effects model), lymph nodes involvement (OR = 1.22: 95% CI: 1.01–1.48; p = 0.04, fixed effects model), and pre-menopausal status (OR = 1.34: 95% CI: 1.08–1.66; p = 0.008, fixed effects model). No association could be found between BRCA1 hypermethylation and tumor histology (OR = 0.78: 95% CI: 0.59–1.03; p = 0.08, fixed effects model), tumor grading (OR = 0.78: 95% CI :0.46–1.32; p = 0.36, fixed effects model), and breast cancer molecular classification (OR = 1.59: 95% CI: 0.68–3.72; p = 0.29, random effects model). Conclusions: hypermethylation of the BRCA1 gene significantly correlates with advanced breast cancer disease, lymph nodes involvement, and pre-menopausal cancer onset.
1. Introduction
Breast cancer gene 1 (BRCA1) encodes a polyfunctional protein responsible for DNA repair, cell cycle control, protein ubiquitinoylation, and chromatin remodeling [1,2]. Germline mutations of BRCA1 are known to cause up to 45% of familial breast cancer, while germline aberrations in BRCA1 gene DNA sequences are involved in only 1% of sporadic breast cancer onsets [3]. Nevertheless, BRCA1 gene silencing was found to be a very frequent event in sporadic breast cancer, and it has been correlated with its progression and overall survival [4]. Methylation of CpG islands is an epigenetic mechanism involved in gene silencing. DNA aberrant hypermethylation was observed to be the major cause of transcriptional silencing of the BRCA1 gene, a phenomenon ranging from 13% to 40% in sporadic breast cancer [5,6].
While the clinicopathological characterization of germline BRCA1 mutated breast cancer has been well defined [7], there is still little known in regards to the pathological signature of BRCA1 gene hypermethylated breast cancer in women. During the last 15 years, several studies attempted to trace a clinical profile of these patients’ subset, but no unique results have been reported. Identifying the clinical features of BRCA1 gene hypermethylation in breast cancer patients is currently considered a major open question, since the definition of the clinical traits of this molecular signature could help oncologists to address methylated patients with dedicated treatment options and follow-up. In 2020, Kawachi et al. published the results of the first clinical trial reporting that human breast cancers with BRCA1 methylation showed a clinical response to PARP-inhibitors [8]. This is in line with what is already known in women with BRCA methylated ovarian cancer, who are currently suitable for treatment with Parp-Inhibitors even in front-line therapy [9]. This evidence has the potential to represent the paradigm shift of breast cancer treatment, by enlarging the cancer population who could benefit from these new targeted agents. In this scenario, the aim of the present meta-analysis was to analyze all available studies reporting clinical characteristics of BRCA1 gene hypermethylated breast cancer in women, and to pool the results in order to provide a unique clinical profile of this cancer setting population.
2. Materials and Methods
2.1. Data Identification and Selection
The present meta-analysis was carried out following the Preferred Reporting Items for Systematic reviews and Meta-Analyses (PRISMA) statement and included all studies without any restriction on publication year. On September 2020, a systematic literature search was performed. Data were retrieved from the electronic databases, PubMed, MEDLINE, and Scopus, by searching the terms: “BRCA*” AND “methyl*” AND “breast”. All English language original reports evaluating the association between BRCA1 gene methylation status and breast cancer patients’ clinicopathological features were considered for inclusion.
The reference list of original reports and reviews already published was also analyzed to identify other potential studies.
Review articles, case reports, editorials, and letters were excluded. Based on inclusion and exclusion criteria, two independent reviewers (IR and MLG) identified and selected the studies. Differences in the studies’ selection were resolved asking a third author (DC).
For each study included in the meta-analysis, the following data were recorded: first author’s information, publication year, study design, detection method, criteria to define BRCA1 gene hypermethylation for dichotomized methylation status (“hyper-” vs. “hypo-methylated”), sample size, percentage of BRCA1 gene hypermethylated cases, tumor histology, stage, grading, molecular classification, lymph nodal status, and patients’ menopausal status (pre- vs. postmenopausal women).
2.2. Endpoints
The primary endpoint was the association between BRCA1 gene methylation status and patients’ clinicopathological features, including tumor histology, stage, grading, lymph nodal status, tumor molecular classification, and patients’ menopausal status.
2.3. Statistical Analysis
The number of BRCA1 gene hypermethylated cases detected in association with each clinicopathological variable were stratified by studies, and the pooled odds ratio (OR) was calculated using a fixed- or a random-effects model. A χ2 test for heterogeneity among proportions was performed to assess the presence of statistical heterogeneity between studies. A fixed-effects model was applied in case statistical heterogeneity was not significant (I2 value ≤ 50%); differently, a random-effects model was used. Graphical representation of each study and pooled analysis were displayed by forest plots. The weight that each study provides in the meta-analysis was graphically reported as squares of different size. Confidence intervals (CIs) for each study were symbolized by the horizontal lines passing through the squares. The pooled OR was represented as a lozenge in the forest plot, and its size corresponded to the 95% CI of the OR. A p value ≤ 0.05 was considered significant.
Statistical analysis was performed using Review Manager 5.4 (http://www.cochrane.org (accessed on 16 March 2021)).
3. Results
In total, 465 studies were retrieved through the literature search. Among these, 13 (2.8%) studies were removed as duplicates. A further 397 papers (85.4%) were excluded after title and abstract evaluation, being non-English-language original reports, studies not regarding breast cancers, studies performed on animals, review papers, or studies not evaluating the BRCA1 gene methylation status. Twenty-five (5.4%) studies were successively excluded after full-text evaluation: 13 were excluded because extrapolation of BRCA1 gene methylation status was not deducible [10,11,12,13,14,15,16,17,18,19,20,21,22]; seven other papers were excluded because no patient’s clinicopathological characteristics were reported [23,24,25,26,27,28,29]; four studies were eliminated for the coexistence of both the previous reasons [30,31,32,33]; one paper was excluded for the reanalysis of a study population that was previously reported [34]. Thirty (6.4%) studies remained for comparison at the end of the selection process. The PRISMA flow chart summarizing the process of evidence acquisition was shown in Figure 1. The flow chart maps out the number of studies identified, screened, included, and excluded, as well as the reasons for exclusions.
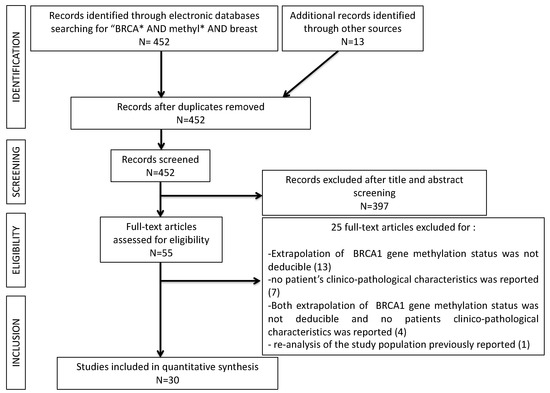
Figure 1.
PRISMA flow-chart of the study selection process.
Globally, the total number of patients included in the meta-analysis was 3985, ranging from 26 to 851 patients per study. Thirteen [35,36,37,38,39,40,41,42,43,44,45,46,47], 12 [36,39,44,45,47,48,49,50,51,52,53,54], 10 [40,42,43,44,47,49,54,55,56,57], 18 [34,36,38,39,41,44,45,46,47,48,49,50,51,52,55,56,58,59], 9 [38,43,45,49,51,52,56,60,61], and 12 [34,35,38,45,47,49,50,51,52,58,62,63] studies reported data regarding the association between BRCA1 gene methylation status and breast cancer histology, disease stage, tumor grading, patients’ lymph nodal status, disease molecular classification, and patients’ menopausal status, respectively.
BRCA1 gene methylation status was evaluated by methylation-specific PCR (MS-PCR) in 23 studies [34,36,37,38,40,41,42,43,44,45,47,48,51,52,53,54,55,56,58,59,61,62,63]. The remaining seven studies adopted Southern Blots [35], PCR [57], MS-MLPA (Methylation-Specific Multiplex Ligation-dependent Probe Amplification) [39], Bisulfite sequencing PCR [50], combined bisulfite and restriction analysis (COBRA) [46], percentage of relative methylation (PRM) [60], and REMS-PCR (Restriction endonuclease-mediated selective PCR) [49] as a gene methylation detecting method, respectively. The main characteristics of the selected studies were listed in Table 1.

Table 1.
Characteristics of the included studies.
3.1. Correlation of BRCA1 Gene Methylation Status with Clinico-Pathological Variables
The pooled analysis data revealed a significant correlation between BRCA1 gene hypermethylation and advanced breast cancer disease stage, (OR = 0.75: 95% CI: 0.58–0.97; p = 0.03, fixed effects model, Figure 2b), lymph nodes involvement (OR = 1.22: 95% CI: 1.01–1.48; p = 0.04, fixed effects model, Figure 2d), and pre-menopausal status (OR = 1.34: 95% CI: 1.08–1.66; p = 0.008, fixed effects model, Figure 2f). On the contrary, no association could be found between BRCA1 gene hypermethylation and tumor histology (OR = 0.78: 95% CI: 0.59–1.03; p = 0.08, fixed effects model, Figure 2a), tumor grading (OR = 0.78: 95% CI: 0.46–1.32; p = 0.36, fixed effects model, Figure 2c), and breast cancer molecular classification (OR = 1.59: 95% CI: 0.68–3.72; p = 0.29, random effects model, Figure 2e).
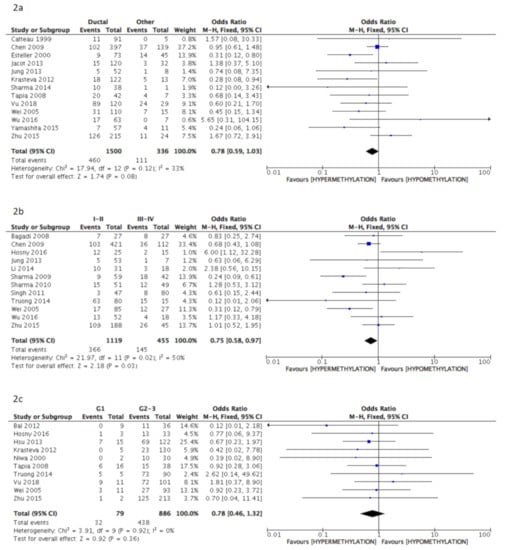
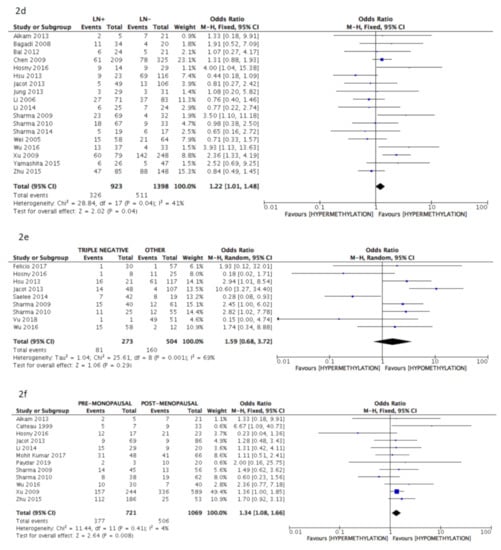
Figure 2.
Pooled results on forest plots about the correlation between breast cancer gene 1 (BRCA1) gene methylation status with breast cancer patients’ clinicopathological characteristics. Events= number of BRCA1 gene hypermethylated cases. Figure shows the correlation between BRCA1 gene methylation and histology (2a), disease stage (2b), tumor grading (2c), lymph nodal status (2d), molecular classification (2e) and menopausal status (2f).
Pooled Results of Studies Adopting Only the Methylation-Specific PCR (MS-PCR) Methodology for Detection of BRCA1 Methylation Status
As MS-PCR was the most adopted (by 23/30 included studies, 76.7%) methodology for the detection of BRCA1 methylation status, a subanalysis pooling only the results obtained by studies applying MS-PCR was carried out, in order to assess potential differences in results due to multiple methylation detection methods.
The pooled data of the subanalysis confirmed a significant correlation between BRCA1 gene hypermethylation and advanced breast cancer disease stage, (OR = 0.66: 95% CI: 0.50–0.87; p = 0.003, fixed effects model, Figure 3b) as well as a positive correlation with patients’ pre-menopausal status (OR = 1.35: 95% CI: 1.08–1.69; p = 0.009, fixed effects model, Figure 3f), but lymph nodes involvement was not observed to be correlated with BRCA1 hypermethylation (OR = 1.18: 95% CI: 0.97–1.45; p = 0.10, fixed effects model, Figure 3d). Additionally, in this case, no association was found between BRCA1 gene hypermethylation and tumor histology (OR = 0.81: 95% CI: 0.60–1.07; p = 0.14, fixed effects model, Figure 3a), tumor grading (OR = 0.80: 95% CI: 0.46–1.39; p = 0.43, fixed effects model, Figure 3c), and breast cancer molecular classification (OR = 1.93: 95% CI: 0.79–4.72; p = 0.15, random effects model, Figure 3e).
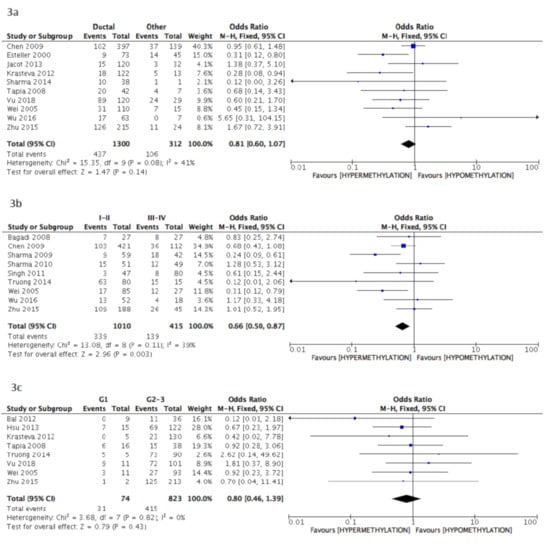
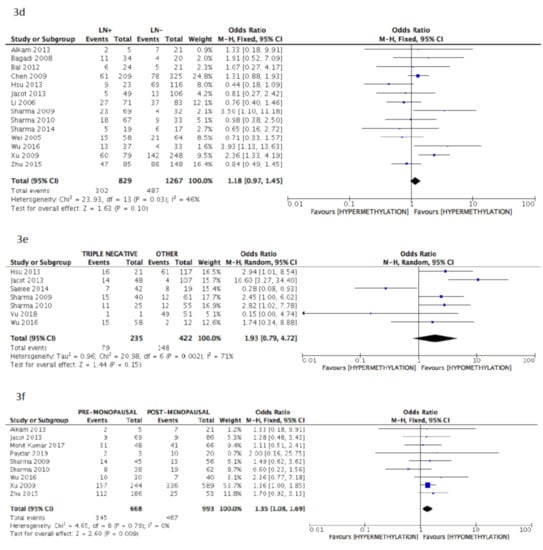
Figure 3.
Pooled results on forest plots about the correlation between BRCA1 gene methylation status with breast cancer patients’ clinicopathological characteristics. In this subanalysis, only studies adopting methylation-specific PCR (MS-PCR) methodology for the detection of BRCA1 methylation status were included. Events= number of BRCA1 gene hypermethylated cases. Figure shows the correlation between BRCA1 gene methylation and histology (3a), disease stage (3b), tumor grading (3c), lymph nodal status (3d), molecular classification (3e) and menopausal status (3f).
4. Discussion
“Molecular tumor boards” have increasing become a reality worldwide for referral multidisciplinary oncologic centers, aiming to personalize cancer treatments for each single patient [64]. In this context, breast cancer has been a pioneering model for integrating surgery, oncology, histology, and genetics with molecular biology, with “Breast Units” being the most successful example of an integrated approach to cancer patients, under the leading principle “from the bench to the bedside”.
So far, huge advances have been carried out in the clinical application of molecular biology, and we are now facing the challenging step of going over cancer biology and cancer genetics by considering epigenetics as an essential part of cancer diagnostics and therapy [65].
Epigenetics groups post-transcriptional modifications of the genetic information undertaken basically through the DNA methylation phenomenon. DNA methylation refers to the addition of a methyl group (CH3) to the cytosine residue of a cytosine–guanidine pair, a CpG dinucleotide, in the DNA sequence. DNA methylation is a pivotal mechanism in early development, the so called “epigenetic reprogramming” event [66]. In adult cells, DNA methylation has been extensively demonstrated to be involved in the onset and progression of cancer, mainly through the silencing of tumor suppressor genes such as BRCA1, ATM, and PALB2 [67,68,69].
Up to now, in breast cancer patients, the investigation of BRCA1/2 gene epigenetic silencing has not routinely been included into the clinical algorithm of patients’ profiling and therapeutic approach. This currently limits the patients’ access to a wider platform of biological treatment, such as parp-inhibitors [8].
Identifying the clinical profile of breast cancer women harboring BRCA1 gene hypermethylation may help oncologists to select the subgroup of patients who may be candidates for BRCA1 methylation assessment, thus, possibly enlarging the cancer population who may benefit from new target-therapy agents.
This meta-analysis showed that BRCA1 gene hypermethylation in breast cancer significantly correlated with advanced disease stage, lymph nodal involvement, and pre-menopausal age at diagnosis. On the contrary, triple negative molecular classification, as well as advanced tumor grading and ductal histology, were not found to be more represented in the pooled group of BRCA1 hypermethylated breast cancer patients.
To our knowledge, the present meta-analysis is the first study which systematically investigates the role of BRCA1 gene hypermethylation in breast cancer patients’ clinicopathological characteristics. Involving 3985 patients, our results can be considered more reliable than those reported in each of single included studies. Nevertheless, our findings may be accompanied by some limitations. First, the definition of “hypermethylated” BRCA1 gene was heterogeneous among studies.
Second, the techniques applied for the detection of BRCA1 methylation status included seven different methodologies (MS-PCR, Southern Blots, PCR, MS-MLPA, Bisulfite sequencing PCR, combined bisulfite and restriction analysis (COBRA), percentage of relative methylation (PRM), and REMS-PCR). As the global debate concerning the attempt to identify a unique technique for the detection BRCA1 methylation status still ongoing, the authors decided to include in the present meta-analysis the studies carried out with all currently adopted technologies, each considering different definitions of a hypermethylated BRCA1 gene. Nevertheless, a subanalysis pooling only the studies adopting MS-PCR, as the methodology applied in 77% of included studies, was carried out, substantially confirming the result of the global pooled analysis. Until the international scientific community defines the most appropriate methodology for the assessment of BRCA methylation status, which will be accompanied by a unique laboratory definition of hypermethylated BRCA, it will not be possible to draw definitive conclusions on the clinical profile of this molecular signature.
Third, the study design of the included reports was retrospective in the vast majority of cases. Prospective studies on BRCA1 gene methylation status on a large cohort of breast cancer women was strongly awaited.
5. Conclusions
The present meta-analysis showed that BRCA1 gene hypermethylation should be suspected in all breast cancer patients with advanced disease stages, positive lymph nodes, and premenopausal age at diagnosis. Multidisciplinary groups treating women with breast cancer should take into account the possibility of addressing patients with these characteristics with a BRCA1 gene methylation status analysis.
No association between BRCA1 methylation status and other clinicopathological variables (such as tumor histology, tumor grading, and tumor molecular classification) was identified.
Larger sample-size prospective studies and a unanimous methodology to determine BRCA gene methylation status, as well as a unique laboratory definition of BRCA1 hypermethylated cases, will help in future to draw definitive conclusions about the clinical signature of BRCA1 gene methylated breast cancer in women.
Author Contributions
Conceptualization, I.R. and M.L.G.; methodology, I.R. and M.L.G.; software, M.P.D.M., F.C. and A.R.B.; validation, A.P., T.K. and O.D.G.; formal analysis, I.R. and M.L.G.; investigation, F.B.; resources, D.C.; data curation, I.R. and M.L.G.; writing—original draft preparation, I.R., M.L.G., M.P.D.M. and F.C.; writing—review and editing, all coauthors.; supervision, D.C.; project administration, D.C. All authors have read and agreed to the published version of the manuscript.
Funding
No funding.
Conflicts of Interest
All authors declare no conflict of interest.
References
- Somasundaram, K. Breast cancer gene 1 (BRCA1): Role in cell cycle regulation and DNA repair—perhaps through transcrip-tion. J. Cell Biochem. 2003, 15, 1084–1091. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Lee, W.-H.; Chew, H.K. Emerging roles of BRCA1 in transcriptional regulation and DNA repair. J. Cell. Physiol. 1999, 181, 385–392. [Google Scholar] [CrossRef]
- Van Der Looij, M.; Szabo, C.; Besznyak, I.; Liszka, G.; Csokay, B.; Pulay, T.; Toth, J.; Devilee, P.; King, M.C.; Olah, E. Preva-lence of founder BRCA1 and BRCA2 mutations among breast and ovarian cancer patients in Hungary. Int. J. Cancer 2000, 86, 737–740. [Google Scholar] [CrossRef]
- Thompson, M.E.; Jensen, R.A.; Obermiller, P.S.; Page, D.L.; Holt, J.T. Decreased expression of BRCA1 accelerates growth and is often present during sporadic breast cancer progression. Nat. Genet. 1995, 9, 444–450. [Google Scholar] [CrossRef] [PubMed]
- Jerónimo, C.; Costa, I.; Martins, M.C.; Monteiro, P.; Lisboa, S.; Palmeira, C.; Henrique, R.; Teixeira, M.R.; Lopes, C. Detection of gene promoter hypermethylation in fine needle washings from breast lesions. Clin. Cancer Res. 2003, 9, 3413–3417. [Google Scholar] [PubMed]
- Jing, F.; Zhang, J.; Tao, J.; Zhou, Y.; Jun, L.; Tang, X.; Wang, Y.; Hai, H. Hypermethylation of tumor suppressor genes BRCA1, p16 and 14–3-3sigma in serum of sporadic breast cancer patients. Onkologie 2007, 30, 14–19. [Google Scholar]
- Kim, E.-K.; Park, S.Y.; Kim, S.-W. Clinicopathological characteristics of BRCA-associated breast cancer in Asian patients. J. Pathol. Transl. Med. 2020, 54, 265–275. [Google Scholar] [CrossRef]
- Kawachi, A.; Yamashita, S.; Okochi-Takada, E.; Hirakawa, A.; Tsuda, H.; Shimomura, A.; Kojima, Y.; Yonemori, K.; Fujiwara, Y.; Kinoshita, T.; et al. BRCA1 promoter methylation in breast cancer patients is associated with response to olaparib/eribulin combination therapy. Breast Cancer Res. Treat. 2020, 181, 323–329. [Google Scholar] [CrossRef]
- Ray-Coquard, I.; Pautier, P.; Pignata, S.; Pérol, D.; González-Martín, A.; Berger, R.; Fujiwara, K.; Vergote, I.; Colombo, N.; Mäenpää, J.; et al. Olaparib plus Bevacizumab as First-Line Maintenance in Ovarian Cancer. N. Engl. J. Med. 2019, 381, 2416–2428. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, Y.; Salas, L.A.; Miller, T.W.; Mark, K.; Marotti, J.D.; Kettenbach, A.N.; Cheng, C.; Christensen, B.C. Molecular and epigenetic profiles of BRCA1-like hormone-receptor-positive breast tumors identified with development and application of a copy-number-based classifier. Breast Cancer Res. 2019, 21, 14. [Google Scholar] [CrossRef]
- Salta, S.; Nunes, S.P.; Fontes-Sousa, M.; Lopes, P.; Freitas, M.; Caldas, M.; Antunes, L.; Castro, F.; Antunes, P.; De Sousa, S.P.; et al. A DNA Methylation-Based Test for Breast Cancer Detection in Circulating Cell-Free DNA. J. Clin. Med. 2018, 7, 420. [Google Scholar] [CrossRef]
- Kozomara, Z.; Supic, G.; Krivokuca, A.; Magic, Z.; Dzodic, R.; Milovanovic, Z.; Brankovic-Magic, M. Promoter hypermethylation of p16, BRCA1 and RASSF1A genes in triple-negative breast cancer patients from Serbia. J. BUON 2018, 23, 684–691. [Google Scholar]
- Shilpi, A.; Bi, Y.; Jung, S.; Patra, S.K.; Davuluri, R.V. Identification of Genetic and Epigenetic Variants Associated with Breast Cancer Prognosis by Integrative Bioinformatics Analysis. Cancer Informatics 2017, 16, CIN-S39783. [Google Scholar] [CrossRef]
- Murria Estal, R.; Palanca Suela, S.; de Juan Jiménez, I.; Alenda Gonzalez, C.; Egoavil Rojas, C.; García-Casado, Z.; López Guerrero, J.A.; Juan Fita, M.J.; Sánchez Heras, A.B.; Segura Huerta, Á.; et al. Relationship of immunohistochemistry, copy number aberrations and epigenetic disorders with BRCAness pattern in hereditary and sporadic breast cancer. Fam. Cancer 2016, 15, 193–200. [Google Scholar] [CrossRef]
- Watanabe, Y.; Maeda, I.; Oikawa, R.; Wu, W.; Tsuchiya, K.; Miyoshi, Y.; Itoh, F.; Tsugawa, K.-I.; Ohta, T. Aberrant DNA methylation status of DNA repair genes in breast cancer treated with neoadjuvant chemotherapy. Genes Cells 2013, 18, 1120–1130. [Google Scholar] [CrossRef]
- Yu, Z.; Xiao, Q.; Zhao, L.; Ren, J.; Bai, X.; Sun, M.; Wu, H.; Liu, X.; Song, Z.; Yan, Y.; et al. DNA methyltransferase 1/3a overexpression in sporadic breast cancer is associated with reduced expression of estrogen receptor-alpha/breast cancer susceptibility gene 1 and poor prognosis. Mol. Carcinog. 2015, 54, 707–719. [Google Scholar] [CrossRef]
- Klajic, J.; Fleischer, T.; Dejeux, E.; Edvardsen, H.; Wärnberg, F.; Bukholm, I.; Lonning, P.E.; Solvang, H.; Børresen-Dale, A.-L.; Tost, J.; et al. Quantitative DNA methylation analyses reveal stage dependent DNA methylation and association to clinico-pathological factors in breast tumors. BMC Cancer 2013, 13, 456. [Google Scholar] [CrossRef]
- Al-Moundhri, M.S.; Al-Ansari, A.; Al-Mawali, K.; Al-Bahrani, B. BRCA1 gene Molecular Alterations in Omani Breast Cancer Patients. Gulf. J. Oncol. 2013, 1, 45–51. [Google Scholar]
- Moelans, C.B.; Van Diest, P.J.; Verschuur-Maes, A.H. Frequent promoter hypermethylation of BRCA2, CDH13, MSH6, PAX5, PAX6 and WT1 in ductal carcinoma in situ and invasive breast cancer. J. Pathol. 2011, 225, 222–231. [Google Scholar] [CrossRef]
- Bae, Y.K.; Brown, A.; Garrett, E.; Bornman, D.; Fackler, M.J.; Sukumar, S.; Herman, J.G.; Gabrielson, E. Hypermethylation in Histologically Distinct Classes of Breast Cancer. Clin. Cancer Res. 2004, 10, 5998–6005. [Google Scholar] [CrossRef]
- Vos, S.; Moelans, C.B.; Van Diest, P.J. BRCA promoter methylation in sporadic versus BRCA germline mutation-related breast cancers. Breast Cancer Res. 2017, 19, 64. [Google Scholar] [CrossRef]
- Gacem, M.; Hachana, S.; Ziadi, K.; Amara, F.; Ksia, F.; Mokni, M.; Trimeche, M. Contribution of epigenetic alteration of BRCA1 and BRCA2 genes in breast carcinomas in Tunisian patients. Cancer Epidemiol. 2012, 2, 190–197. [Google Scholar] [CrossRef]
- Scott, C.M.; Joo, J.E.; O’Callaghan, N.; Buchanan, D.D.; Clendenning, M.; Giles, G.G.; Hopper, J.L.; Wong, E.M.; Southey, M.C. Methylation of Breast Cancer Predisposition Genes in Early-Onset Breast Cancer: Australian Breast Cancer Family Registry. PLoS ONE 2016, 11, e0165436. [Google Scholar] [CrossRef]
- Daniels, S.L.; Burghel, G.J.; Chambers, P.; Al-Baba, S.; Connley, D.D.; Brock, I.W.; Cramp, H.E.; Dotsenko, O.; Wilks, O.; Wyld, L.; et al. Levels of DNA Methylation Vary at CpG Sites across the BRCA1 Promoter, and Differ According to Triple Negative and “BRCA-Like” Status, in Both Blood and Tumour DNA. PLoS ONE 2016, 11, e0160174. [Google Scholar] [CrossRef]
- Judes, G.; Dagdemir, A.; Karsli-Ceppioglu, S.; Lebert, A.; Echegut, M.; Ngollo, M.; Bignon, Y.-J.; Penault-Llorca, F.; Bernard-Gallon, D. H3K4 acetylation, H3K9 acetylation and H3K27 methylation in breast tumor molecular subtypes. Epigenomics 2016, 8, 909–924. [Google Scholar] [CrossRef]
- Romagnolo, D.F.; Papoutsis, A.J.; Laukaitis, C.; Selmin, O.I. Constitutive expression of AhR and BRCA-1 promoter CpG hy-permethylation as biomarkers of ERα-negative breast tumorigenesis. BMC Cancer 2015, 15, 1–11. [Google Scholar] [CrossRef]
- Magdinier, F.; Ribieras, S.; Lenoir, G.M.; Frappart, L.; Dante, R. Down-regulation of BRCA1 in human sporadic breast cancer; analysis of DNA methylation patterns of the putative promoter region. Oncogene 1998, 17, 3169–3176. [Google Scholar] [CrossRef]
- Stefansson, O.A.; Jonasson, J.G.; Olafsdottir, K.; Hilmarsdottir, H.; Olafsdottir, G.; Esteller, M.; Johannsson, O.T.; Eyfjord, J.E. CpG island hypermethylation ofBRCA1and loss of pRb as co-occurring events in basal/triple-negative breast cancer. Epigenetics 2011, 6, 638–649. [Google Scholar] [CrossRef]
- Cai, F.F.; Chen, S.; Wang, M.H.; Lin, X.Y.; Zhang, L.; Zhang, J.X.; Wang, L.X.; Yang, J.; Ding, J.H.; Pan, X.; et al. Pyrosequencing quantified methylation level of BRCA1 promoter as prognostic factor for survival in breast cancer patient. Oncotarget 2016, 7, 27499–27510. [Google Scholar] [CrossRef]
- Huang, K.T.; Mikeska, T.; Li, J.; Takano, E.A.; Millar, E.K.; Graham, P.H.; Boyle, S.E.; Campbell, I.G.; Speed, T.P.; Dobrovic, A.; et al. Assessment of DNA methylation profiling and copy number variation as indications of clonal relationship in ip-silateral and contralateral breast cancers to distinguish recurrent breast cancer from a second primary tumour. BMC Cancer 2015, 15, 1–11. [Google Scholar] [CrossRef][Green Version]
- Moelans, C.B.; Vlug, E.J.; Ercan, C.; Bult, P.; Buerger, H.; Cserni, G.; van Diest, P.J.; Derksen, P.W. Methylation biomarkers for pleomorphic lobular breast cancer—a short report. Cell. Oncol. 2015, 38, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Murria, R.; Palanca, S.; De Juan, I.; Alenda, C.; Egoavil, C.; Seguí, F.J.; García-Casado, Z.; Juan, M.J.; Sánchez, A.B.; Segura, Á.; et al. Immunohistochemical, genetic and epigenetic profiles of hereditary and triple negative breast cancers. Relevance in personalized medicine. Am. J. Cancer Res. 2015, 5, 2330–2343. [Google Scholar] [PubMed]
- Li, Y.; Melnikov, A.A.; Levenson, V.; Guerra, E.; Simeone, P.; Alberti, S.; Deng, Y. A seven-gene CpG-island methylation panel predicts breast cancer progression. BMC Cancer 2015, 15, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Gammon, M.D.; Zhang, Y.; Cho, Y.H.; Wetmur, J.G.; Bradshaw, P.T.; Garbowski, G.; Hibshoosh, H.; Teitelbaum, S.L.; Neugut, A.I.; et al. Gene promoter methylation is associated with increased mortality among women with breast cancer. Breast Cancer Res. Treat. 2010, 121, 685–692. [Google Scholar] [CrossRef][Green Version]
- Catteau, A.; Harris, W.H.; Xu, C.-F.; Solomon, E. Methylation of the BRCA1 promoter region in sporadic breast and ovarian cancer: Correlation with disease characteristics. Oncogene 1999, 18, 1957–1965. [Google Scholar] [CrossRef]
- Chen, Y.; Zhou, J.; Xu, Y.; Li, Z.; Wen, X.; Yao, L.; Xie, Y.; Deng, D. BRCA1 promoter methylation associated with poor sur-vival in Chinese patients with sporadic breast cancer. Cancer Sci. 2009, 100, 1663–1667. [Google Scholar] [CrossRef]
- Esteller, M.; Silva, J.M.; Dominguez, G.; Bonilla, F.; Matias-Guiu, X.; Lerma, E.; Bussaglia, E.; Prat, J.; Harkes, I.C.; Repasky, E.A.; et al. Promoter Hypermethylation and BRCA1 Inactivation in Sporadic Breast and Ovarian Tumors. J. Natl. Cancer Inst. 2000, 92, 564–569. [Google Scholar] [CrossRef]
- Jacot, W.; Thezenas, S.; Senal, R.; Viglianti, C.; Laberenne, A.-C.; Lopez-Crapez, E.; Bibeau, F.; Bleuse, J.-P.; Romieu, G.; Lamy, P.-J. BRCA1 promoter hypermethylation, 53BP1 protein expression and PARP-1 activity as biomarkers of DNA repair deficit in breast cancer. BMC Cancer 2013, 13, 523. [Google Scholar] [CrossRef]
- Jung, E.-J.; Kim, I.-S.; Lee, E.Y.; Kang, J.-E.; Lee, S.-M.; Kim, D.C.; Kim, J.-Y.; Park, S.-T. Comparison of Methylation Profiling in Cancerous and Their Corresponding Normal Tissues from Korean Patients with Breast Cancer. Ann. Lab. Med. 2013, 33, 431–440. [Google Scholar] [CrossRef]
- Krasteva, M.E.; Bozhanov, S.S.; Antov, G.G.; Gospodinova, Z.I.; Angelova, S.G. Breast cancer patients with hypermethylation in the promoter of BRCA1 gene exhibit favorable clinical status. Neoplasma 2012, 59, 85–91. [Google Scholar] [CrossRef]
- Sharma, P.; Stecklein, S.R.; Kimler, B.F.; Sethi, G.; Petroff, B.K.; Phillips, T.A.; Tawfik, O.W.; Godwin, A.K.; Jensen, R.A. The prognostic value of BRCA1 promoter methylation in early stage triple negative breast cancer. J. Cancer Ther. Res. 2014, 3, 2–11. [Google Scholar] [CrossRef]
- Tapia, T.; Smalley, S.V.; Kohen, P.; Muñoz, A.; Solis, L.M.; Corvalan, A.; Faundez, P.; Devoto, L.; Camus, M.; Alvarez, M.; et al. Promoter hypermethylation of BRCA1 correlates with absence of expression in hereditary breast cancer tumors. Epigenetics 2008, 3, 157–163. [Google Scholar] [CrossRef]
- Vu, T.L.; Nguyen, T.T.; Doan, V.T.H.; Vo, L.T.T. Methylation Profiles of BRCA1, RASSF1A and GSTP1 in Vietnamese Women with Breast Cancer. Asian Pac. J. Cancer Prev. 2018, 19, 1887–1893. [Google Scholar]
- Wei, M.; Grushko, T.A.; Dignam, J.; Hagos, F.; Nanda, R.; Sveen, L.; Xu, J.; Fackenthal, J.; Tretiakova, M.; Das, S.; et al. BRCA1 promoter methylation in sporadic breast cancer is associated with reduced BRCA1 copy number and chromo-some 17 aneusomy. Cancer Res. 2005, 65, 10692–10699. [Google Scholar] [CrossRef]
- Wu, L.; Shen, Y.; Peng, X.; Zhang, S.; Wang, M.; Xu, G.; Zheng, X.; Wang, J.; Lu, C. Aberrant promoter methylation of can-cer-related genes in human breast cancer. Oncol. Lett. 2016, 12, 5145–5155. [Google Scholar] [CrossRef]
- Yamashita, N.; Tokunaga, E.; Kitao, H.; Hitchins, M.; Inoue, Y.; Tanaka, K.; Hisamatsu, Y.; Taketani, K.; Akiyoshi, S.; Okada, S.; et al. Epigenetic nactivation of BRCA1 Through Promoter Hypermethylation and Its Clinical Importance in Triple-Negative Breast Cancer. Clin. Breast Cancer 2015, 15, 498–504. [Google Scholar] [CrossRef]
- Zhu, X.; Shan, L.; Wang, J.; Wang, F.; Shen, G.; Liu, X.; Wang, B.; Yuan, Y.; Ying, J.; Yang, H. Hypermethylation of BRCA1 gene: Implication for prognostic biomarker and therapeutic target in sporadic primary triple-negative breast cancer. Breast Cancer Res. Treat. 2015, 150, 479–486. [Google Scholar] [CrossRef]
- Bagadi, S.A.; Prasad, C.P.; Kaur, J.; Srivastava, A.; Prashad, R.; Gupta, S.D.; Ralhan, R. Clinical significance of promoter hypermethylation of RASSF1A, RARbeta2, BRCA1 and HOXA5 in breast cancers of Indian patients. Life Sci. 2008, 82, 1288–1292. [Google Scholar] [CrossRef]
- Hosny, M.M.; Sabek, N.A.; El-Abaseri, T.B.; Hassan, F.M.; Farrag, S.H. Promoter Methylation Status of Breast Cancer Susceptibility Gene 1 and 17 Beta Hydroxysteroid Dehydrogenase Type 1 Gene in Sporadic Breast Cancer Patients. Int. J. Breast Cancer 2016, 2016, 9545241. [Google Scholar] [CrossRef][Green Version]
- Li, Q.; Huawei, Y.; Jiang, Y.I.; Yang, H.; Liu, J. Promoter methylation and expression changes of BRCA1 in cancerous tissues of patients with sporadic breast cancer. Oncol. Lett. 2015, 9, 1807–1813. [Google Scholar] [CrossRef]
- Sharma, G.; Mirza, S.; Parshad, R.; Srivastava, A.; Gupta, S.D.; Pandya, P.; Ralhan, R. Clinical significance of promoter hy-permethylation of DNA repair genes in tumor and serum DNA in invasive ductal breast carcinoma patients. Life Sci. 2010, 87, 83–91. [Google Scholar] [CrossRef]
- Sharma, G.; Mirza, S.; Yang, Y.-H.; Parshad, R.; Hazrah, P.; Gupta, S.D.; Ralhan, R. Prognostic Relevance of Promoter Hypermethylation of Multiple Genes in Breast Cancer Patients. Anal. Cell. Pathol. 2009, 31, 487–500. [Google Scholar] [CrossRef]
- Singh, A.K.; Pandey, A.; Tewari, M.; Shukla, H.S.; Pandey, H.P. Epigenetic silencing of BRCA1 gene associated with demographic and pathologic factors in sporadic breast cancer: A study of an Indian population. Eur. J. Cancer Prev. 2011, 20, 478–483. [Google Scholar] [CrossRef]
- Truong, P.K.; Lao, T.D.; Doan, T.P.; Le, T.A. BRCA1 promoter hypermethylation signature for early detection of breast can-cer in the Vietnamese population. Asian Pac. J. Cancer Prev. 2014, 15, 9607–9610. [Google Scholar] [CrossRef]
- Bal, A.; Verma, S.; Joshi, K.; Singla, A.; Thakur, R.; Arora, S.; Singh, G. BRCA1-methylated sporadic breast cancers are BRCA-like in showing a basal phenotype and absence of ER expression. Virchows Arch. Pathol. Anat. Physiol. Klin. Med. 2012, 461, 305–312. [Google Scholar] [CrossRef]
- Hsu, N.C.; Huang, Y.F.; Yokoyama, K.K.; Chu, P.Y.; Chen, F.M.; Hou, M.F. Methylation of BRCA1 promoter region is associated with unfavorable prognosis in women with early-stage breast cancer. PLoS ONE 2013, 8, e56256. [Google Scholar] [CrossRef]
- Niwa, Y.; Oyama, T.; Nakajima, T. BRCA1 Expression Status in Relation to DNA Methylation of theBRCA1Promoter Region in Sporadic Breast Cancers. Jpn. J. Cancer Res. 2000, 91, 519–526. [Google Scholar] [CrossRef]
- Alkam, Y.; Mitomi, H.; Nakai, K.; Himuro, T.; Saito, T.; Takahashi, M.; Arakawa, A.; Yao, T.; Saito, M. Protein expression and methylation of DNA repair genes hMLH1, hMSH2, MGMT and BRCA1 and their correlation with clinicopathological parameters and prognosis in basal-like breast cancer. Histopathology 2013, 63, 713–725. [Google Scholar]
- Li, S.; Rong, M.; Iacopetta, B. DNA hypermethylation in breast cancer and its association with clinicopathological features. Cancer Lett. 2006, 237, 272–280. [Google Scholar] [CrossRef]
- Felicio, P.S.; Melendez, M.E.; Arantes, L.M.R.B.; Kerr, L.M.; Carraro, D.M.; Grasel, R.S.; Campacci, N.; Scapulatempo-Neto, C.; Fernandes, G.C.; De Carvalho, A.C.; et al. Genetic and epigenetic characterization of the BRCA1 gene in Brazilian women at-risk for hereditary breast cancer. Oncotarget 2016, 8, 2850–2862. [Google Scholar] [CrossRef]
- Saelee, P.; Chaiwerawattana, A.; Ogawa, K.; Cho, Y.-M.; Tiwawech, D.; Suktangman, V. Clinicopathological Significance of BRCA1 Promoter Hypermethylation in Thai Breast Cancer Patients. Asian Pac. J. Cancer Prev. 2015, 15, 10585–10589. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kumar, M.; Sahu, R.K.; Goyal, A.; Sharma, S.; Kaur, N.; Mehrotra, R.; Singh, U.R.; Hedau, S. BRCA1 Promoter Methylation and Expression-Associations with ER+, PR+ and HER2+ Subtypes of Breast Carcinoma. Asian Pac. J. Cancer Prev. 2017, 18, 3293–3299. [Google Scholar]
- Paydar, P.; Asadikaram, G.; Nejad, H.Z.; Akbari, H.; Abolhassani, M.; Moazed, V.; Nematollahi, M.H.; Ebrahimi, G.; Fallah, H. Epigenetic modulation of BRCA-1 and MGMT genes, and histones H4 and H3 are associated with breast tumors. J. Cell. Biochem. 2019, 120, 13726–13736. [Google Scholar] [CrossRef] [PubMed]
- Danesi, R.; Fogli, S.; Indraccolo, S.; Del Re, M.; Tos, A.D.; Leoncini, L.; Antonuzzo, L.; Bonanno, L.; Guarneri, V.; Pierini, A.; et al. Druggable targets meet oncogenic drivers: Opportunities and limitations of target-based classification of tumors and the role of Molecular Tumor Boards. ESMO Open 2021, 6, 100040. [Google Scholar] [CrossRef] [PubMed]
- Zahir, N.; Sun, R.; Gallahan, D.; Gatenby, R.A.; Curtis, C. Characterizing the ecological and evolutionary dynamics of cancer. Nat. Genet. 2020, 52, 759–767. [Google Scholar] [CrossRef] [PubMed]
- Cantone, I.; Fisher, A.G. Epigenetic programming and reprogramming during development. Nat. Struct. Mol. Biol. 2013, 20, 282–289. [Google Scholar] [CrossRef]
- Iwamoto, T.; Yamamoto, N.; Taguchi, T.; Tamaki, Y.; Noguchi, S. BRCA1 promoter methylation in peripheral blood cells is associated with increased risk of breast cancer with BRCA1 promoter methylation. Breast Cancer Res. Treat. 2011, 129, 69–77. [Google Scholar] [CrossRef]
- Brennan, K.; Garcia-Closas, M.; Orr, N.; Fletcher, O.; Jones, M.; Ashworth, A.; Swerdlow, A.; Thorne, H.; Riboli, E.; Vineis, P.; et al. Intragenic ATM Methylation in Peripheral Blood DNA as a Biomarker of Breast Cancer Risk. Cancer Res. 2012, 72, 2304–2313. [Google Scholar] [CrossRef]
- Potapova, A.; Hoffman, A.M.; Godwin, A.K.; Al-Saleem, T.; Cairns, P. Promoter hypermethylation of the PALB2 susceptibil-ity gene in inherited and sporadic breast and ovarian cancer. Cancer Res. 2008, 68, 998–1002. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).