Abstract
There is extensive and unequivocal evidence that secondary metabolism in filamentous fungi and plants is associated with oxidative stress. In support of this idea, transcription factors related to oxidative stress response in yeast, plants, and fungi have been shown to participate in controlling secondary metabolism. Aflatoxin biosynthesis, one model of secondary metabolism, has been demonstrated to be triggered and intensified by reactive oxygen species buildup. An oxidative stress-related bZIP transcription factor AtfB is a key player in coordinate expression of antioxidant genes and genes involved in aflatoxin biosynthesis. Recent findings from our laboratory provide strong support for a regulatory network comprised of at least four transcription factors that bind in a highly coordinated and timely manner to promoters of the target genes and regulate their expression. In this review, we will focus on transcription factors involved in co-regulation of aflatoxin biosynthesis with oxidative stress response in aspergilli, and we will discuss the relationship of known oxidative stress-associated transcription factors and secondary metabolism in other organisms. We will also talk about transcription factors that are involved in oxidative stress response, but have not yet been demonstrated to be affiliated with secondary metabolism. The data support the notion that secondary metabolism provides a secondary line of defense in cellular response to oxidative stress.
1. Introduction
Living organisms including fungi and plants use a variety of signal transduction mechanisms to sense and respond to different forms of environmental stress. This review is focused on those signaling pathways, which are activated in response to reactive oxygen species (ROS) such as hydrogen peroxide and superoxide anion. The classical view of the response to oxidative stress was developed based on studies in yeast which demonstrated that modulation of transcription of defense-related antioxidant genes assists in the survival of the organism. Like yeast, filamentous fungi must respond to oxidative stress, but due to the variety of environmental conditions with which they cope, their response is more robust and complicated than that of yeast. One factor in particular contributes to the complexity of filamentous fungal response to oxidative stress. Recent studies provide solid support for the notion that regulation of secondary metabolism is closely linked to the cellular response to oxidative stress in filamentous fungi [,,,,]. The available data strongly suggest that several transcription factors associated with the stress activated protein kinase/mitogen-activated protein kinase (SAPK/MAPK) pathway coordinate the timing and level of expression of target genes including antioxidant and secondary metabolism genes, thus controlling metabolic processes with cellular stress response. In aspergilli, it was proposed that antioxidant enzymes represent the first line of defense against excessive ROS formation and that synthesis of secondary metabolites functions as a second line of defense from ROS damages [,].
In this review, we focus on oxidative stress-related transcription factors, which have been shown to contribute in co-regulation of secondary metabolism, in particular aflatoxin biosynthesis, with oxidative stress response in the aspergilli. Several of these transcription factors are directly coupled to the SAPK/MAPK pathway. In addition, based on functional analyses of transcription factors, such as the CCAAT binding complex and Myb transcription factors, we make several predictions for the involvement of specific transcription factors, not yet reported, in co-regulation of aflatoxin biosynthesis with oxidative stress.
2. Stress Activated Signaling Pathways in Yeast and Filamentous Fungi
The signaling pathways that participate in response to a number of stresses (osmotic, UV irradiation, high temperature) including oxidative stress were comprehensively dissected in yeast and were found to be evolutionally conserved in filamentous fungi. These conserved signaling pathways incorporate a multistep phosphorelay system module (an analog of the bacterial two-component system), which relays the signal to the stress activated protein kinase/mitogen-activated protein kinase (SAPK/MAPK) pathway module, and this modulates activity of an array of specific oxidative/osmotic stress related transcription factors (Figure 1) [,,,,]. The transcription factors, depending on their activation state, alter expression of the target genes involved in the cellular response to the stress signals. The components of these signaling cascades and the mechanisms of the signal transduction have been extensively explored and documented [,,,,,].
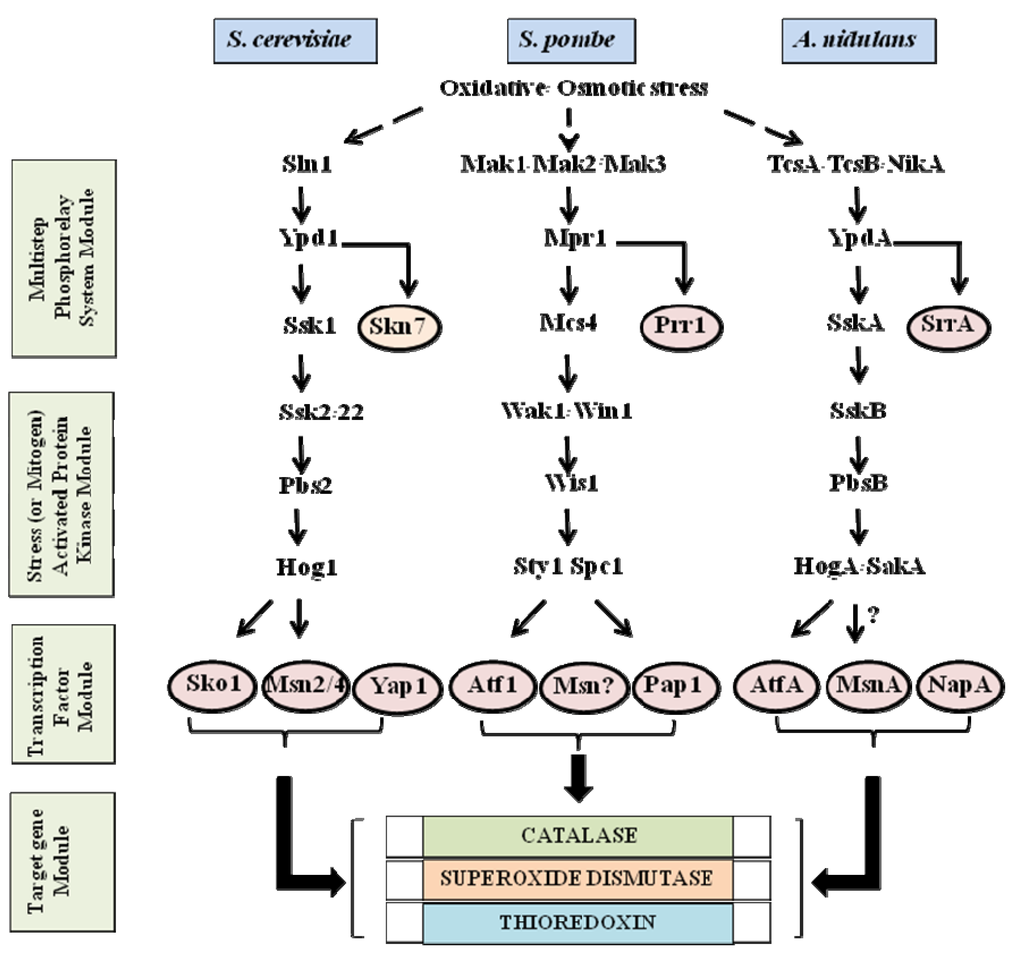
Figure 1.
Stress activated signaling pathways in Saccharomyces cerevisiae, S. pombe, and Aspergillus nidulans. In S. pombe and A. nidulans, Sty1/Spc1 (HogA/SakA) is activated by environmental stress such as oxidative and osmotic stress and induces transcription factors such as Atf1 (AtfA) for target gene expression. On the other hand, Hog1 activation for Sko1 in S. cerevisiae is dependent on osmotic stress. The TcsA/TcsB/NikA sensor kinases transmit oxidative stress signals to the HogA/SakA SAPK/MAPK cascade through YpdA and SskA in A. nidulans while Sln1 sensor kinase transmits osmotic stress signals to the Hog1 SAPK/MAPK cascade through Ypd1 and Ssk1 in S. cerevisiae. The Skn7 response regulator is also under the Sln1-Ypd1 signal transduction, but oxidative stress appears to activate Skn7 independently of the phosphorelay system. The solid lined arrows indicate signal transduction between two proteins and the dotted arrows indicate signal transduction from oxidative and/or osmotic stress. The thick lined arrows designate binding of transcription factors to target gene promoters. The circles around the proteins indicate transcription factors. Unknown signal transduction between two proteins and unidentified transcription factors are shown by question marks.
The essential elements of the multistep phosphorelay system module include sensor hybrid histidine kinases (Sln1/Mak/Tcs/NikA), histidine-containing phosphotransfer intermediates (Ypd1/Mpr1/YpdA), and response regulators (Ssk1/Mcs4/SskA) [,]; these signaling elements convey the phospho-signal from a conserved histidine to an aspartate residue thus relaying the signal to the SAPK/MAPK module. The SAPK/MAPK signaling module is highly conserved between yeast, filamentous fungi, and higher eukaryotes [,,,,]. The module consists of a major MAPK (Mitogen-Activated Protein Kinase), which phosphorylates target transcription factors and other cytosolic proteins on serine and threonine residues within a consensus motif. MAPK is activated via dual phosphorylation of conserved threonine and tyrosine residues, located in the activation loop, by MAPK kinase (MAPKK, MEK). MAPKK, in turn, is activated by MAPKK kinase (MAPKKK, MEKK).
The bZIP transcription factors that belong to the CREB/ATF (cAMP response element binding protein/activating transcription factor) family are highly conserved between yeast and filamentous fungi in structure (a basic motif for DNA binding and a leucine-zipper motif for dimerization) and regulatory mechanisms. bZIPs are controlled not only by MAPK, but also by cAMP-PKA (Protein Kinase A) activity. Atf1 and Pap1 are homologous to human ATF2 and c-Jun, respectively. Most target genes of these transcription factors are antioxidant-related genes such as catalase, thioredoxin, and superoxide dismutase which mediate cellular defense against oxidative stress [].
A ROS signal can be incorporated into the signaling pathway at different levels. For example, extra- and intra-cellular ROS can be perceived by the membranous and cytosolic sensor histidine kinases; it has been suggested that PAS/PAC and GAF domains adjacent to the histidine kinase domain, are likely involved in ROS sensing [,]. In Schizosaccharomyces pombe, an oxidative stress signal can be transmitted to the SAPK/MAPK module independently of the phosphorelay system module suggesting a direct ROS activation mechanism on Wak1/Win1 MEKK or involvement of a novel zinc-finger transcription factor Hsr1 signaling circuit [,]. Several transcription factors such as AP-1 homologs and CCAAT-binding transcription factors (see below) can be directly engaged in signaling by redox stimulus [,] or by modulation of AP-1 activity with the involvement of the thioredoxin system [,]. This highly conserved regulatory network that includes signaling modules, transcription factors, and target genes provides a robust defense system against oxidative stress in yeast and filamentous fungi.
Despite conservation of the major signaling modules, filamentous fungi, in contrast to yeast, have additional mechanisms to cope with ROS, such as the presence of a larger number of sensor histidine kinases [], antioxidant enzymes, and production of secondary metabolites with antioxidant function []. In Aspergillus parasiticus, it has been shown that there is a correlation between ROS formation, activation of oxidative stress-related transcription factors (ApyapA, AtfB, and MsnA), antioxidant enzyme activation, and production of the secondary metabolite aflatoxin, suggesting that oxidative stress triggers aflatoxin production [,,,,]. Several oxidative stress-related transcription factors (Apapy1, AtfB, Nap1) associated with the SAPK/MAPK signaling cascade, regulate secondary metabolism in aspergilli [,,]. These data indicate that incorporation of secondary metabolism into oxidative stress response occurs at least in part at the level of the transcription factors. Therefore, transcription factors (transcription factor module) are of special interest as they define metabolic decisions during stress response.
3. Transcription Factors Related to Oxidative Stress, Which Are Involved in Regulation of Secondary Metabolism
Existence of transcription factors with dual functionality (toward ROS signaling and secondary metabolism) serves as a strong support to the statement that secondary metabolism is a crucial part of the cellular response to oxidative stress in filamentous fungi; these transcription factors are the primary focus of this chapter. We will also present an interesting example of a transcriptional regulator, CCTTA transcription factor complex, which possesses a set of characteristics that provide the complex with the potential to serve as a co-regulator of aflatoxin biosynthesis and oxidative stress.
3.1. AP-1
AP-1 (Activating Protein 1), a bZIP transcription factor in yeast and filamentous fungi, is a transcriptional activator expressed in response to oxidative stress [,,]. AP-1 is one of the immediate targets for regulation by SAPK/MAPK. AP-1 binds to an AP-1 binding site (5'TGAC/GTCA3', 5'TT/GACTAA3'), which is highly similar to CRE (5'TG/TACGTC/AA3'), in the promoters of target genes and carries out multiple functions as a red-ox regulator [,].
Fungal AP-1 transcription factors contain N-terminal and C-terminal cysteine rich domains []. The AP-1 activation mechanism is well known for the yeast Yap1. Upon exposure to H2O2, 2 or more cysteines in the C-terminal cysteine rich domain (c-CRD) of Yap1 undergo direct oxidation and form intramolecular disulfide bond []. The oxidized form of Yap1 is transcriptionally active and is retained in the nucleus due to conformational changes that prevent its interaction with the export receptor Crm1/Xpo1 and removal from the nucleus. Yap1 deactivation involves enzymatic reduction of the oxidized form of Yap1 by thioredoxin whose transcription is increased by activation of Yap1 [].
The yeast AP-1 family of transcription factors includes Yap1 of Saccharomyces cerevisiae, Pap1 of S. pombe, Cap1 of Candida albicans, and Kap1 of Kluyveromyces lactis []. These homologs regulate the expression of a number of genes involved in oxidative stress response, including CTT1 (cytosolic catalase), TRX2 (thioredoxin), TRR1 (thioredoxin reductase), SOD1 (Cu/Zn superoxide dismutase), TSA1 (thioredoxin peroxidase), and GLR1 (glutathione reductase) [,]. It was reported that Yap1 binds to the Yap1 response element (5'TT/GAC/GT/AAA3'; YRE) in target genes [,,]. Similarly, Toda and co-workers reported that Pap1 binds to the sequence 5'TTAGTCA3' in target genes []. It is also known that Yap1-dependent transcriptional activity is inhibited by cAMP-dependent PKA [].
In aspergilli, Yap1 orthologs were identified in A. nidulans, A. fumigatus, and A. parasiticus. In A. nidulans, NapA (a Yap1 ortholog) was reported to play an important role in cellular defense against oxidative stress such as hydrogen peroxide and superoxide radicals []. NapA was shown to be a transcriptional activator of oxidative stress associated genes such as catB (mycelia-specific catalase), trxB (thioredoxin reductase), thiO (thioredoxin), and glrA (glutathione reductase) in response to hydrogen peroxide []. Of particular importance, NapA functioned coordinately with SrrA and/or SskA response regulators in response to oxidative stress in similar fashion as Yap1 cooperates with Skn7 in yeast [,]. In addition, overexpression of napA resulted in an increased tolerance to oxidative stress and decreased secondary metabolite production in A. nidulans, including sterigmatocystin, emericellin, asperthecin, shamixanthone, and epishamixanthone [].
Deletion of ApyapA (A. parasiticus ortholog of YAP1) in A. parasiticus resulted in an increased susceptibility to extracellular oxidants, precocious ROS and aflatoxin accumulation, and premature conidia formation as compared to the wild type strain []. In addition, the ApyapA disruptant produced more hydroperoxides and aflatoxin in maize seeds compared to the wild type strain, suggesting a correlation between oxidative stress and aflatoxin biosynthesis [].
Similar to the ApyapA disruptant, deletion of Aoyap1 in A. ochraceus resulted in increased ochratoxin production and higher amounts of ROS formation compared to the wild type strain []. In addition, expression of catA (conidia-specific catalase) and Cu, Zn sod1 (Cu, Zn-superoxide dismutase) in the Aoyap1 deletion mutant was down-regulated.
3.2. AtfA
The bZIP transcription factor AtfA in A. nidulans regulates oxidative and osmotic stress responses and activates expression of catB by binding to CRE sites in its promoter in response to H2O2 []. In addition to being regulated by SAPK/MAPK, AtfA is also controlled by cAMP/PKA. It was reported that AtfA plays a critical role in regulation of catA during fungal development []. AtfA was also shown to play an important role in tolerance of conidia to oxidative stress via interaction with SakA while it played a minor role in a mycelium resistance to H2O2 [,]. Interestingly, Lara-Rojas and co-workers suggested that AtfA and AtfB may interact to regulate some genes []. Considering the interaction of AtfA and AtfB in oxidative stress response, it is reasonable to anticipate that AtfA may have a possible connection with secondary metabolism.
In a plant pathogen Botrytis cinerea, an A. nidulans AtfA ortholog BcAtf1 is partially targeted by SAPK cascade signaling and is induced at transcription level upon exposure to H2O2 independently of SAPK []. Similar to A. nidulans Atf1, BcAtf1 controls expression of catalase B but is not involved in overall tolerance to osmotic and oxidative stress as measured by colony growth. Interestingly, bcatf1 deletion mutant accumulates 2 to 10 fold higher levels of secondary metabolites (polyketide botcininA, sesquiterpene botrydial, and botryendial) []. Accumulation of phytotoxins correlated with up-regulation of expression of genes involved in toxin biosynthesis suggesting that BcAtf1functions as a repressor of genes involved in biosynthesis of secondary metabolites in B. cinerea. Although a yeast one-hybrid analysis did not show a direct interaction of BcAtf1 with the promoters of biosynthetic genes, the data strongly demonstrate indirect influence of BcAtf1 on gene expression likely through interaction with other regulators (see above and below).
In S. pombe, Atf1 (an ortholog of AtfA in A. nidulans) is activated via phosphorylation by Sty1 in the SAPK signaling cascade. Atf1 induces expression of ctt1 (cytosolic catalase), gpx1 (glutathione peroxidase), and trr1 (thioredoxin reductase) in response to oxidative stress [,,]. Atf1 was shown to activate the transcription of some target genes by forming a heterodimer with a small bZIP transcription factor Pcr1, and Atf1 and Pcr1 have similar roles in response to oxidative stress [,]. In contrast to A. nidulans and S. pombe, Sko1 (an ortholog of Atf1 in S. pombe) in S. cerevisiae binds to CRE sites in the promoters of target genes and acts as a transcriptional repressor under normal growth conditions but induces expression of target genes after activation by phosphorylation in response to osmotic stress []. In this regard, filamentous fungi are closer to fission yeast than to budding yeast.
3.3. AtfB
AtfB is a member of the CREB/ATF family that binds CRE sites (5'TG/TACGTC/AA3'). An atfB disruptant of Aspergillus oryzae showed decreased transcription of antioxidant target genes such as catA (conidia-specific catalase) and produced conidia sensitive to H2O2 []. atfB was expressed in the late growth phase during conidiation on solid media. In A. parasiticus, AtfB was shown to bind to promoters of aflatoxin biosynthetic genes including nor-1, fas-1, ver-1, and omtA, which carry CRE sites [,]. Mutations in the CRE binding site in the nor-1 promoter reduced nor-1 transcription upon induction by exogenous cAMP. These data indicated that cAMP/PKA pathway regulates aflatoxin biosynthesis at least in part through AtfB []. Expression of atfB correlated with aflatoxin gene expression under aflatoxin-inducing conditions, suggesting that AtfB activates the aflatoxin gene promoters carrying CRE sites. AtfB has been demonstrated to be an important co-regulator that binds to the promoters of many aflatoxin genes including fas-1 and ver-1 as well as stress response genes such as mycelia-specific cat1 and mitochondria-specific Mn sod []. Thus, AtfB regulates coordinate expression of the aflatoxin genes and antioxidant genes. Each of the promoters that bind AtfB carries CRE sites whereas the vbs promoter (an aflatoxin gene), which lacks a CRE site, does not bind AtfB []. Recent data also support the existence of a transcription factor regulatory network that consists of AtfB, AP-1, MsnA, SrrA, and AflR, and this network, together with other signaling components, promotes fungal response to oxidative stress [,]. We proposed that increased levels of intracellular ROS activate antioxidant and aflatoxin genes via two signal transduction pathways. First, ROS down-regulate the cAMP-PKA pathway, which results in MsnA binding to the target promoter and activation of the antioxidant gene. Simultaneously, ROS up-regulate the SAPK/MAPK signaling cascade through the multistep phosphorelay system, which results in AtfB and SrrA binding to the promoters and induction of the antioxidant genes. MsnA, AtfB, and SrrA (SrrA recruits AP-1) then bind to the promoters of aflatoxin biosynthetic genes to assist in their induction by zinc binuclear cluster (C6) transcription factor AflR (Figure 2) [].
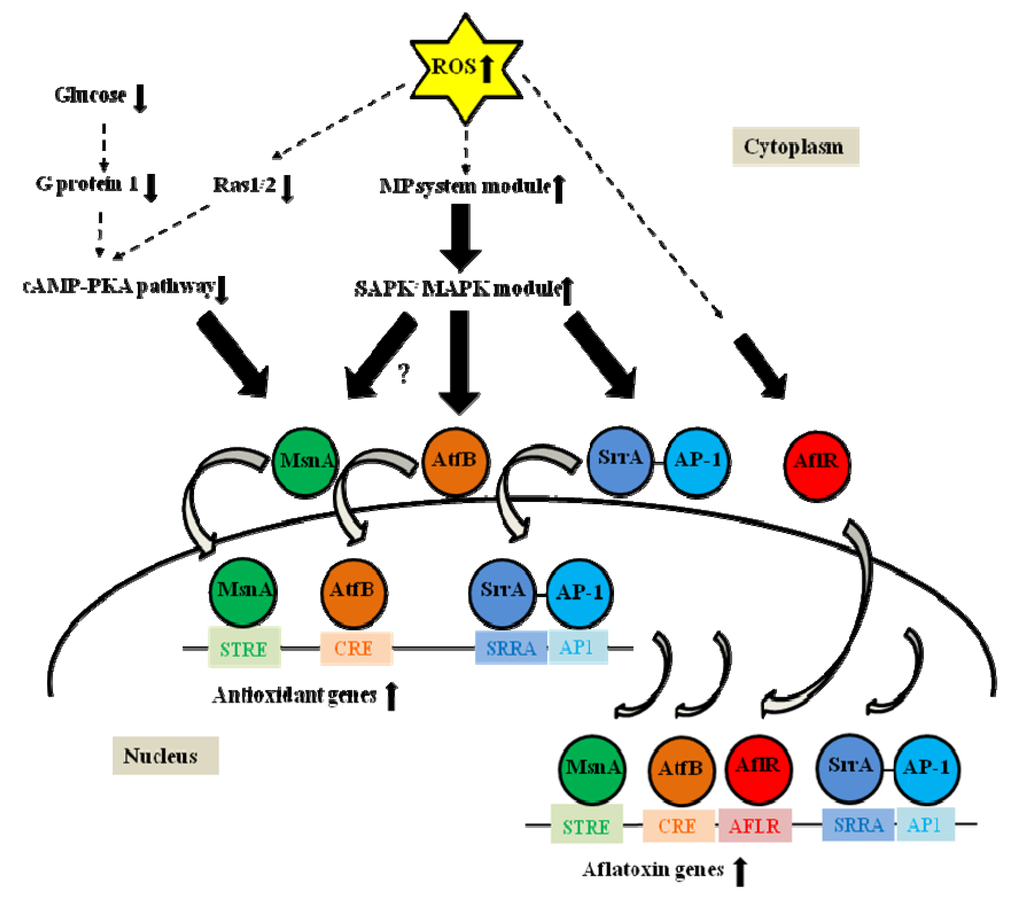
Figure 2.
A model for transcriptional activation of antioxidant and aflatoxin biosynthetic genes by oxidative stress-related transcription factors. Based on available experimental evidence, the model proposes that increased levels of intracellular reactive oxygen species (ROS) in fungal cells down-regulate the cAMP-PKA signaling pathway. This promotes MsnA binding to STRE sites in promoters of antioxidant genes for their activation. Simultaneously, ROS up-regulate the stress activated protein kinase/mitogen activated protein kinase (SAPK/MAPK) signaling cascades through the multistep phosphorelay system. Activation of the SAPK/MAPK cascade promotes AtfB and SrrA binding (SrrA recruits AP-1) to corresponding CRE, SRRA, and AP1 sites in promoters of the antioxidant genes for their induction. Then MsnA, AtfB, and SrrA bind (SrrA recruits AP-1) to corresponding STRE, CRE, SRRA, and AP1 sites in promoters of aflatoxin genes for their activation due to excessive levels of ROS. AflR assists in induction of aflatoxin biosynthetic genes by binding to AFLR sites in the aflatoxin gene promoters. The dotted arrows indicate signal transduction from ROS and the solid lined arrows indicate signal transduction pathways. The curved arrows designate entering of transcription factors from cytoplasm to nucleus and binding of the transcription factors to the corresponding recognition motifs. Undefined yet signal transduction between MsnA and SAPK/MAPK module is shown by a question mark. PKA, protein kinase A; MP, multistep phosphorelay; SAPK/MAPK, stress activated protein kinase/mitogen activated protein kinase.
3.4. MsnA
MsnA2 and MsnA4 in S. cerevisiae are Cys2His2 zinc finger transcription factors that are induced as part of the cellular response to oxidative stress as well as other types of stresses such as carbon starvation, heat shock, and osmotic stress []. MsnA2 and MsnA4 activate transcription of stress response genes such as CTT1 (catalase T) and HSP12 (heat shock protein 12) by binding to the stress response element (5'AGGGG3') in their promoters. It is known that Msn2/4 activity is inhibited by cAMP-dependent protein kinase (PKA).
Deletion of msnA (an ortholog of MSNA2) in A. parasiticus and in A. flavus resulted in growth inhibition but increased production of conidia, ROS, aflatoxin, and kojic acid []. msnA disruption up-regulated expression of genes encoding enzymes that protect against ROS (e.g., superoxide dismutase, putative catalase, and cytochrome c peroxidase in A. parasiticus and catalase A in A. flavus). These findings may imply that deletion of msnA in A. parasiticus and in A. flavus down-regulates expression of mycelial catalase (in A. parasiticus) and of catalase B (in A. flavus) which in turn up-regulates expression of other antioxidant genes including the putative catalase and superoxide dismutase for fungal defense to compensate for the decreased expression of the mycelial catalase and catalase B and to cope with higher levels of ROS.
3.5. StuA
Incomplete data exist on the helix-loop-helix developmental regulator StuA in regard to its involvement in co-regulation of secondary metabolism and oxidative stress response. Expression of the cpeA, a gene that encodes for a Hülle cell-specific bifunctional enzyme catalase-peroxidase, is under positive influence of StuA in A. nidulans []. On the other hand, in A. fumigatus, six secondary metabolic gene clusters were found to be under control of StuA [].
3.6. CCAAT-Binding Transcription Factor Complex
Eukaryotes including fungi, plants, and mammals possess evolutionally conserved orthologs of a multi-subunit Hap (Heme Activator Protein) complex consisting of at least three subunits in filamentous fungi, plants, and vertebrates, and four subunits in yeast [,,,,]. The complex binds specifically to a CCAAT motif within promoters of target genes involved in primary metabolism, cell-cycle, development, stress response, and particularly oxidative stress, and controls expression of the target genes (see below). In plants and vertebrates, each histone-like component of the heterotrimeric complex (known as NF-Y, Nuclear Factor Y, or CBF, CCAAT-binding factor) is required for subunit interactions, CCAAT binding through histone fold motifs, and bifunctional regulation of the target gene expression [,,]. In mammalian cells, it has been shown that NF-Y affects the histone acetylation landscape in gene promoters, thus influencing gene transcription []. The Hap complex binding motif CCAAT was first identified in yeast [,]. The yeast Hap complex plays a role as a transcriptional regulator during respiration and oxidative stress response and induces genes such as CYC1 (cytochrome c-iso-1), CYC7 (cytochrome c iso-2), CYT1 (cytochrome c1), and CTT1 (catalase). The heterotrimer consisting of Hap2p, Hap3p, and Hap5p, is essential for DNA binding, whereas a fourth component, Hap4p, carries a transcription activation domain.
In the filamentous fungus Aspergillus nidulans, at least HapC, HapE, and HapB are required for assembly of a heterotrimeric complex called AnCF/PenR1/AnCP which is able to bind the consensus CCAAT motif and regulate gene transcription [,,]. HapB, the only subunit that carries a nuclear localization signal sequence, is responsible for the translocation of the entire heterotrimeric complex to the nucleus [,].
The A. nidulans Hap-like complex(s) AnCF/PenR1 has been reported to control expression of genes involved in penicillin biosynthesis including ipnA, acvA, aatA, and aatB [,,]. The CCAAT binding complex AnCF (A. nidulans CCAAT binding factor) exerts a positive influence on ipnA, aatA, and aatB transcription, whereas a negative effect on transcription of acvA was documented. A basic-region helix-loop-helix transcription factor AnBH1 has been found to bind to an asymmetric E-box 5'-AATCACAGG-3' which partially overlaps the CCAAT motif within the aatA promoter; these findings suggest competitive interactions between these transcription factors for binding to the promoter [].
AnCF serves as a necessary signaling component in the cellular response to oxidative stress []. In particular, AnCF fulfills the role of a redox sensor in the cell. Three cysteine residues are located in conserved positions within the histone fold motif in all HapC subunits from fungi to humans; only a form of HapC that possesses all 3 cysteines in a reduced state, is able to assemble into a functional heterotrimeric AnCF complex. Although there currently is no direct indication for the involvement of an AnCF complex in the SAPK/MAPK pathway, the AnCF complex may regulate oxidative stress response indirectly in concert with NapA (A. nidulans ortholog of S. cereviseae Yap1), which is regulated by SAPK/MAPK []. Specifically, AnCF binds the consensus binding motif in promoters of napA and NapA target genes such as catB (mycelial catalase B), trxA (thioredoxin), and prxA (thioredoxin-dependent peroxidase), thus maintaining glutathione homeostasis [].
In Arabidopsis thaliana, over-expression of one of the subunits of the NF-Y complex, NFYA5, affected expression of 130 genes including a number of genes involved in oxidative stress response, e.g., a subunit of cytochrome b6-f complex, glutathione S-transferase, peroxidases, and an oxidoreductase family protein []. However, no connection between the A. thaliana NF-Y complex and secondary metabolism was reported yet.
In A. parasiticus, a CCAAT motif was identified in promoters of many genes involved in aflatoxin biosynthesis, except for aflR (a pathway specific regulator) and ver-1 (encoding a middle enzyme in the aflatoxin biosynthetic pathway) []. The intergenic region between the divergently transcribed fas1 and fas2 genes (encoding enzymes that catalyze hexanoate formation during the first step in aflatoxin biosynthesis) does not possess an AflR recognition site. Thus, it is not clear how Aspergillus coordinates fas gene transcription with the transcription of the other genes in the cluster. While missing the AflR recognition site, the fas1/fas2 intergenic region does possess a Hap-like transcription complex binding site CCAAT and AtfB binding motifs []. Hap complex and AtfB represent strong candidates for playing a role in the mechanism that initiates transcription of the fas1 and fas2 genes and that coordinates their expression with transcription of other genes in the aflatoxin cluster as a part of the response to oxidative stress.
In other fungal species, CCAAT-binding complexes have been shown to regulate a number of genes encoding polysaccharide degrading enzymes such as Taka-amylase A (taaG2) in A. oryzae [], xylan- and cellulose-degrading enzymes in Trichoderma reesei [,], and glutamate dehydrogenase in Neurospora crassa []. In A. fumigatus, a mutation in the CCAAT-binding transcription factor complex subunit HapE underlies a mechanism of resistance to azole, the primary antifungal agent for patients with mycoses caused by A. fumigatus []. Although the CCAAT-binding complex in these fungi has not been shown to participate in secondary metabolism or oxidative stress response regulation or to be associated with SAPK/MAPK signaling pathway, the complex represents a strong extrapolative element in the regulatory network.
4. Transcription Factors with Strong Potential to Coordinate Oxidative Stress and Secondary Metabolism
A complexity of the transcription factor regulatory network that coordinates secondary metabolism with oxidative stress response has just begun to unravel. In this chapter we focus on a group of transcription factors that have a strong potential for the role in this network despite currently accessible data are incomplete. A number of transcription factors have been shown to regulate response to oxidative stress such as Hsf1, SrrA/Skn7/Prr1, Pcr1, and Myb. These transcription factors may play a direct or indirect regulatory role in regulation of secondary metabolism and as transcriptional coordinators although the validating data are not immediately available. For example, the transcription factor SrrA (an Aspergillus ortholog of yeast Skn7/Prr1) that has a prominent function in oxidative stress response in aspergilli and yeast, has been suggested as a regulator in aflatoxin biosynthesis [] (see below).
Undoubtedly, one of the greatest challenges of fungal biology is to understand the entire regulatory network that coordinates secondary metabolism and cellular response to oxidative stress. Making the entire picture even more intricate, the essential elements of this network must comprise a heterotrimeric complex VeA-LaeA-VelB, the light-responsive global regulator of secondary metabolism. The Velvet complex proteins are not transcription factors but function as global regulators of many aspects of fungal biology including secondary metabolism such as aflatoxin gene activation. Involvement of VeA in cellular response to oxidative stress has now begun to be elucidated. In ongoing studies, VeA has been shown to be involved in transcriptional activation of genes involved in antioxidant response, cat1 (mycelial catalase) and trxA (thioredoxin).
4.1. Hsf1
In S. cerevisiae Hsf1 (heat shock factor 1) binds as a homotrimer to the heat shock element (HSE) found in the promoters of the heat shock genes, the genes which are expressed under heat shock conditions causing damage of proteins such as denaturation. The element consists of tandem inverted repeats of the sequence 5'AGAAN3' (where N is any nucleotide) []. The Hsf1 undergoes phosphorylation, which activates transcription of CUP1 (copper metallothionein) for cellular protection in response to oxidative stress, in particular superoxide anion []. In the presence of hydrogen peroxide, Hsf1 cooperates with Skn7 to induce heat shock gene expression []. Activation of Hsf1 in response to superoxide anion is inhibited by cAMP-dependent PKA activity []. It seems that heat shock gene activation via Hsf1 is involved in refolding the damaged proteins during recovery from severe oxidative stress []. Considering the involvement of Hsf1 in oxidative stress response and crosstalk between transcription factors in the transcriptional network such as cooperation of Hsf1 with Skn7, it is reasonable to anticipate that Hsf1 may have a possible connection with secondary metabolism.
4.2. SrrA
A. nidulans SrrA (an ortholog of S. cerevisiae Skn7 and S. pombe Prr1) is a member of the multicomponent histidine-to-aspartate phosphorelay system (analogous to the two-component phosphorelay system in prokaryotes) in a signal transduction pathway that mediates cellular response to environmental stimuli [,]. This response regulator is a transcription factor that contains a mammalian heat shock factor (HSF)-like DNA binding domain adjacent to a receiver domain, which is essential for phosphorelay function [,]. Disruption of srrA in A. nidulans showed hypersensitivity to oxidative stresses such as hydrogen peroxide and decreased levels of expression of mycelia-specific catB (catalase B) [,].
Skn7 in S. cerevisiae is a winged helix-turn-helix transcription factor that has been shown to physically interact with either Yap1 (an ortholog of A. nidulans NapA) or Hsf1 (heat shock factor 1) before it binds to target promoters in response to oxidative stress and/or heat shock []. He and Fassler reported that Skn7 binds to the oxidative stress response element (OSRE) (5'GGCNNGGC3', 5'GGCNGGC3', 5'GGCNAGA3', or 5'GGCNNAGA3') in the promoters of CTT1 (cytosolic catalase), CCP1 (mytochondrial cytochrome c peroxidase), TSA1 (cytosolic thioredoxin peroxidase), AHP1 (alkyl hydroperoxide reductase) as target genes and induces their expression in response to H2O2. Morgan and co-workers reported that Skn7 binds to the sequence 5'CCGAAA3' in TRX2 (thioredoxin) promoter [,]. Skn7 binds to the heat shock element (HSE) in heat shock genes by cooperation with Hsf1 especially in response to hydrogen peroxide []. It seems that Skn7 binds to the promoters of the specific target genes by interaction with different partner proteins in response to different stress conditions.
Similar to Skn7, Pap1 and Prr1 (S. pombe orthologs of Yap1 and Skn7) are required for activation of the ctt1 and trr1 in S. pombe [].
Interaction of Skn7 with AP-1 orthologs implies an involvement of Skn7, although indirect, in regulation of secondary metabolism. Indeed, Hong and co-workers found a conserved 5-base motif 5'AAGCC3', a recognition site for SrrA, in promoters of the antioxidant genes cat1 and Mn sod, as well as in the promoters of aflatoxin biosynthetic genes, fas-1 and ver-1 in A. parasiticus []. Location of this site in close proximity to ATG translation initiation codon and to AP-1 binding sites in the promoter suggests the physical interaction between SrrA and AP-1, and their involvement in regulation of aflatoxin gene expression.
A. nidulans SskA is one of several response regulators in a signal transduction pathway that mediates cellular response to environmental stimuli [,]. Despite not being a transcription factor, disruption of sskA in A. nidulans showed not only a similar phenotype to srrA deletion mutants (hypersensitivity to hydrogen peroxide and decreased levels of expression of catB) but also hypersensitivity of conidia to hydrogen peroxide and reduced germination efficiency []. In contrast to observations by Hagiwara and co-workers, Vargas-Perez and co-workers reported that sskA knock-out mutants of A. nidulans do not show sensitivity to hydrogen peroxide and that sskA is not required for expression of catB []. These observations pointed out at two alternative signaling routes, which function simultaneously and/or in an alternate fashion. Because SrrA is an upstream component in a redox signaling pathway, there is the possible connection for this protein to secondary metabolism.
4.3. Pcr1
Pcr1, a bZIP transcription factor, was shown to bind to CRE sites and regulated sexual development by cAMP-PKA in S. pombe []. Watanabe and co-workers suggested that Pcr1 may form a heterodimer with another bZIP transcription factor Atf1 in S. pombe []. Pcr1 was required for full response to oxidative stress or heat shock in liquid culture []. Similar to Atf1, Pcr1 was required for transcription of most of stress-dependent genes such as gpx1 (glutathione peroxidase) and ctt1 (cytosolic catalase). Sanso and co-workers demonstrated that Pcr1 is a phosphoprotein and its dephosphorylation occurs in response to oxidative stress in a Sty1-dependent manner [].
4.4. Myb
Myb transcription factors are conserved in animals, plants, algae, and fungi. Myb proteins are often encoded by multiple genes in an individual organism, possess imperfect helix-turn-helix repeats, and are among a group of transcriptional regulators involved in eukaryotic cell cycle progression; c-myb is an oncogene in mammals []. In the plant Arabidopsis thaliana, Myb transcription factors are involved in phenylpropanoid and flavonoid biosynthesis [,,] and in a number of responses to different stressors including phosphorous, salinity, cold, dehydration, and osmotic shock [].
An A. nidulans Myb transcription factor FlbD is a specific regulator of asexual and sexual development; regulation of sexual development by FlbD is veA-independent [,]. Myb is also able to perceive cellular red-ox state []. In the plant pathogen Phytophthora sojae, the Myb transcription factor PsMYB1 is required for zoospore development and zoospore-mediated plant infection []. In addition, it has been shown that activation of the SAPK/MAPK pathway directly or indirectly increases transcript levels of PsMYB1 [].
Given the established stress response involvement of Myb transcription factors, their connection with SAPK/MAPK pathway, and involvement in regulation of secondary metabolism in plants and development in fungi, it is reasonable to anticipate that uncharacterized yet Myb transcription factors may coordinate secondary metabolism and oxidative stress response.
5. Conclusions and Future Research
(1) Signaling pathways utilized to sense oxidative stress and to regulate target gene expression are evolutionary well conserved between filamentous fungi and yeast. Yet, there are particular significant differences between species with regard to their response to damaging levels of ROS. Findings from our laboratory and others indicate that in filamentous fungi, in contrast to yeast, cellular response to excessive ROS incorporates secondary metabolism. Of special importance, a network of oxidative stress-related transcription factors helps coordinate the expression of many downstream target genes including antioxidant genes and genes involved in secondary metabolism. The network transmits signals that arise from a variety of sources through different signaling routes, thus showing a high degree of cross-talk.
(2) Individual transcription factors fulfill distinct but often overlapping signaling functions. For instance, Yap-1, Nap-1, and APyap1 - all belong to the AP-1 family of transcription factors. These transcription factors show overlapping functions related to ROS signaling in yeast and filamentous fungi and activation of antioxidant target genes. On the other hand, in filamentous fungi these factors perform an additional unique task, which is co-regulation of secondary metabolism. What structural/functional features provide them with this uniqueness? One common characteristic may be the ability to interact with the variety of other transcription factors.
(3) The connection between ROS, secondary metabolism, and fungal development is a well-known phenomenon but was out of the scope in this review. Nevertheless, it is likely that the same collection of transcription factors discussed above orchestrates regulation of developmental processes during response to oxidative stress. The network may include recently described transcription factors NsdC and NsdD []. These zinc-finger transcription factors have been shown to participate in regulation of asexual development and aflatoxin biosynthesis in A. flavus, although their involvement in oxidative stress response is currently unclear. Research efforts focused to explore regulatory ties between ROS, secondary metabolism, and fungal development help us to view secondary metabolism interconnected with essential processes in the fungal cell.
(4) Despite strong evidence for a close regulatory connection between secondary metabolism and antioxidant defense in response to elevated ROS levels, a question arises: “Could secondary metabolism be regulated independently from the oxidative stress”? Deletion or overexpression of RsmA, a bZIP transcription factor, which is a Yap3 ortholog in A. nidulans, has been shown to specifically affect sterigmatocystin biosynthesis with no effect on tolerance/sensitivity to ROS under investigated conditions [,]. Therefore, it is likely that such pathway(s) exist, but this remains an interesting and still open question.
(5) A better understanding of the transcriptional regulatory network that coordinates secondary metabolism and stress response will provide new insights into secondary metabolism in fungi and may help us to develop stress tolerant industrial fungal strains for beneficial secondary metabolite production and to find new targets for eliminating detrimental secondary metabolite production from fungi.
References
- Reverberi, M.; Zjalic, S.; Ricelli, A.; Fabbri, A.A.; Fanelli, C. Oxidant/antioxidant balance in Aspergillus parasiticus affects aflatoxin biosynthesis. Mycotoxin Res. 2006, 22, 39–47. [Google Scholar] [CrossRef]
- Reverberi, M.; Gazzetti, K.; Punelli, F.; Scarpari, M.; Zjalic, S.; Ricelli, A.; Fabbri, A.A.; Fanelli, C. Aoyap1 regulates ota synthesis by controlling cell redox balance in Aspergillus ochraceus. Appl. Microbiol. Biotechnol. 2012, 95, 1293–1304. [Google Scholar] [CrossRef]
- Roze, L.V.; Chanda, A.; Wee, J.; Awad, D.; Linz, J.E. Stress-related transcription factor ATFB integrates secondary metabolism with oxidative stress response in aspergilli. J. Biol. Chem. 2011, 286, 35137–35148. [Google Scholar]
- Yin, W.B.; Amaike, S.; Wohlbach, D.J.; Gasch, A.P.; Chiang, Y.M.; Wang, C.C.; Bok, J.W.; Rohlfs, M.; Keller, N.P. An Aspergillus nidulans bZIP response pathway hardwired for defensive secondary metabolism operates through aflR. Mol. Microbiol. 2012, 83, 1024–1034. [Google Scholar] [CrossRef]
- Hong, S.Y.; Roze, L.V.; Wee, J.; Linz, J.E. Evidence that a transcription factor regulatory network coordinates oxidative stress response and secondary metabolism in aspergilli. Microbiol. Open 2013, 2, 144–160. [Google Scholar]
- Miskei, M.; Karanyi, Z.; Pocsi, I. Annotation of stress-response proteins in the aspergilli. Fungal Genet. Biol. 2009, 46, S105–S120. [Google Scholar] [CrossRef]
- Toone, W.M.; Morgan, B.A.; Jones, N. Redox control of AP-1-like factors in yeast and beyond. Oncogene 2001, 20, 2336–2346. [Google Scholar]
- Ikner, A.; Shiozaki, K. Yeast signaling pathways in the oxidative stress response. Mutat. Res. 2005, 569, 13–27. [Google Scholar] [CrossRef]
- Toone, W.M.; Jones, N. Stress-activated signalling pathways in yeast. Genes Cells 1998, 3, 485–498. [Google Scholar]
- Bahn, Y.S.; Xue, C.; Idnurm, A.; Rutherford, J.C.; Heitman, J.; Cardenas, M.E. Sensing the environment: Lessons from fungi. Nat. Rev. 2007, 5, 57–69. [Google Scholar] [CrossRef]
- Banuett, F. Signalling in the yeasts: An informational cascade with links to the filamentous fungi. Microbiol. Mol. Biol. Rev. 1998, 62, 249–274. [Google Scholar]
- Aguirre, J.; Hansberg, W.; Navarro, R. Fungal responses to reactive oxygen species. Med. Mycol. 2006, 44, S101–S107. [Google Scholar] [CrossRef]
- Bahn, Y.S. Master and commander in fungal pathogens: The two-component system and the hog signaling pathway. Eukaryot. Cell 2008, 7, 2017–2036. [Google Scholar] [CrossRef]
- Eaton, C.J.; Jourdain, I.; Foster, S.J.; Hyams, J.S.; Scott, B. Functional analysis of a fungal endophyte stress-activated MAP kinase. Curr. Genet. 2008, 53, 163–174. [Google Scholar] [CrossRef]
- Segmuller, N.; Ellendorf, U.; Tudzynski, B.; Tudzynski, P. BcSak1, a stress-activated mitogen-activated protein kinase, is involved in vegetative differentiation and pathogenicity in Botrytis cinerea. Eukaryot. Cell 2007, 6, 211–221. [Google Scholar] [CrossRef]
- Heller, J.; Ruhnke, N.; Espino, J.J.; Massaroli, M.; Collado, I.G.; Tudzynski, P. The mitogen-activated protein kinase BcSak1 of Botrytis cinerea is required for pathogenic development and has broad regulatory functions beyond stress response. Mol. Plant Microbe Interact. 2012, 25, 802–816. [Google Scholar] [CrossRef]
- Nguyen, T.V.; Schafer, W.; Bormann, J. The stress-activated protein kinase FgOS-2 is a key regulator in the life cycle of the cereal pathogen Fusarium graminearum. Mol. Plant Microbe Interact. 2012, 25, 1142–1156. [Google Scholar] [CrossRef]
- Moye-Rowley, W.S. Transcription factors regulating the response to oxidative stress in yeast. Antioxid. Redox Signal. 2002, 4, 123–140. [Google Scholar] [CrossRef]
- Zhulin, I.B.; Taylor, B.L.; Dixon, R. PAS domain S-boxes in Archaea, Bacteria and sensors for oxygen and redox. Trends Biochem. Sci. 1997, 22, 331–333. [Google Scholar] [CrossRef]
- Nguyen, A.N.; Lee, A.; Place, W.; Shiozaki, K. Multistep phosphorelay proteins transmit oxidative stress signals to the fission yeast stress-activated protein kinase. Mol. Biol. Cell 2000, 11, 1169–1181. [Google Scholar]
- Chen, D.; Wilkinson, C.R.; Watt, S.; Penkett, C.J.; Toone, W.M.; Jones, N.; Bahler, J. Multiple pathways differentially regulate global oxidative stress responses in fission yeast. Mol. Biol. Cell 2008, 19, 308–317. [Google Scholar] [CrossRef]
- Thon, M.; Al Abdallah, Q.; Hortschansky, P.; Scharf, D.H.; Eisendle, M.; Haas, H.; Brakhage, A.A. The CCAAT-binding complex coordinates the oxidative stress response in eukaryotes. Nucleic Acids Res. 2010, 38, 1098–1113. [Google Scholar] [CrossRef]
- Karimpour, S.; Lou, J.; Lin, L.L.; Rene, L.M.; Lagunas, L.; Ma, X.; Karra, S.; Bradbury, C.M.; Markovina, S.; Goswami, P.C.; et al. Thioredoxin reductase regulates AP-1 activity as well as thioredoxin nuclear localization via active cysteines in response to ionizing radiation. Oncogene 2002, 21, 6317–6327. [Google Scholar] [CrossRef]
- Thon, M.; Al-Abdallah, Q.; Hortschansky, P.; Brakhage, A.A. The thioredoxin system of the filamentous fungus Aspergillus nidulans: Impact on development and oxidative stress response. J. Biol. Chem. 2007, 282, 27259–27269. [Google Scholar] [CrossRef]
- Reverberi, M.; Zjalic, S.; Ricelli, A.; Punelli, F.; Camera, E.; Fabbri, C.; Picardo, M.; Fanelli, C.; Fabbri, A.A. Modulation of antioxidant defense in Aspergillus parasiticus is involved in aflatoxin biosynthesis: A role for the ApyapA gene. Eukaryot. Cell 2008, 7, 988–1000. [Google Scholar] [CrossRef]
- Reverberi, M.; Zjalic, S.; Punelli, F.; Ricelli, A.; Fabbri, A.A.; Fanelli, C. Apyap1 affects aflatoxin biosynthesis during Aspergillus parasiticus growth in maize seeds. Food Addit. Contam. 2007, 24, 1070–1075. [Google Scholar]
- Yin, W.B.; Reinke, A.W.; Szilagyi, M.; Emri, T.; Chiang, Y.M.; Keating, A.E.; Pocsi, I.; Wang, C.C.; Keller, N.P. bZIP transcription factors affecting secondary metabolism, sexual development and stress responses in Aspergillus nidulans. Microbiology 2013, 159, 77–88. [Google Scholar] [CrossRef]
- Wu, A.L.; Moye-Rowley, W.S. GSH1, which encodes gamma-glutamylcysteine synthetase, is a target gene for yAP-1 transcriptional regulation. Mol. Cell. Biol. 1994, 14, 5832–5839. [Google Scholar] [CrossRef]
- Karin, M.; Liu, Z.; Zandi, E. AP-1 function and regulation. Curr. Opin. Cell biol. 1997, 9, 240–246. [Google Scholar] [CrossRef]
- Fernandes, L.; Rodrigues-Pousada, C.; Struhl, K. Yap, a novel family of eight bZIP proteins in Saccharomyces cerevisiae with distinct biological functions. Mol. Cell. Biol. 1997, 17, 6982–6993. [Google Scholar]
- Delaunay, A.; Isnard, A.D.; Toledano, M.B. H2O2 sensing through oxidation of the Yap1 transcription factor. Embo J. 2000, 19, 5157–5166. [Google Scholar] [CrossRef]
- Lee, J.; Godon, C.; Lagniel, G.; Spector, D.; Garin, J.; Labarre, J.; Toledano, M.B. Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J. Biol. Chem. 1999, 274, 16040–16046. [Google Scholar]
- Abate, C.; Patel, L.; Rauscher, F.J., 3rd; Curran, T. Redox regulation of fos and jun DNA-binding activity in vitro. Science 1990, 249, 1157–1161. [Google Scholar]
- He, X.J.; Fassler, J.S. Identification of novel Yap1p and Skn7p binding sites involved in the oxidative stress response of Saccharomyces cerevisiae. Mol. Microbiol. 2005, 58, 1454–1467. [Google Scholar] [CrossRef]
- Toda, T.; Shimanuki, M.; Saka, Y.; Yamano, H.; Adachi, Y.; Shirakawa, M.; Kyogoku, Y.; Yanagida, M. Fission yeast pap1-dependent transcription is negatively regulated by an essential nuclear protein, crm1. Mol. Cell. Biol. 1992, 12, 5474–5484. [Google Scholar]
- Asano, Y.; Hagiwara, D.; Yamashino, T.; Mizuno, T. Characterization of the bZip-type transcription factor NapA with reference to oxidative stress response in Aspergillus nidulans. Biosci. Biotechnol. Biochem. 2007, 71, 1800–1803. [Google Scholar] [CrossRef]
- Mulford, K.E.; Fassler, J.S. Association of the Skn7 and Yap1 transcription factors in the Saccharomyces cerevisiae oxidative stress response. Eukaryot. Cell 2011, 10, 761–769. [Google Scholar] [CrossRef]
- Balazs, A.; Pocsi, I.; Hamari, Z.; Leiter, E.; Emri, T.; Miskei, M.; Olah, J.; Toth, V.; Hegedus, N.; Prade, R.A.; et al. AtfA bZIP-type transcription factor regulates oxidative and osmotic stress responses in Aspergillus nidulans. Mol. Genet. Genomics 2010, 283, 289–303. [Google Scholar] [CrossRef]
- Lara-Rojas, F.; Sanchez, O.; Kawasaki, L.; Aguirre, J. Aspergillus nidulans transcription factor AtfA interacts with the MAPK SakA to regulate general stress responses, development and spore functions. Mol. Microbiol. 2011, 80, 436–454. [Google Scholar] [CrossRef]
- Hagiwara, D.; Asano, Y.; Yamashino, T.; Mizuno, T. Characterization of bZip-type transcription factor AtfA with reference to stress responses of conidia of Aspergillus nidulans. Biosci. Biotechnol. Biochem. 2008, 72, 2756–2760. [Google Scholar] [CrossRef]
- Temme, N.; Oeser, B.; Massaroli, M.; Heller, J.; Simon, A.; Collado, I.G.; Viaud, M.; Tudzynski, P. Bcatf1, a global regulator, controls various differentiation processes and phytotoxin production in Botrytis cinerea. Mol. Plant Pathol. 2012, 13, 704–718. [Google Scholar] [CrossRef]
- Watanabe, Y.; Yamamoto, M. Schizosaccharomyces pombe pcr1+ encodes a CREB/ATF protein involved in regulation of gene expression for sexual development. Mol. Cell. Biol. 1996, 16, 704–711. [Google Scholar]
- Sanso, M.; Gogol, M.; Ayte, J.; Seidel, C.; Hidalgo, E. Transcription factors Pcr1 and Atf1 have distinct roles in stress- and sty1-dependent gene regulation. Eukaryot. Cell 2008, 7, 826–835. [Google Scholar] [CrossRef]
- Proft, M.; Struhl, K. Hog1 kinase converts the Sko1-Cyc8-Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol. Cell 2002, 9, 1307–1317. [Google Scholar] [CrossRef]
- Sakamoto, K.; Iwashita, K.; Yamada, O.; Kobayashi, K.; Mizuno, A.; Akita, O.; Mikami, S.; Shimoi, H.; Gomi, K. Aspergillus oryzae atfA controls conidial germination and stress tolerance. Fungal Genet. Biol. 2009, 46, 887–897. [Google Scholar] [CrossRef]
- Roze, L.V.; Miller, M.J.; Rarick, M.; Mahanti, N.; Linz, J.E. A novel cAMP-response element, CRE1, modulates expression of nor-1 in Aspergillus parasiticus. J. Biol. Chem. 2004, 279, 27428–27439. [Google Scholar]
- Martinez-Pastor, M.T.; Marchler, G.; Schuller, C.; Marchler-Bauer, A.; Ruis, H.; Estruch, F. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J. 1996, 15, 2227–2235. [Google Scholar]
- Chang, P.K.; Scharfenstein, L.L.; Luo, M.; Mahoney, N.; Molyneux, R.J.; Yu, J.; Brown, R.L.; Campbell, B.C. Loss of msnA, a putative stress regulatory gene, in Aspergillus parasiticus and Aspergillus flavus increased production of conidia, aflatoxins and kojic acid. Toxins 2011, 3, 82–104. [Google Scholar] [CrossRef]
- Scherer, M.; Wei, H.; Liese, R.; Fischer, R. Aspergillus nidulans catalase-peroxidase gene (cpea) is transcriptionally induced during sexual development through the transcription factor StuA. Eukaryot. Cell 2002, 1, 725–735. [Google Scholar] [CrossRef]
- Twumasi-Boateng, K.; Yu, Y.; Chen, D.; Gravelat, F.N.; Nierman, W.C.; Sheppard, D.C. Transcriptional profiling identifies a role for BrlA in the response to nitrogen depletion and for StuA in the regulation of secondary metabolite clusters in Aspergillus fumigatus. Eukaryot. Cell 2009, 8, 104–115. [Google Scholar] [CrossRef]
- Brakhage, A.A.; Andrianopoulos, A.; Kato, M.; Steidl, S.; Davis, M.A.; Tsukagoshi, N.; Hynes, M.J. HAP-like CCAAT-binding complexes in filamentous fungi: Implications for biotechnology. Fungal Genet. Biol. 1999, 27, 243–252. [Google Scholar] [CrossRef]
- Romier, C.; Cocchiarella, F.; Mantovani, R.; Moras, D. The NF-YB/NF-YC structure gives insight into DNA binding and transcription regulation by CCAAT factor NF-Y. J. Biol. Chem. 2003, 278, 1336–1345. [Google Scholar]
- Laloum, T.; de Mita, S.; Gamas, P.; Baudin, M.; Niebel, A. CCAAT-box binding transcription factors in plants: Y so many? Trends Plant Sci. 2013, 18, 157–166. [Google Scholar] [CrossRef]
- Dolfini, D.; Gatta, R.; Mantovani, R. Nf-y and the transcriptional activation of CCAAT promoters. Crit. Rev. Biochem. Mol. Biol. 2012, 47, 29–49. [Google Scholar] [CrossRef]
- Petroni, K.; Kumimoto, R.W.; Gnesutta, N.; Calvenzani, V.; Fornari, M.; Tonelli, C.; Holt, B.F., 3rd; Mantovani, R. The promiscuous life of plant nuclear factor y transcription factors. Plant Cell 2012, 24, 4777–4792. [Google Scholar] [CrossRef]
- Gatta, R.; Mantovani, R. NF-Y affects histone acetylation and H2A.Z deposition in cell cycle promoters. Epigenetics 2011, 6, 526–534. [Google Scholar] [CrossRef]
- Hahn, S.; Guarente, L. Yeast HAP2 and HAP3: Transcriptional activators in a heteromeric complex. Science 1988, 240, 317–321. [Google Scholar]
- Forsburg, S.L.; Guarente, L. Identification and characterization of HAP4: A third component of the CCAAT-bound HAP2/HAP3 heteromer. Genes Dev. 1989, 3, 1166–1178. [Google Scholar] [CrossRef]
- Huber, E.M.; Scharf, D.H.; Hortschansky, P.; Groll, M.; Brakhage, A.A. DNA minor groove sensing and widening by the CCAAT-binding complex. Structure 2012, 20, 1757–1768. [Google Scholar] [CrossRef]
- Kato, M. An overview of the CCAAT-box binding factor in filamentous fungi: Assembly, nuclear translocation, and transcriptional enhancement. Biosci. Biotechnol. Biochem. 2005, 69, 663–672. [Google Scholar] [CrossRef]
- Steidl, S.; Tuncher, A.; Goda, H.; Guder, C.; Papadopoulou, N.; Kobayashi, T.; Tsukagoshi, N.; Kato, M.; Brakhage, A.A. A single subunit of a heterotrimeric CCAAT-binding complex carries a nuclear localization signal: Piggy back transport of the pre-assembled complex to the nucleus. J. Mol. Biol. 2004, 342, 515–524. [Google Scholar] [CrossRef]
- Goda, H.; Nagase, T.; Tanoue, S.; Sugiyama, J.; Steidl, S.; Tuncher, A.; Kobayashi, T.; Tsukagoshi, N.; Brakhage, A.A.; Kato, M. Nuclear translocation of the heterotrimeric CCAAT binding factor of Aspergillus oryzae is dependent on two redundant localising signals in a single subunit. Arch. Microbiol. 2005, 184, 93–100. [Google Scholar] [CrossRef]
- Litzka, O.; Then Bergh, K.; Brakhage, A.A. The Aspergillus nidulans penicillin-biosynthesis gene aat (penDE) is controlled by a CCAAT-containing DNA element. Eur. J. Biochem. 1996, 238, 675–682. [Google Scholar]
- Litzka, O.; Papagiannopolous, P.; Davis, M.A.; Hynes, M.J.; Brakhage, A.A. The penicillin regulator PENR1 of Aspergillus nidulans is a HAP-like transcriptional complex. Eur. J. Biochem. 1998, 251, 758–767. [Google Scholar]
- Sprote, P.; Hynes, M.J.; Hortschansky, P.; Shelest, E.; Scharf, D.H.; Wolke, S.M.; Brakhage, A.A. Identification of the novel penicillin biosynthesis gene aatb of Aspergillus nidulans and its putative evolutionary relationship to this fungal secondary metabolism gene cluster. Mol. Microbiol. 2008, 70, 445–461. [Google Scholar]
- Caruso, M.L.; Litzka, O.; Martic, G.; Lottspeich, F.; Brakhage, A.A. Novel basic-region helix-loop-helix transcription factor (AnBH1) of Aspergillus nidulans counteracts the CCAAT-binding complex AnCF in the promoter of a penicillin biosynthesis gene. J. Mol. Biol. 2002, 323, 425–439. [Google Scholar]
- Li, W.X.; Oono, Y.; Zhu, J.; He, X.J.; Wu, J.M.; Iida, K.; Lu, X.Y.; Cui, X.; Jin, H.; Zhu, J.K. The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell 2008, 20, 2238–2251. [Google Scholar] [CrossRef]
- Zeilinger, S.; Schmoll, M.; Pail, M.; Mach, R.L.; Kubicek, C.P. Nucleosome transactions on the Hypocrea jecorina (Trichoderma reesei) cellulase promoter cbh2 associated with cellulase induction. Mol. Genet. Genomics 2003, 270, 46–55. [Google Scholar] [CrossRef]
- Rauscher, R.; Wurleitner, E.; Wacenovsky, C.; Aro, N.; Stricker, A.R.; Zeilinger, S.; Kubicek, C.P.; Penttila, M.; Mach, R.L. Transcriptional regulation of xyn1, encoding xylanase I, in Hypocrea jecorina. Eukaryot. Cell 2006, 5, 447–456. [Google Scholar]
- Chen, H.; Crabb, J.W.; Kinsey, J.A. The Neurospora aab-1 gene encodes a CCAAT binding protein homologous to yeast HAP5. Genetics 1998, 148, 123–130. [Google Scholar]
- Camps, S.M.; Dutilh, B.E.; Arendrup, M.C.; Rijs, A.J.; Snelders, E.; Huynen, M.A.; Verweij, P.E.; Melchers, W.J. Discovery of a hapE mutation that causes azole resistance in Aspergillus fumigatus through whole genome sequencing and sexual crossing. PLoS One 2012, 7, e50034. [Google Scholar]
- Fernandes, M.; Xiao, H.; Lis, J.T. Fine structure analyses of the Drosophila and Saccharomyces heat shock factor—Heat shock element interactions. Nucleic Acids Res. 1994, 22, 167–173. [Google Scholar] [CrossRef]
- Liu, X.D.; Thiele, D.J. Oxidative stress induced heat shock factor phosphorylation and HSF-dependent activation of yeast metallothionein gene transcription. Genes Dev. 1996, 10, 592–603. [Google Scholar] [CrossRef]
- Raitt, D.C.; Johnson, A.L.; Erkine, A.M.; Makino, K.; Morgan, B.; Gross, D.S.; Johnston, L.H. The Skn7 response regulator of Saccharomyces cerevisiae interacts with Hsf1 in vivo and is required for the induction of heat shock genes by oxidative stress. Mol. Biol. Cell 2000, 11, 2335–2347. [Google Scholar]
- Yamamoto, A.; Ueda, J.; Yamamoto, N.; Hashikawa, N.; Sakurai, H. Role of heat shock transcription factor in Saccharomyces cerevisiae oxidative stress response. Eukaryot. Cell 2007, 6, 1373–1379. [Google Scholar] [CrossRef]
- Hagiwara, D.; Asano, Y.; Marui, J.; Furukawa, K.; Kanamaru, K.; Kato, M.; Abe, K.; Kobayashi, T.; Yamashino, T.; Mizuno, T. The SskA and SrrA response regulators are implicated in oxidative stress responses of hyphae and asexual spores in the phosphorelay signaling network of Aspergillus nidulans. Biosci. Biotechnol. Biochem. 2007, 71, 1003–1014. [Google Scholar] [CrossRef]
- Vargas-Perez, I.; Sanchez, O.; Kawasaki, L.; Georgellis, D.; Aguirre, J. Response regulators SrrA and SskA are central components of a phosphorelay system involved in stress signal transduction and asexual sporulation in Aspergillus nidulans. Eukaryot. Cell 2007, 6, 1570–1583. [Google Scholar] [CrossRef]
- Fassler, J.S.; West, A.H. Fungal Skn7 stress responses and their relationship to virulence. Eukaryot. Cell 2011, 10, 156–167. [Google Scholar] [CrossRef]
- Morgan, B.A.; Banks, G.R.; Toone, W.M.; Raitt, D.; Kuge, S.; Johnston, L.H. The Skn7 response regulator controls gene expression in the oxidative stress response of the budding yeast Saccharomyces cerevisiae. EMBO J. 1997, 16, 1035–1044. [Google Scholar] [CrossRef]
- Ohmiya, R.; Kato, C.; Yamada, H.; Aiba, H.; Mizuno, T. A fission yeast gene (prr1+) that encodes a response regulator implicated in oxidative stress response. J. Biochem. 1999, 125, 1061–1066. [Google Scholar] [CrossRef]
- Lippold, F.; Sanchez, D.H.; Musialak, M.; Schlereth, A.; Scheible, W.R.; Hincha, D.K.; Udvardi, M.K. AtMyb41 regulates transcriptional and metabolic responses to osmotic stress in arabidopsis. Plant Physiol. 2009, 149, 1761–1772. [Google Scholar]
- Borevitz, J.O.; Xia, Y.; Blount, J.; Dixon, R.A.; Lamb, C. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 2000, 12, 2383–2394. [Google Scholar]
- Scheible, W.R.; Morcuende, R.; Czechowski, T.; Fritz, C.; Osuna, D.; Palacios-Rojas, N.; Schindelasch, D.; Thimm, O.; Udvardi, M.K.; Stitt, M. Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol. 2004, 136, 2483–2499. [Google Scholar] [CrossRef]
- Peel, G.J.; Pang, Y.; Modolo, L.V.; Dixon, R.A. The LAP1 MYB transcription factor orchestrates anthocyanidin biosynthesis and glycosylation in medicago. Plant J. 2009, 59, 136–149. [Google Scholar] [CrossRef]
- Wieser, J.; Adams, T.H. FlbD encodes a Myb-like DNA-binding protein that coordinates initiation of Aspergillus nidulans conidiophore development. Genes Dev. 1995, 9, 491–502. [Google Scholar] [CrossRef]
- Arratia-Quijada, J.; Sanchez, O.; Scazzocchio, C.; Aguirre, J. FlbD, a Myb transcription factor of Aspergillus nidulans, is uniquely involved in both asexual and sexual differentiation. Eukaryot. Cell 2012, 11, 1132–1142. [Google Scholar] [CrossRef]
- Zhang, M.; Lu, J.; Tao, K.; Ye, W.; Li, A.; Liu, X.; Kong, L.; Dong, S.; Zheng, X.; Wang, Y. A Myb transcription factor of Phytophthora sojae, regulated by MAP kinase PsSAK1, is required for zoospore development. PloS One 2012, 7, e40246. [Google Scholar]
- Cary, J.W.; Harris-Coward, P.Y.; Ehrlich, K.C.; Mack, B.M.; Kale, S.P.; Larey, C.; Calvo, A.M. NsdC and NsdD affect Aspergillus flavus morphogenesis and aflatoxin production. Eukaryot. Cell 2012, 11, 1104–1111. [Google Scholar] [CrossRef]
- Shaaban, M.I.; Bok, J.W.; Lauer, C.; Keller, N.P. Suppressor mutagenesis identifies a velvet complex remediator of Aspergillus nidulans secondary metabolism. Eukaryot. Cell 2010, 9, 1816–1824. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).