Abstract
Saxitoxin (STX) is one of the most potent marine neurotoxins, produced by several species of freshwater cyanobacteria and marine dinoflagellates. Although omics-based approaches have advanced our understanding of STX biosynthesis in recent decades, the origin, regulation, and ecological drivers of STX in dinoflagellates remain poorly resolved. Specifically, dinoflagellate STX biosynthetic genes (sxt) are extremely fragmented, inconsistently expressed, and unevenly distributed between toxic and non-toxic taxa. Environmental studies further report inconsistent relationships between abiotic factors and STX production, suggesting regulation across multiple genomic, transcriptional, post-transcriptional, and epigenetic levels. These gaps prevent a comprehensive understanding of STX biosynthesis in dinoflagellates and limit the development of accurate predictive models for harmful algal blooms (HABs) and paralytic shellfish poisoning (PSP). Artificial intelligence (AI), including machine learning and deep learning, offers new opportunities in ecological pattern recognition, molecular annotation, and data-driven prediction. This review explores the current state of knowledge and persistent knowledge gaps in dinoflagellate STX research and proposes an AI-integrated multi-omics framework highlighting recommended models for sxt gene identification (e.g., DeepFRI, ProtTrans, ESM-2), evolutionary reconstruction (e.g., PhyloGAN, GNN, PhyloVAE, NeuralNJ), molecular regulation (e.g., MOFA+, LSTM, GRU, DeepMF), and toxin prediction (e.g., XGBoost, LightGBM, LSTM, ConvLSTM). By integrating AI with diverse biological datasets, this novel framework outlines how AI can advance fundamental understanding of STX biosynthesis and inform future applications in HAB monitoring, seafood safety, and PSP risk management in aquaculture and fisheries.
Keywords:
machine learning integration; multi-omics analysis; deep learning; paralytic shellfish poisoning (PSP); harmful algal blooms; saxitoxin; dinoflagellates Key Contribution:
This review provides the first integrated framework for applying AI and multi-omics approaches to unravel the origin, regulation, and ecological dynamics of dinoflagellate saxitoxin, offering a novel pathway toward predictive toxin monitoring and HAB forecasting.
1. Introduction
The UN Decade of Ocean Science for Sustainable Development (2021–2030) calls for innovative and timely scientific approaches to ensure a safe and sustainable ocean, one where human health, ecosystems, and economies coexist in balance [1]. Yet, despite significant progress in marine research, the ocean continues to present natural hazards with profound biological and socioeconomic consequences [2]. Among these, marine biotoxins remain a persistent and complex threat to food security, coastal livelihoods, and global public health [3,4].
Saxitoxins (STXs) are among the most potent natural biotoxins known, produced primarily by certain marine dinoflagellates and freshwater cyanobacteria [5,6,7,8]. These toxins are the primary cause of paralytic shellfish poisoning (PSP), a severe neurological disorder resulting from the consumption of contaminated seafood [8,9]. Globally, approximately 2000 cases of PSP are reported annually, with mortality rates reaching up to 15%, underscoring the continuing public health and economic burden associated with harmful algal blooms (HABs) [10,11,12,13,14].
The growing incidence of HAB-related poisonings is increasingly linked to climate change, intensifying the urgency to investigate toxin-producing species [2,15,16]. Rising temperatures and shifting ocean conditions are expanding the range and frequency of HAB events worldwide [2]. Evidence indicates that climate-driven alterations in temperature, salinity, and nutrient dynamics shape the growth and toxicity of PSP-producing taxa such as the marine dinoflagellate Alexandrium [17,18,19,20,21]. Early warnings on toxicity would require knowing the biochemical processes responsible for the synthesis of STX-related compounds in dinoflagellates in response to environmental changes [5,6]. Given these emerging risks, a comprehensive understanding of STX biosynthesis, regulation, and distribution remains critical for ensuring both ocean and public health security.
Despite decades of research, significant gaps remain in understanding the molecular and ecological mechanisms underlying STX biosynthesis in dinoflagellates. Although extensive genetic and transcriptomic data are now available [22], their complexity continues to obscure the evolutionary dynamics of saxitoxin biosynthesis genes (sxt), patterns of gene expression, and climate-driven influences on toxin production, a paradox where each discovery seems to reveal even deeper layers of complexity. These challenges emphasize the urgent need for innovative computational and integrative approaches to unravel the complex nature of STX biosynthesis.
In this sense, machine learning (ML) and artificial intelligence (AI) have become powerful tools, providing new capabilities in ecological pattern recognition, molecular annotation, and data-driven prediction [23,24,25]. AI has the potential to completely change how we identify, model, and comprehend saxitoxin-producing organisms by combining genomics, transcriptomics, proteomics, metabolomics, and environmental datasets [25,26,27,28,29,30]. This review examines the current state and persistent knowledge gaps in dinoflagellate saxitoxin research and proposes the integration of multi-omics data and AI to advance the field. Furthermore, it introduces an innovative research framework that applies AI to sxt gene identification, evolutionary inference, molecular regulation, and toxin prediction. With this integration, saxitoxin research is moving from decades of contradictory results into a new era of comprehensive understanding of dinoflagellate toxins. The AI integration bridges the gap between basic science and practical applications in marine toxin management by unifying fragmented data, elucidating biological mechanisms, and turning knowledge into action, positioning these advancements within the broader context of ocean health and public health security.
2. Saxitoxin Biosynthesis: Cyanobacteria vs. Dinoflagellates
The biosynthesis of saxitoxins was first elucidated in freshwater cyanobacteria (Raphidiopsis brookii, Anabaena circinalis, Aphanizomenon sp., Cylindrospermopsis raciborskii, and Microseira wollei), where a well-characterized sxt gene cluster comprising approximately 30 genes (sxtA-sxtZ) orchestrates modular enzymatic steps [8,20,31,32]. These include core genes (sxtA, sxtB, sxtD, sxtG, sxtH/T, sxtI, sxtS, and sxtU) and various tailoring and transporting enzymes responsible for the structural diversity of STX and its derivatives, such as GTX2, GTX3, and neosaxitoxin (NeoSTX) [32,33]. Cyanobacterial STX biosynthesis serves as a foundational model for understanding the genetic and enzymatic logic of saxitoxin production [8,34].
In contrast, dinoflagellates exhibit a highly fragmented and complex distribution of sxt homologs and produce a broader diversity of STX derivatives, including sulfated forms such as C1/C2, GTX4, and GTX6, which are only rarely produced by cyanobacteria [35,36,37]. Among dinoflagellates, Alexandrium spp. (approximately ten toxic species), Pyrodinium bahamense, Gymnodinium catenatum, and Centrodinium punctatum are the only confirmed saxitoxin (STX)-producing taxa [37,38,39,40,41,42]. Core genes such as sxtA (particularly the sxtA4 domain) and sxtG have been identified across most toxic species [36,39,42,43]. In contrast, non-toxic species such as Alexandrium fragae, A. fraterculus, Scrippsiella trochoidea, Margalefidinium polykrikoides, and freshwater Palatinus apiculatus typically lack these core genes [42,44,45,46]. However, the distribution of sxt genes does not always correspond to toxicity status. For example, C. punctatum reportedly lacks sxtA4 [47], and G. catenatum lacks sxtB [39]. Moreover, complete sxt gene sets have been detected in non-toxic strains of Alexandrium spp. [37,48,49,50] and in the non-toxic G. smaydae [51]. These inconsistencies highlight the complex evolutionary dynamics of sxt homologs in dinoflagellates [52], which seem to be different from cyanobacteria [53] and raise a critical question: which specific genes and pathways ultimately determine saxitoxin biosynthesis in this group?
Furthermore, comparative transcriptomic analyses further reveal substantial variation in sxt gene expression across dinoflagellate species. Nevertheless, the relationship between sxt gene expression and actual toxin production remains inconsistent. For example, some studies reported that sxtA4 and sxtG expression in Alexandrium species were significantly reduced in non-toxic strains [49,54,55,56]. In contrast, other studies observed no significant difference in sxt gene expression between toxic and non-toxic individuals, suggesting that STX biosynthesis in dinoflagellates may be regulated beyond transcription, potentially at post-transcriptional or translational levels [37,57,58].
In addition, although several abiotic stressors, such as nutrient limitation, salinity shifts, and temperature or light fluctuations, have been linked to changes in STX production [17,18,19,21,59,60], current evidence does not yet support a consistent, predictive model connecting environmental triggers, sxt gene expressions, and toxin yield [19,61,62,63]. This inconsistency implies that additional regulatory layers, including epigenetic modulation [64,65] or microbial community interactions [66], may play key roles in mediating STX biosynthesis [5,22]. Collectively, these observations highlight a critical knowledge gap in our understanding of how sxt genes, environmental factors, and toxin production interact.
3. Evolutionary Perspective
The inconsistent relationship between sxt gene expression, environmental drivers, and STX production suggests that sxt gene evolution in dinoflagellates involves complex mechanisms beyond simple vertical inheritance [34,36]. Early hypotheses proposed that sxt genes were acquired in dinoflagellates via horizontal gene transfer (HGT) from cyanobacteria to a common ancestor of Alexandrium and P. bahamense, with G. catenatum later acquiring them from Alexandrium secondarily via HGT [36]. Phylogenetic analyses appeared to support this, as the STX-producing P. bahamense and C. punctatum consistently cluster within the Alexandrium lineage, whereas G. catenatum forms a more distant branch [47,51]. However, recent studies suggest that cyanobacteria and dinoflagellates may have acquired sxt genes independently rather than through a single ancient HGT event [34,67]. Consistently, G. catenatum forms a distinct lineage in sxt phylogenies, contradicting the idea of secondary acquisition from Alexandrium [39,67].
In dinoflagellates, current evidence suggests that sxt genes were acquired early in dinoflagellate evolution and subsequently shaped by lineage-specific loss, duplication, and divergence [42,67]. This has produced the current patchy distribution of sxt homologs, in which some non-toxic dinoflagellate species retain partial or non-functional remnants of the sxt genes [34,36,39,45,46,50,51,67]. An alternative hypothesis is that the sxt gene cluster may have originated independently in multiple dinoflagellate lineages through a complex history of horizontal gene transfer (HGT), potentially involving bacterial donors such as Proteobacteria or Actinobacteria, like the recurrent HGT events observed in cyanobacteria [33].
All these interpretations remain highly speculative. The evolutionary genomic origins of saxitoxin production and the underlying sxt gene clusters in dinoflagellates remain an open question. Did STX biosynthesis arise once or independently multiple times in dinoflagellates? Do retained sxt homologs represent active enzymes or evolutionary relics? What ecological and environmental forces govern the retention, silencing, or loss of these genes across lineages? Until the evolutionary history of sxt genes is clearly resolved, our understanding of saxitoxin biosynthesis will remain incomplete, limiting predictive capacity and constraining efforts to anticipate or mitigate its ecological and public health impacts.
4. Knowledge Gaps and Challenges in Dinoflagellates
Despite remarkable advances in sequencing technologies and comparative omics, significant gaps remain in our understanding of STX biosynthesis in dinoflagellates. Five critical gaps remain in saxitoxin research:
- Incomplete gene clusters: No dinoflagellate genome has yet revealed a complete sxt gene cluster due to extensive genome fragmentation, repetitive content, and multiple isoforms.
- Regulatory complexity; Mechanisms involving transcriptional regulation, alternative splicing, and epigenetic control are poorly characterized, limiting accurate inference of toxin biosynthesis from sequence data.
- Functional validation: Experimental confirmation of sxt gene function remains difficult because of the extraordinarily large, polyploid, and repetitive genomes of dinoflagellates, combined with a lack of robust genetic tools.
- Environmental modulation: Toxin production varies under multiple interacting stressors (temperature, salinity, nutrient availability, and light), yet the molecular links between environmental cues and toxin biosynthesis remain unclear.
- Unresolved evolutionary origins and diversification; The evolutionary history of sxt genes in dinoflagellates remains poorly resolved.
These uncertainties are compounded by the extraordinarily large and highly repetitive genomes of dinoflagellates, characterized by extensive gene duplication and alternative splicing [68,69,70], which present major challenges for accurately reconstructing sxt gene clusters and elucidating their regulatory networks [69]. Additionally, field observations add further complexity, as dinoflagellate blooms arise under multifactorial environmental conditions and are modulated by intricate ecological interactions such as microbiome composition, grazing pressure, and nutrient competition, factors that are difficult to replicate in controlled laboratory settings [17,71]. Table 1 below summarizes the major knowledge gaps in sxt gene cluster architecture, regulation, environmental modulation, and evolutionary origins in dinoflagellates.

Table 1.
Summary of inconsistent findings and major knowledge gaps in dinoflagellate saxitoxin (STX) research.
Collectively, these challenges underscore why conventional approaches alone have been insufficient to fully resolve the evolutionary dynamics and functional complexity of sxt genes, despite decades of research [22,52]. Addressing these complexities requires analytical strategies capable of integrating heterogeneous, high-dimensional data across genomic, transcriptomic, environmental, and evolutionary scales. In this context, artificial intelligence and machine learning offer powerful tools to combine multi-omics datasets that can lay the groundwork for predictive, data-driven frameworks that not only advance our understanding of dinoflagellate toxins but also enhance risk assessment, forecasting, and early-warning capabilities in an ocean under rapid climate change [23,30,94].
5. Artificial Intelligence: A Future Tool in Dinoflagellate Saxitoxin Research
Artificial intelligence (AI) offers transformative opportunities for resolving long-standing challenges in saxitoxin (STX) research [30,95], particularly considering the highly complex dinoflagellate genomes, fragmented and poorly conserved sxt gene clusters, and the multifactorial environmental drivers that regulate toxin production. While conventional genomics and transcriptomics have provided foundational insights, AI-based approaches uniquely enable the discovery of hidden patterns, the prediction of biological outcomes, the integration of heterogeneous datasets, and the reconstruction of latent molecular structures that remain inaccessible through traditional methods [96,97,98,99]. By synthesizing multiple omics layers, including genomics, transcriptomics, proteomics, metabolomics, and environmental data, AI has the capacity to reveal previously obscured regulatory mechanisms, predict the dynamics of harmful algal blooms, and illuminate the molecular basis of saxitoxin biosynthesis with unprecedented precision (Figure 1).
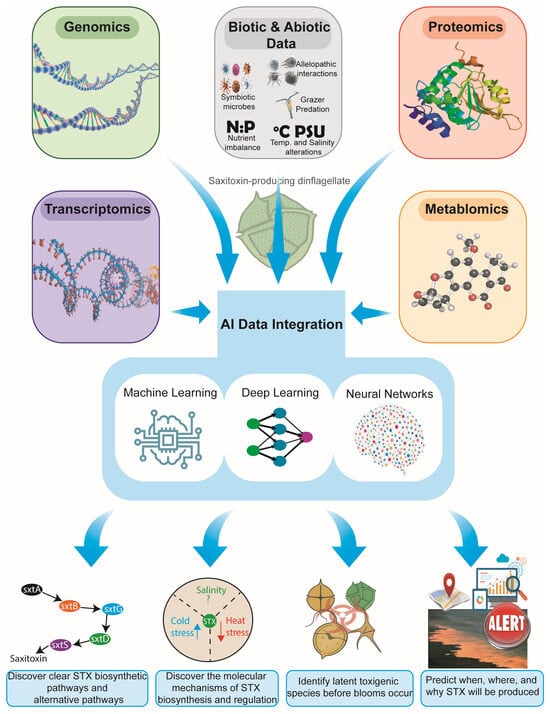
Figure 1.
Integrated multi-omics and environmental data framework for AI-driven saxitoxin research in dinoflagellates. The figure illustrates how genomics, transcriptomics, proteomics, metabolomics, and biotic/abiotic ecological data are integrated through AI approaches, including machine learning, deep learning, and neural networks, to uncover biosynthetic pathways, elucidate molecular regulatory mechanisms, identify latent toxigenic species, and predict the timing, location, and drivers of STX production.
Below, we review key methodological challenges and provide experimental strategies, paired with AI methods, to address them.
5.1. AI for Accurate Molecular Identification of sxt Genes in Dinoflagellate
Accurate identification of sxt genes in dinoflagellates remains one of the major challenges in saxitoxin research. Unlike cyanobacteria, where sxt genes occur in compact and well-defined clusters [32], dinoflagellate genomes are huge, repetitive, and rich in duplicated PKS-like sequences, obscuring gene boundaries and inflating homolog counts [52,69]. To date, several core sxt genes have been isolated and functionally characterized in Alexandrium such as sxtA, sxtB, sxtG, sxtI, and sxtU. Similarly, sxt homologs are reported across many dinoflagellate species primarily from transcriptomic data; however, in all cases these genes remain partial fragments, or expanded paralog families resulting in high false-positive rates [18,46,52,100,101]. Although definitive confirmation requires high-quality genome assemblies, full-length transcripts, and biochemical validation, such rigorous datasets are rarely available across species [5,22,52]. This limitation highlights the need for an AI-guided identification pipeline.
Deep learning–based gene prediction models, including convolutional neural networks (CNN) and transformer architectures [102,103,104,105], can detect coding signatures and functional motifs within highly repetitive or fragmented dinoflagellate sequences that traditional HMM- or BLAST-based workflows fail to distinguish [50]. Protein language models such as ESM-2, ProtT5, ProtBERT, and AlphaFold + DeepFRI generate high-dimensional embeddings that capture biochemical properties, structural constraints, and evolutionary signals [104,105,106,107] enabling functional annotation of remote homologs with limited sequence identity [104]. Specifically, DeepFRI and ProtTrans models have been shown to substantially improve Gene Ontology and enzyme function prediction accuracy in poorly annotated genomes, while maintaining robust performance under sparse training data conditions [108]. Similarly, DeepBGC has demonstrated enhanced sensitivity in detecting cryptic and atypical biosynthetic gene clusters compared with rule-based and profile-based methods, including clusters missed by antiSMASH and HMM-only pipelines [109].
These outcomes are particularly relevant to dinoflagellate sxt genes, which exhibit extensive fragmentation and share conserved domains with polyketide synthase (PKS) and fatty acid synthase (FAS) paralogs. Embedding-based classifiers and structural feature integration have been shown to discriminate functionally distinct enzyme families despite high sequence similarity, supporting their application for separating true sxt enzymes from PKS- or FAS-like homologs [110,111]. In parallel, graph neural networks (GNNs) [112] can classify complex multidomain architectures characteristic of dinoflagellate toxin genes, while ensemble pipelines integrating HMM profiles, structural predictions, and deep-learning embeddings will provide robust cross-validation [102,112]. This will make high-confidence sxt gene identification possible even in the absence of complete genomes or biochemical assays, transforming a previously slow, error-prone, and species-specific process into a scalable, reproducible pipeline [26,111,113]. In short, AI could shift sxt gene discovery from reactive validation to predictive, high-throughput discovery, fundamentally changing our ability to map toxin biosynthesis across dinoflagellate diversity. Collectively, these advances highlight a transformative opportunity to finally resolve the long-standing challenges associated with accurate sxt gene detection in dinoflagellates.
5.2. Integrative Phylogenomics and Phylogenetics of sxt Genes with AI-Enhanced Approaches for Understanding Their Evolution in Dinoflagellates
At the phylogenetic level, sxt genes in dinoflagellates often form clades incongruent with organismal trees, indicating recurrent horizontal acquisitions, retentions of ancestral duplicates, or differential losses across lineages [36,39,42,50]. This mosaic evolution complicates ortholog inference but also highlights the evolutionary flexibility of toxin biosynthesis pathways. Understanding the evolutionary history of sxt genes in dinoflagellates is essential for reconstructing the origins of STX biosynthesis and explaining the remarkable variability in toxicity across species and strains. AI-driven phylogenomic and phylogenetic approaches are proving increasingly transformative [114,115,116]. For instance, machine learning models (e.g., Random Forests, Support Vector Machines), deep learning frameworks (e.g., PhyloGAN, GNN, PhyloVAE, NeuralNJ), and reinforcement learning have been successfully applied to reconstruct phylogenies and unravel complex evolutionary processes, such as horizontal gene transfer, in other species, including bacteria [117,118].
However, in dinoflagellates, to map the evolutionary dynamics of sxt genes, an integrative framework is proposed, combining genome- or transcriptome-based phylogenomics with phylogenetic reconstruction of all identified sxt genes, augmented by AI and ML methods. However, transcriptome-based phylogenomics [119] is more practical because of the incredibly large genome of STX-producing dinoflagellates [68]. Additionally, a number of dinoflagellate species’ transcriptome data are currently accessible on public databases such as GenBank of the National Center for Biotechnology Information (NCBI). Thus, phylotranscriptomics could provide a species tree backbone using conserved single-copy genes, contextualizing gene-specific evolution [119]. AI approaches, such as embedding-based orthology inference with protein language models (ESM, ProtTrans) and GNN, can resolve fragmented or divergent transcripts, distinguishing orthologs from paralogs and improving alignments [116,120,121,122]. For sxt gene phylogenetics, sequences could be first curated by deep-learning classifiers (e.g., embeddings + DL-based selection), before building maximum-likelihood or Bayesian trees. Then, AI-based reconciliation [123,124] could integrate gene and species trees to detect horizontal gene transfer, gene gain/loss, and lineage-specific expansions, a strategy analogous to recent ML-driven reconciliation methods and reconciliation frameworks that model duplication, transfer, and loss events [123,124,125,126,127]. Furthermore, variational autoencoders (VAEs) and other generative deep-learning models can construct low-dimensional latent representations of protein sequence space that reflect evolutionary divergence, selective constraints, and functional relationships; such spaces have been used to model fitness landscapes and predict mutational effects among paralogs or novel variants [128,129,130,131]. Together, these AI-driven and phylogenomic approaches offer a scalable framework to resolve fragmented sxt gene clusters, distinguish orthologs from paralogs, infer horizontal gene transfer, duplication, and loss events, and link sequence evolution to ecological adaptation and toxin diversity, thus providing a predictive roadmap for understanding saxitoxin evolution even in the absence of complete genomes or extensive experimental validation
5.3. Decoding the Regulatory Mechanisms of STX Biosynthesis in Dinoflagellates Using Multi-Omics Data and an AI-Integrated Approach
Deciphering the regulation of STX biosynthesis in dinoflagellates remains a major challenge due to the complex and layered control of gene expression in these organisms [22,52]. Previous studies provide inconsistent evidence of transcriptional activity that regulates STX production [37,52,56,57]. Therefore, emerging evidence strongly suggests post-transcriptional mechanisms, including alternative transcript isoforms, RNA editing, and variations in untranslated regions (UTRs), as critical modulators of toxin production [37,64,76,132]. At present, the regulatory mechanisms of STX synthesis in dinoflagellates are not known. Additionally, omics-based studies in dinoflagellates focusing on saxitoxin biosynthesis are still sporadic and far from coming to a concrete closure [22,65,133]. This highlights the need for an integrated multi-omics (genomics, transcriptomics, proteomics, and metabolomics), e.g., [134] and an AI approach capable of simultaneously capturing transcriptional dynamics, post-transcriptional modifications, and translational output, providing a comprehensive framework to uncover the regulatory architecture of sxt gene expression.
Machine learning–based multi-omics integration has already proven powerful in fields such as disease diagnosis, treatment prediction, and gene regulatory network inference [27,30,99,135,136,137,138]. For example, the mechanism of salt tolerance in plants was revealed by integrating the KANMB Machine Learning Model with metabolomic and transcriptomic data [139]. These same frameworks can be leveraged for dinoflagellate saxitoxin research by combining multi-omics datasets [22,134,140]. RNA-Seq under time-course and environmental treatments could provide high-resolution profiles of transcript abundance, while ribosome profiling (Ribo-Seq) and polysome fractionation can identify which sxt transcripts are actively translated [134]. Furthermore, small RNA sequencing and nanopore direct RNA sequencing could reveal regulatory RNAs, RNA editing, alternative splice variants, and trans-splicing events that shape sxt gene expression beyond transcription; for example, SL trans-splicing is widespread in dinoflagellates [141] and nanopore DRS has been used in other eukaryotes to identify full-length isoforms, mRNA modifications, and novel splice events [142,143]. Complementing these data, AI and machine learning models can predict translational efficiency, detect dynamic regulatory patterns, and infer causal post-transcriptional networks [26,144]. Temporal deep learning models (LSTM, GRU) capture time-resolved expression dynamics, while graph-based models and Bayesian networks integrate multi-omics data to uncover hidden regulatory relationships [140]. VAEs and other integrative frameworks (e.g., MOFA+, DeepMF) can cluster regulatory states and model latent control mechanisms [145,146,147]. By combining multi-omics experiments with AI-driven analysis, it becomes possible to identify sxt transcripts under translational control, predict regulatory interactions, and generate scalable, predictive models of toxin biosynthesis under variable environmental conditions. Figure 2 provides an overview of the omics experimental methods, AI techniques, and expected outcomes.
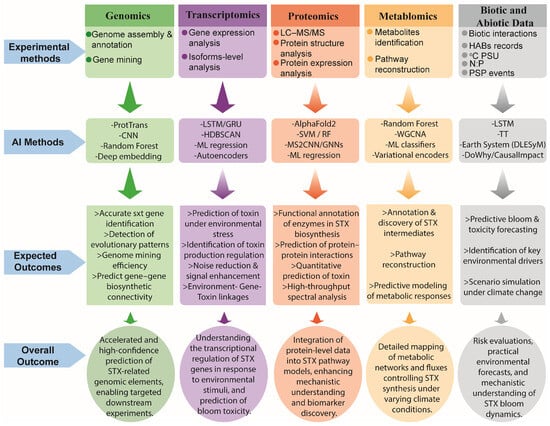
Figure 2.
Multi-omics experimental methods, Artificial Intelligence (AI) approaches, and expected outcomes in dinoflagellate saxitoxin research. The figure summarizes how genomics, transcriptomics, proteomics, metabolomics, and biotic/abiotic environmental data are analyzed using diverse AI, machine-learning, and deep learning techniques to generate predictive, mechanistic, and functional insights. Expected outcomes include improved gene annotation, toxin-regulation modeling, enzyme function prediction, metabolic pathway reconstruction, and environmental forecasting.
5.4. AI-Driven Reconstruction of Saxitoxin Gene Clusters and Biosynthetic Pathways
Reconstructing the saxitoxin biosynthetic pathway in dinoflagellates remains particularly challenging because meaningful pathway inference depends on both accurate gene identification and a clear understanding of how these genes are regulated. Unlike cyanobacteria, where sxt genes form compact, conserved clusters with identified pathways [32,33], dinoflagellates show fragmented, dispersed, plastid-associated, and paralog-rich sxt architectures [34,36,48]. Moreover, increasing evidence suggests that STX biosynthesis may be species-specific, with different dinoflagellates using distinct subsets of sxt genes [39,51], or compensating for missing steps using unrelated PKS-like enzymes. Thus, the pathway may not be universally conserved across the group, making generalization difficult without species-level resolution.
To resolve the STX biosynthetic pathway in dinoflagellates with potential species-specific architectures, a combined experimental and computational strategy will be essential [109,148,149,150]. Long-read sequencing combined with Hi-C scaffolding has been shown to resolve complex gene neighborhoods, reveal micro-clusters, and detect lineage-specific structural rearrangements in highly repetitive genomes [151,152]. Furthermore, co-expression network approaches such as WGCNA [153], together with more advanced integrative Gene Regulatory Network (iGRN) inference methods, will help identify species-specific functional modules and regulatory circuits associated with toxin biosynthesis [136,149,153]. LC-MS/MS metabolomics will allow direct comparison of saxitoxin intermediates to determine differences in pathway completeness, while stable isotope labeling can validate enzyme functions and pathway flux in a species-dependent context [65,154]. Protein–protein interaction assays (Y2H, co-IP, AP-MS) will further uncover enzyme complexes that may differ among lineages. AI-driven methods further enhance cluster reconstruction: Graph Neural Networks (GNNs) can infer hidden biosynthetic modules from fragmented scaffolds [120], while deep-learning metabolomics frameworks (DeepMET, DeepMass) associate metabolites with candidate genes to resolve missing steps in species-specific pathways [155,156]. Finally, multi-omics integrative tools such as DeepPath [157] and NICEpath [158] can then model alternative biosynthetic routes, enabling prediction of lineage-specific sxt gene cluster organization and pathway variants across dinoflagellates. Together, this framework could enable the first realistic reconstruction of sxt gene clusters, whether canonical, fragmented, or species-specific, across diverse toxic dinoflagellates.
5.5. Predicting Toxicity in a Changing Ocean: AI Solutions to Understanding Saxitoxin Environmental Drivers
In the context of rapid climate change, the frequency, intensity, and geographic distribution of saxitoxin-producing harmful algal blooms (HABs) and paralytic shellfish poisoning (PSP) events are expanding into new areas, posing increasing risks to human health, fisheries, and marine ecosystems [159,160,161,162]. As HABs often arise under multifactorial environmental conditions, recent advances across aquatic toxin research, including cyanobacteria and dinoflagellate toxins, employ AI integration of biological and environmental data to understand the dynamics of HABs and forecast HABs, identify key environmental triggers, and predict bloom toxicity with high accuracy (e.g., 84%) [96,98,163,164,165,166]. In dinoflagellates, saxitoxin biosynthesis regulation is indeed a complex process, controlled by several environmental cues that are yet to be clearly defined [5,22]. Based on these, we suggest that machine learning (ML) and artificial intelligence (AI) based frameworks are well-suited for the integration and analysis of biological and environmental data from naturally occurring HABs, and that such an approach would greatly improve our ability to understand saxitoxin and predict HABs under a changing climate [167,168]. For example, deep learning models that capture interactions among environmental variables have improved riverine HAB prediction [169], while multi-horizon architectures enable robust long-term bloom forecasting by resolving complex temporal dependencies [170]. At broader spatial scales, the integration of remote sensing with AI has enhanced early detection and prediction of inland-water HABs [167], and machine-learning approaches have successfully linked environmental variability to blooms of toxin-producing taxa such as Pseudo-nitzschia in coastal systems [168]. Collectively, these advances demonstrate how AI can support predictive frameworks relevant to the monitoring and management of toxin-producing algal blooms.
In dinoflagellate, the effective AI-driven prediction of saxitoxin risk must be grounded in experimental and field-based observations that capture the biochemical and ecological controls of toxin production. Controlled laboratory experiments manipulating nutrients, temperature, pH, or metal concentrations provide mechanistic insight into how environmental stressors influence saxitoxin (STX) production [18,60,62,77], while co-culture and microbiome studies reveal the role of biological interactions, such as competition and symbiosis [66]. High-frequency field sampling of natural blooms, combined with environmental and community metadata, including nutrient levels, light, hydrodynamics, and microbiome composition, could enable in situ assessment of STX variability [17,96,171]. Quantitative toxin measurements (LC–MS/MS) alongside transcriptomic and metabolomic profiling could link both abiotic and biotic factors to molecular and biochemical responses [134,154]. In addition, extensive datasets on species-specific toxin analog profiles and their geographic distributions, derived from long-term monitoring programs and published toxin surveys, provide critical phenotypic labels that can be integrated with molecular and environmental data for AI model training, validation, and cross-regional generalization [172,173]. Artificial intelligence and machine-learning approaches can then integrate these heterogeneous datasets to predict toxin production and identify key drivers. Algorithms such as Random Forests, gradient boosting models (XGBoost, LightGBM), and Support Vector Regression can model sxt responses to both abiotic and biotic variables [174], while spatiotemporal neural networks, including LSTMs, ConvLSTMs, temporal transformers, and Earth system AI models like neural operators, would enable bloom-scale forecasting under dynamic environmental and ecological conditions [96,175,176]. Causal inference frameworks, such as DoWhy [177] and CausalImpact [178], allow researchers to distinguish correlation from causation, revealing which environmental or biological factors directly trigger or amplify STX production. By combining controlled experiments, field data, and AI-driven predictive modeling, this integrative approach provides a comprehensive framework for understanding and forecasting saxitoxin dynamics in a changing and biologically interactive ocean.
6. Potential Impact and Ecological Implications
The integration of AI into saxitoxin research has the potential to be truly transformative, redefining decades of traditional studies and accelerating discovery in ways previously unattainable. By rapidly identifying latent toxigenic species and predicting harmful algal bloom (HAB) dynamics, AI can revolutionize monitoring, early warning systems, and risk assessment for human and ecosystem health [96]. On the molecular and evolutionary front, AI can illuminate the diversification, retention, and horizontal transfer of sxt genes across dinoflagellate lineages, resolving longstanding uncertainties about saxitoxin biosynthesis and regulatory mechanisms. Coupled with environmental and biological interaction data, predictive AI models can forecast toxin production under changing climate and nutrient conditions, enabling proactive ecosystem management and informed policy decisions.
Beyond immediate advances in monitoring and prediction, the integration of AI introduces a paradigm shift in how saxitoxin research is conceptualized and operationalized. AI-driven frameworks enable continuous learning from expanding global datasets, allowing models to evolve alongside changing ocean conditions and emerging toxin profiles. This adaptive capacity supports the translation of fundamental molecular insights into real-world applications, including dynamic risk mapping, decision-support tools for fisheries and public health agencies, and the prioritization of surveillance efforts in data-limited regions. By unifying molecular processes with large-scale environmental variability, AI establishes a scalable research infrastructure that bridges discovery science and applied management, positioning saxitoxin research within a broader system of global ocean intelligence and long-term resilience planning [1].
7. Current Limitations, Challenges, and Future Perspectives
Despite its transformative potential, the application of AI to saxitoxin research faces several major limitations [30,135]. One of the most fundamental is the lack of accurate identification of sxt genes in dinoflagellates, which remains extremely challenging due to fragmented transcripts, extensive paralog expansion, and limited biochemical validation. However, AI frameworks, such as DeepBGC for biosynthetic gene cluster detection, DeepARG for fragmented gene classification, and protein language model–based tools like ESM-2, ProtT5, and DeepFRI, demonstrate that deep learning can reliably detect complex or divergent biosynthetic genes in other organisms [104,106,120]. Building on these approaches, we propose a dedicated AI-driven platform, available as a web or standalone tool, capable of predicting sxt genes from transcriptomic or genomic data with far higher accuracy than current methods. Leveraging deep-learning classifiers, protein language models, and embedding-based orthology inference, it would distinguish true biosynthetic genes from homologs or partial fragments, assign orthologous and paralogous relationships, and provide confidence scores [104]. This scalable system would enable rapid, standardized annotation across species, integrate with evolutionary analyses, and reduce reliance on labor-intensive validation, accelerating saxitoxin research and improving reproducibility.
Furthermore, the genomic investigation in dinoflagellates remains constrained, due to the scarcity of high-quality, well-annotated multi-omics datasets for dinoflagellates [22], with most discoveries limited to a few characterized genes [18,100] with others identified through scattered transcriptomic approaches [39,43,52]. Aside from a few draft genomes within Symbiodinium [179,180], comprehensive genomic resources for other dinoflagellates, especially regarding the saxitoxin biosynthesis, are largely lacking. Overall, the extremely large, repetitive genomes, scattered transcriptomes, and unexplored proteomes and metabolomes restrict the ability of AI models to learn accurate biological or biochemical patterns [181]. In addition, these challenges are compounded by the inherent difficulty of sequencing dinoflagellates, slowing the development of reference-quality genomic resources needed for pathway and gene cluster inference. However, as omics technologies advance and more data enter public repositories, there is an opportunity to decode saxitoxin biosynthesis. Therefore, instead of relying on isolated omics studies, a fully integrated multi-omics framework across multiple species is needed [22]. Using unified extraction and processing pipelines will generate comparable transcriptomics, proteomics, metabolomics, and environmental datasets from the same material. Applying advanced AI and machine-learning tools to these combined datasets will reveal the regulatory architecture of saxitoxin biosynthesis and enable predictive modeling of species-specific pathways and environmental drivers of toxicity [135,140].
Harmful algal blooms (HABs) are inherently complex ecological phenomena shaped by interacting with biotic and abiotic factors, generating highly multidimensional datasets [182,183]. Yet many coastal regions still lack long-term, high-frequency monitoring programs, and existing datasets often contain substantial gaps, such as missing information on nutrients, hydrodynamics, pollutants, or microbiome structure. Although new sensing technologies and data platforms are emerging, they frequently lack the historical depth required for reliable HAB forecasting [184]. These limitations create severe data imbalance and restrict model generalizability, causing AI systems to perform well in one region or species but fail when applied to different ecosystems, taxa, or climate-driven scenarios. Importantly, such limitations directly affect the reliability of food safety risk assessments, the prediction of paralytic shellfish poisoning (PSP) events, and the timely implementation of aquaculture harvesting restrictions and area closures.
However, despite the complexity of HABs and persistent gaps in biological and toxin-specific datasets, the rapid expansion of open-access physical and chemical ocean databases offers a path forward [16]. Global platforms such as NASA OceanColor (MODIS/VIIRS), NOAA ERDDAP, and the World Ocean Database now provide high-resolution, long-term data on temperature, chlorophyll, nutrients, and circulation that can be systematically integrated with biological, molecular, and toxin-profile datasets [185,186,187]. Future HAB research will increasingly rely on AI frameworks capable of fusing these heterogeneous datasets, handling data imbalance, and transferring knowledge across regions and species. Such integrative approaches will enhance model generalizability, facilitate region-independent forecasting, and support early warning systems for toxin-producing blooms under ongoing climate change. Ultimately, these advances will strengthen food safety surveillance, enhance HAB prediction under climate change, and provide actionable decision-support tools for marine resource management and aquaculture risk prevention.
Importantly, a lot of these issues are related to financial and resource constraints. Sustained financial investment has been repeatedly identified as a critical prerequisite for effective HAB monitoring, multi-omics integration, and AI-driven forecasting, particularly for maintaining long-term datasets and computational infrastructure [23,28,181]. High-performance computing, sequencing budgets, specialized monitoring programs, and steady funding streams are all unavailable to many research groups where HABs are growing quickly [16]. Furthermore, the process of creating genomic data is time-consuming and labor-intensive by nature, and it is likely that the goals and resources of the staff members involved in their collection do not include the use of ML and AI for HAB modeling [188,189]. This leads to disparities in the ability to use AI and create predictive STX frameworks. Addressing current financial and resource constraints will be critical for advancing AI-driven HAB and saxitoxin research. Future progress will likely depend on coordinated international funding, shared infrastructure, and open-access multi-omics and environmental databases that reduce duplication of effort and lower entry barriers for under-resourced regions. Advances in cost-efficient sequencing, cloud-based high-performance computing, and automated AI workflows are expected to make large-scale analyses more accessible.
AI models, particularly deep learning architectures, can face several challenges in multi-omics studies [189]. High-dimensional data with limited samples increases the risk of overfitting, reducing the model’s ability to generalize to new datasets [190]. Small sample sizes, common in dinoflagellate studies, further exacerbate overfitting and can produce spurious or unstable predictions [191]. Additionally, many AI models act as “black boxes,” limiting interpretability and making it difficult to extract biologically meaningful insights [192]. These issues can be mitigated through careful feature selection, regularization, cross-validation, data augmentation, transfer learning, and the use of explainable AI techniques [193]. However, efficient application of these technologies demands interdisciplinary partnerships. Therefore, establishing strong collaborations among biologists, data scientists, and AI/ML specialists is essential to address the unique challenges at the interface of AI and multi-omics in dinoflagellate toxins
Finally, barriers related to expertise, standardized workflows, computational access, and data-sharing policies further slow progress. Overall, overcoming these challenges will require sustained funding, international collaboration, and coordinated efforts to expand datasets, improve interpretability, and ensure that AI tools capture the biological and ecological realities of saxitoxin production to fully realize the potential of AI-driven research in unraveling decades of unknowns of the most potent marine toxins amid a changing climate.
8. Conclusions
As climate change increases, the intensity of HABs is expected to rise, increasing the risk of PSP caused by dinoflagellate saxitoxin, with profound impacts on socio-economic systems, ecosystem health, and human well-being [4,15]. In this review, we summarized the current knowledge on dinoflagellate STX biosynthesis and highlighted persistent knowledge gaps, such as fragmented genomic data, unknown gene origins, uncertain annotations, unresolved biosynthetic steps, and inconsistent environmental observations, that have slowed progress toward an understanding of STX evolution, biosynthesis, and ecological dynamics. In this perspective, we suggest the application of AI and machine learning to overcome these limitations, offering a critical advancement for saxitoxin research and HAB forecasting. By integrating advanced AI tools with experimental biology, researchers can establish a comprehensive, scalable, and predictive framework to understand and anticipate saxitoxin production in a rapidly changing ocean, marking a transition from decades of inconsistent findings to a new era of comprehensive insight, where AI bridges the gap between data, mechanism, and actionable knowledge.
Author Contributions
Conceptualization, methodology, B.L.M.; validation, B.L.M. and H.-S.K.; investigation, B.L.M.; resources, B.L.M., H.-S.K., I.A. and H.A.S.; writing—original draft preparation, B.L.M.; writing—review and editing, B.L.M., H.-S.K., I.A., H.A.S. and J.-S.K.; supervision, J.-S.K.; funding acquisition, J.-S.K. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (RS-2024-00354842).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| UN | United Nations |
| STX | Saxitoxin |
| STXs | Saxitoxin and its analogs (STX, GTX, C1, dSTX, etc.) |
| PSP | Paralytic Shellfish Poisoning |
| HABs | Harmful Algal Blooms |
| AI | Artificial Intelligence |
| ML | Machine Learning |
| sxt | Saxitoxin biosynthesis gene(s) |
| HGT | Horizontal Gene Transfer |
| CNN | Convolutional Neural Network |
| PKS | Polyketide Synthase |
| FAS | Fatty Acid Synthase |
| ESM-2 | Evolutionary Scale Modeling version 2 |
| ProtT5 | Protein T5 |
| ProtBERT | Protein BERT |
| DeepFRI | Deep Functional Residue Identification |
| GNN | Graph Neural Network |
| VAEs | Variational Autoencoders |
| iGRN | Integrative Gene Regulatory Network |
| LC–MS/MS | Liquid Chromatography–Tandem Mass Spectrometry |
| Y2H | Yeast Two-Hybrid |
| co-IP | Co-Immunoprecipitation |
| AP-MS | Affinity Purification Mass Spectrometry |
| DeepMET | Deep Learning Metabolomics Framework |
| DeepMass | Deep Learning Mass Spectrometry Framework |
| MOFA+ | Multi-Omics Factor Analysis Plus |
| DRS | Direct RNA Sequencing |
| LSTM | Long Short-Term Memory |
| GRU | Gated Recurrent Unit |
| NASA | National Aeronautics and Space Administration |
| MODIS | Moderate Resolution Imaging Spectroradiometer |
| VIIRS | Visible Infrared Imaging Radiometer Suite |
| NOAA | National Oceanic and Atmospheric Administration |
| ERDDAP | Environmental Research Division’s Data Access Program |
| WOD | World Ocean Database |
| PST | Paralytic Shellfish Toxins |
| N:P | Nitrogen-Phosphorous ratio |
| ESM | Evolutionary Scale Modeling |
| ProtTrans | Protein Transformers |
| RNN | RNN |
| XGBoost | XGBoost |
| HDBSCAN | Hierarchical Density-Based Spatial Clustering of Applications with Noise |
| SHAP | SHapley Additive exPlanations |
| ESMFold | Evolutionary Scale Modeling Fold |
| SVM | Support Vector Machine |
| RF | Random Forest |
| MS2CNN | Mass Spectrometry to Convolutional Neural Network |
| WGCNA | Weighted Gene Co-expression Network Analysis |
| GG Models | Gaussian Graphical Models |
| ConvLSTM | Convolutional Long Short-Term Memory |
| EL | Ensemble learning |
| RL | Reinforcement learning |
| TT | Temporal Transformers |
References
- The United Nations Decade of Ocean Science for Sustainable Development (2021–2030): Implementation Plan. UNESCO Digital Library. 2021. Available online: https://unesdoc.unesco.org/ark:/48223/pf0000377082 (accessed on 21 November 2025).
- Venegas, R.M.; Acevedo, J.; Treml, E.A. Three Decades of Ocean Warming Impacts on Marine Ecosystems: A Review and Perspective. Deep Sea Res. Part II Top. Stud. Oceanogr. 2023, 212, 105318. [Google Scholar] [CrossRef]
- Visciano, P.; Schirone, M.; Berti, M.; Milandri, A.; Tofalo, R.; Suzzi, G. Marine Biotoxins: Occurrence, Toxicity, Regulatory Limits and Reference Methods. Front. Microbiol. 2016, 7, 207058. [Google Scholar] [CrossRef] [PubMed]
- Berdalet, E.; Fleming, L.E.; Gowen, R.; Davidson, K.; Hess, P.; Backer, L.C.; Moore, S.K.; Hoagland, P.; Enevoldsen, H. Marine Harmful Algal Blooms, Human Health and Wellbeing: Challenges and Opportunities in the 21st Century. J. Mar. Biol. Assoc. United Kingd. 2016, 96, 61–91. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Shang, X.; Zhu, H.; Huang, N.; Wang, L.; Sun, M. Saxitoxin: A Comprehensive Review of Its History, Structure, Toxicology, Biosynthesis, Detection, and Preventive Implications. Mar. Drugs 2025, 23, 277. [Google Scholar] [CrossRef]
- Cusick, K.D.; Sayler, G.S. An Overview on the Marine Neurotoxin, Saxitoxin: Genetics, Molecular Targets, Methods of Detection and Ecological Functions. Mar. Drugs 2013, 11, 991–1018. [Google Scholar] [CrossRef]
- Wiese, M.; D’Agostino, P.M.; Mihali, T.K.; Moffitt, M.C.; Neilan, B.A. Neurotoxic Alkaloids: Saxitoxin and Its Analogs. Mar. Drugs 2010, 8, 2185–2211. [Google Scholar] [CrossRef]
- Wang, D.Z.; Zhang, S.F.; Zhang, Y.; Lin, L. Paralytic Shellfish Toxin Biosynthesis in Cyanobacteria and Dinoflagellates: A Molecular Overview. J. Proteom. 2016, 135, 132–140. [Google Scholar] [CrossRef]
- De Carvalho, M.; Jacinto, J.; Ramos, N.; De Oliveira, V.; Pinho E Melo, T.; De Sá, J. Paralytic Shellfish Poisoning: Clinical and Electrophysiological Observations. J. Neurol. 1998, 245, 551–554. [Google Scholar] [CrossRef]
- Guillotin, S.; Delcourt, N. Marine Neurotoxins’ Effects on Environmental and Human Health: An OMICS Overview. Mar. Drugs 2021, 20, 18. [Google Scholar] [CrossRef]
- Gribble, M.O.; Bennett, B.J.; Liddie, J.M.; Borchert, W.; Pfluger, B.A.; Segars, J.S.; Keast, J.M.; Hans, A.; Kikkeri, N.S.; Shin, C.; et al. Global Epidemiology of Paralytic Shellfish Poisoning: A Systematic Search Literature Review. Lancet Planet. Health 2025, 9, 101271. [Google Scholar] [CrossRef]
- Kibler, S.R.; Litaker, R.W.; Matweyou, J.A.; Hardison, D.R.; Wright, B.A.; Tester, P.A. Paralytic Shellfish Poisoning Toxins in Butter Clams (Saxidomus gigantea) from the Kodiak Archipelago, Alaska. Harmful Algae 2022, 111, 102165. [Google Scholar] [CrossRef]
- Temple, C.; Hughes, A. A Case of Fatal Paralytic Shellfish Poisoning in Alaska. Clin. Toxicol. 2022, 60, 414–415. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, R.; Oikawa, H.; Tsunemitsu, T.; Miyahara, K.; Ozawa, M.; Numano, S.; Uchida, H.; Matsushima, R.; Suzuki, T. A Case of Paralytic Shellfish Poisoning Caused by Consumption of Visceral Balls from Geoduck Panopea japonica in Japan. Toxicon 2024, 243, 107738. [Google Scholar] [CrossRef] [PubMed]
- Moore, S.K.; Trainer, V.L.; Mantua, N.J.; Parker, M.S.; Laws, E.A.; Backer, L.C.; Fleming, L.E. Impacts of Climate Variability and Future Climate Change on Harmful Algal Blooms and Human Health. Environ. Health 2008, 7, S4. [Google Scholar] [CrossRef] [PubMed]
- Wells, M.L.; Trainer, V.L.; Smayda, T.J.; Karlson, B.S.O.; Trick, C.G.; Kudela, R.M.; Ishikawa, A.; Bernard, S.; Wulff, A.; Anderson, D.M.; et al. Harmful Algal Blooms and Climate Change: Learning from the Past and Present to Forecast the Future. Harmful Algae 2015, 49, 68–93. [Google Scholar] [CrossRef]
- Bui, Q.T.N.; Pradhan, B.; Kim, H.S.; Ki, J.S. Environmental Factors Modulate Saxitoxins (STXs) Production in Toxic Dinoflagellate Alexandrium: An Updated Review of STXs and Synthesis Gene Aspects. Toxins 2024, 16, 210. [Google Scholar] [CrossRef]
- Abassi, S.; Kim, H.S.; Bui, Q.T.N.; Ki, J.S. Effects of Nitrate on the Saxitoxins Biosynthesis Revealed by Sxt Genes in the Toxic Dinoflagellate Alexandrium pacificum (Group IV). Harmful Algae 2023, 127, 102473. [Google Scholar] [CrossRef]
- Bui, Q.T.N.; Kim, H.; Park, H.; Ki, J.S. Salinity Affects Saxitoxins (STXs) Toxicity in the Dinoflagellate Alexandrium pacificum, with Low Transcription of Sxt-Biosynthesis Genes SxtA4 and SxtG. Toxins 2021, 13, 733. [Google Scholar] [CrossRef]
- Cirés, S.; Delgado, A.; González-Pleiter, M.; Quesada, A. Temperature Influences the Production and Transport of Saxitoxin and the Expression of Sxt Genes in the Cyanobacterium Aphanizomenon gracile. Toxins 2017, 9, 322. [Google Scholar] [CrossRef]
- Kim, H.; Park, H.; Wang, H.; Yoo, H.Y.; Park, J.; Ki, J.S. Low Temperature and Cold Stress Significantly Increase Saxitoxins (STXs) and Expression of STX Biosynthesis Genes SxtA4 and SxtG in the Dinoflagellate Alexandrium catenella. Mar. Drugs 2021, 19, 291. [Google Scholar] [CrossRef]
- Akbar, M.A.; Yusof, N.Y.M.; Tahir, N.I.; Ahmad, A.; Usup, G.; Sahrani, F.K.; Bunawan, H. Biosynthesis of Saxitoxin in Marine Dinoflagellates: An Omics Perspective. Mar. Drugs 2020, 18, 103. [Google Scholar] [CrossRef] [PubMed]
- Dara, M.; Dianatpour, M.; Azarpira, N.; Tanideh, N. The Transformative Role of Artificial Intelligence in Genomics: Opportunities and Challenges. Gene Rep. 2025, 41, 102314. [Google Scholar] [CrossRef]
- Zhong, S.; Zhang, K.; Bagheri, M.; Burken, J.G.; Gu, A.; Li, B.; Ma, X.; Marrone, B.L.; Ren, Z.J.; Schrier, J.; et al. Machine Learning: New Ideas and Tools in Environmental Science and Engineering. Environ. Sci. Technol. 2021, 55, 12741–12754. [Google Scholar] [CrossRef] [PubMed]
- Libbrecht, M.W.; Noble, W.S. Machine Learning Applications in Genetics and Genomics. Nat. Rev. Genet. 2015, 16, 321–332. [Google Scholar] [CrossRef]
- Cheng, Y.; Xu, S.M.; Santucci, K.; Lindner, G.; Janitz, M. Machine Learning and Related Approaches in Transcriptomics. Biochem. Biophys. Res. Commun. 2024, 724, 150225. [Google Scholar] [CrossRef]
- Cai, Z.; Poulos, R.C.; Liu, J.; Zhong, Q. Machine Learning for Multi-Omics Data Integration in Cancer. iScience 2022, 25, 103798. [Google Scholar] [CrossRef]
- Gupta, S.; Janu, N.; Nawal, M.; Goswami, A. Genomics and Machine Learning: ML Approaches, Future Directions and Challenges in Genomics. In Genomics at the Nexus of AI, Computer Vision, and Machine Learning; John Wiley & Sons: Hoboken, NJ, USA, 2024; pp. 437–457. [Google Scholar] [CrossRef]
- Cheng, Y.; Bi, X.; Xu, Y.; Liu, Y.; Li, J.; Du, G.; Lv, X.; Liu, L. Machine Learning for Metabolic Pathway Optimization: A Review. Comput. Struct. Biotechnol. J. 2023, 21, 2381. [Google Scholar] [CrossRef]
- Yetgin, A. Revolutionizing Multi-Omics Analysis with Artificial Intelligence and Data Processing. Quant. Biol. 2025, 13, e70002. [Google Scholar] [CrossRef]
- Ramos, T.K.; Costa, L.D.F.; Yunes, J.S.; Resgalla, C.; Barufi, J.B.; de Oliveira Bastos, E.; Horta, P.A.; Rörig, L.R. Saxitoxins from the Freshwater Cyanobacterium Raphidiopsis raciborskii Can Contaminate Marine Mussels. Harmful Algae 2021, 103, 102004. [Google Scholar] [CrossRef]
- Kellmann, R.; Mihali, T.K.; Young, J.J.; Pickford, R.; Pomati, F.; Neilan, B.A. Biosynthetic Intermediate Analysis and Functional Homology Reveal a Saxitoxin Gene Cluster in Cyanobacteria. Appl. Environ. Microbiol. 2008, 74, 4044–4053. [Google Scholar] [CrossRef]
- Stern, D.B.; Raborn, R.T.; Lovett, S.P.; Boise, N.R.; Carrasquilla, L.; Enke, S.; Radune, D.; Woodruff, D.L.; Wahl, K.L.; Rosovitz, M.J. Novel Toxin Biosynthetic Gene Cluster in Harmful Algal Bloom-Causing Heteroscytonema crispum: Insights into the Origins of Paralytic Shellfish Toxins. Genome Biol. Evol. 2025, 17, evae248. [Google Scholar] [CrossRef] [PubMed]
- Hackett, J.D.; Wisecaver, J.H.; Brosnahan, M.L.; Kulis, D.M.; Anderson, D.M.; Bhattacharya, D.; Gerald Plumley, F.; Erdner, D.L. Evolution of Saxitoxin Synthesis in Cyanobacteria and Dinoflagellates. Mol. Biol. Evol. 2013, 30, 70–78. [Google Scholar] [CrossRef]
- Stüken, A.; Orr, R.J.S.; Kellmann, R.; Murray, S.A.; Neilan, B.A.; Jakobsen, K.S. Discovery of Nuclear-Encoded Genes for the Neurotoxin Saxitoxin in Dinoflagellates. PLoS ONE 2011, 6, e20096. [Google Scholar] [CrossRef] [PubMed]
- Orr, R.J.S.; Stüken, A.; Murray, S.A.; Jakobsen, K.S. Evolution and Distribution of Saxitoxin Biosynthesis in Dinoflagellates. Mar. Drugs 2013, 11, 2814–2828. [Google Scholar] [CrossRef] [PubMed]
- Mary, L.; Quere, J.; Latimier, M.; Rovillon, G.A.; Hégaret, H.; Réveillon, D.; Gac, M. Le Genetic Association of Toxin Production in the Dinoflagellate Alexandrium minutum. Microb. Genom. 2022, 8, 000879. [Google Scholar] [CrossRef]
- Shin, H.H.; Li, Z.; Réveillon, D.; Rovillon, G.A.; Mertens, K.N.; Hess, P.; Kim, H.J.; Lee, J.; Lee, K.W.; Kim, D.; et al. Centrodinium punctatum (Dinophyceae) Produces Significant Levels of Saxitoxin and Related Analogs. Harmful Algae 2020, 100, 101923. [Google Scholar] [CrossRef]
- Muhammad, B.L.; Kim, H.S.; Bui, Q.T.N.; Ki, J.S. Transcriptomic Comparison Unveils Saxitoxin Biosynthesis Genes in the Marine Dinoflagellate Gymnodinium catenatum. Harmful Algae 2025, 147, 102872. [Google Scholar] [CrossRef]
- Yang, I.; John, U.; Beszteri, S.; Glöckner, G.; Krock, B.; Goesmann, A.; Cembella, A.D. Comparative Gene Expression in Toxic versus Non-Toxic Strains of the Marine Dinoflagellate Alexandrium minutum. BMC Genom. 2010, 11, 248. [Google Scholar] [CrossRef]
- Landsberg, J.H.; Hall, S.; Johannessen, J.N.; White, K.D.; Conrad, S.M.; Abbott, J.P.; Flewelling, L.J.; Richardson, R.W.; Dickey, R.W.; Jester, E.L.E.; et al. Saxitoxin Puffer Fish Poisoning in the United States, with the First Report of Pyrodinium bahamense as the Putative Toxin Source. Environ. Health Perspect. 2006, 114, 1502–1507. [Google Scholar] [CrossRef]
- Bui, Q.T.N.; Kim, H.S.; Ki, J.S. Polyphyletic Origin of Saxitoxin Biosynthesis Genes in the Marine Dinoflagellate Alexandrium Revealed by Comparative Transcriptomics. Harmful Algae 2024, 134, 102620. [Google Scholar] [CrossRef]
- Wang, H.; Kim, H.; Ki, J.S. Transcriptome Survey and Toxin Measurements Reveal Evolutionary Modification and Loss of Saxitoxin Biosynthesis Genes in the Dinoflagellates Amphidinium carterae and Prorocentrum micans. Ecotoxicol. Environ. Saf. 2020, 195, 110474. [Google Scholar] [CrossRef]
- Wang, H.; Guo, R.; Lim, W.A.; Allen, A.E.; Ki, J.S. Comparative Transcriptomics of Toxin Synthesis Genes between the Non-Toxin Producing Dinoflagellate Cochlodinium polykrikoides and Toxigenic Alexandrium pacificum. Harmful Algae 2020, 93, 101777. [Google Scholar] [CrossRef] [PubMed]
- Cooper, J.T.; Sinclair, G.A.; Wawrik, B. Transcriptome Analysis of Scrippsiella trochoidea CCMP 3099 Reveals Physiological Changes Related to Nitrate Depletion. Front. Microbiol. 2016, 7, 191902. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, B.L.; Thi, Q.; Bui, N.; Kim, H.-S.; Ki, J.-S. Transcriptomic Insights into Polyketides and Toxin Biosynthesis Genes in Freshwater Dinoflagellates. Sci. Rep. 2026. under review (submitted). [Google Scholar]
- Li, Z.; Mertens, K.N.; Nézan, E.; Chomérat, N.; Bilien, G.; Iwataki, M.; Shin, H.H. Discovery of a New Clade Nested Within the Genus Alexandrium (Dinophyceae): Morpho-Molecular Characterization of Centrodinium punctatum (Cleve) F.J.R. Taylor. Protist 2019, 170, 168–186. [Google Scholar] [CrossRef] [PubMed]
- Leong, W.; Anton, A.; Rodrigues, F.; Lie, W. Gene expression in the biosynthesis of paralytic shellfish poisoning (PSP) toxins in dinoflagellates: A mini review. Trans. Sci. Technol. 2016, 3, 374–381. [Google Scholar]
- Geffroy, S.; Lechat, M.M.; Le Gac, M.; Rovillon, G.A.; Marie, D.; Bigeard, E.; Malo, F.; Amzil, Z.; Guillou, L.; Caruana, A.M.N. From the SxtA4 Gene to Saxitoxin Production: What Controls the Variability Among Alexandrium minutum and Alexandrium pacificum Strains? Front. Microbiol. 2021, 12, 613199. [Google Scholar] [CrossRef]
- Orr, R.J.S.; Stüken, A.; Murray, S.A.; Jakobsen, K.S. Evolutionary Acquisition and Loss of Saxitoxin Biosynthesis in Dinoflagellates: The Second “Core” Gene, SxtG. Appl. Environ. Microbiol. 2013, 79, 2128–2136. [Google Scholar] [CrossRef]
- Muhammad, B.L.; Kim, H.-S.; Ki, J.-S. Saxitoxin Biosynthesis Genes in Gymnodinium Species: Transcriptomic Insights and Their Environmental Implications. Harmful Algae 2026. under review (submitted). [Google Scholar]
- Verma, A.; Barua, A.; Ruvindy, R.; Savela, H.; Ajani, P.A.; Murray, S.A. The Genetic Basis of Toxin Biosynthesis in Dinoflagellates. Microorganisms 2019, 7, 222. [Google Scholar] [CrossRef]
- Cho, Y.; Hiramatsu, K.; Ogawa, M.; Omura, T.; Ishimaru, T.; Oshima, Y. Non-toxic and toxic subclones obtained from a toxic clonal culture of Alexandrium tamarense (Dinophyceae): Toxicity and molecular biological feature. Harmful Algae 2008, 7, 740–751. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, S.F.; Lin, L.; Wang, D.Z. Comparative Transcriptome Analysis of a Toxin-Producing Dinoflagellate Alexandrium catenella and Its Non-Toxic Mutant. Mar. Drugs 2014, 12, 5698–5718. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.; Hidema, S.; Omura, T.; Koike, K.; Koike, K.; Oikawa, H.; Konoki, K.; Oshima, Y.; Yotsu-Yamashita, M. SxtA Localizes to Chloroplasts and Changes to Its 3′UTR May Reduce Toxin Biosynthesis in Non-Toxic Alexandrium catenella (Group I). Harmful Algae 2021, 101, 101972. [Google Scholar] [CrossRef] [PubMed]
- Touzet, N.; Franco, J.M.; Raine, R. Characterization of nontoxic and toxin-producing strains of Alexandrium minutum (Dinophyceae) in Irish coastal waters. Appl. Environ. Microbiol. 2007, 73, 3333–3342. [Google Scholar] [CrossRef]
- Perini, F.; Galluzzi, L.; Dell’Aversano, C.; Iacovo, E.D.; Tartaglione, L.; Ricci, F.; Forino, M.; Ciminiello, P.; Penna, A. SxtA and SxtG Gene Expression and Toxin Production in the Mediterranean Alexandrium minutum (Dinophyceae). Mar. Drugs 2014, 12, 5258–5276. [Google Scholar] [CrossRef]
- Roy, S.; Morse, D. A Full Suite of Histone and Histone Modifying Genes Are Transcribed in the Dinoflagellate Lingulodinium. PLoS ONE 2012, 7, e34340. [Google Scholar] [CrossRef]
- Band-Schmidt, C.J.; Bustillos-Guzmán, J.J.; Hernández-Sandoval, F.E.; Núñez-Vázquez, E.J.; López-Cortés, D.J. Effect of Temperature on Growth and Paralytic Toxin Profiles in Isolates of Gymnodinium catenatum (Dinophyceae) from the Pacific Coast of Mexico. Toxicon 2014, 90, 199–212. [Google Scholar] [CrossRef]
- Aboualaalaa, H.; Rijal Leblad, B.; Elkbiach, M.L.B.; Ibghi, M.; Boutaib, R.; Maamour, N.; Savar, V.; Masseret, E.; Abadie, E.; Rolland, J.L.; et al. Effect of Temperature, Salinity and Nutrients on the Growth and Toxin Content of the Dinoflagellate Gymnodinium catenatum from the Southwestern Mediterranean. Sci. Total Environ. 2024, 945, 174094. [Google Scholar] [CrossRef]
- Salgado, P.; Vázquez, J.A.; Riobó, P.; Franco, J.M.; Figueroa, R.I.; Kremp, A.; Bravo, I. A Kinetic and Factorial Approach to Study the Effects of Temperature and Salinity on Growth and Toxin Production by the Dinoflagellate Alexandrium ostenfeldii from the Baltic Sea. PLoS ONE 2015, 10, e0143021. [Google Scholar] [CrossRef]
- Sung, J.; Choi, D.H.; Lee, Y.; Kim, J.H.; Shin, H.H.; Kim, Y.E.; Choi, J.H.; Noh, J.H.; Gobler, C.J.; Park, B.S. Temperature-Driven Intraspecific Diversity in Paralytic Shellfish Toxin Profiles of the Dinoflagellate Alexandrium pacificum and Intragenic Variation in the Saxitoxin Biosynthetic Gene, SxtA4. Microb. Ecol. 2025, 88, 87. [Google Scholar] [CrossRef]
- Han, M.; Lee, H.; Anderson, D.M.; Kim, B. Paralytic Shellfish Toxin Production by the Dinoflagellate Alexandrium pacificum (Chinhae Bay, Korea) in Axenic, Nutrient-Limited Chemostat Cultures and Nutrient-Enriched Batch Cultures. Mar. Pollut. Bull. 2016, 104, 34. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.; Tsuchiya, S.; Omura, T.; Koike, K.; Konoki, K.; Oshima, Y.; Yotsu-Yamashita, M. Metabolic Inhibitor Induces Dynamic Changes in Saxitoxin Biosynthesis and Metabolism in the Dinoflagellate Alexandrium pacificum (Group IV) under In Vivo Labeling Condition. Harmful Algae 2023, 122, 102372. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.; Tsuchiya, S.; Omura, T.; Koike, K.; Oikawa, H.; Konoki, K.; Oshima, Y.; Yotsu-Yamashita, M. Metabolomic Study of Saxitoxin Analogues and Biosynthetic Intermediates in Dinoflagellates Using 15N-Labelled Sodium Nitrate as a Nitrogen Source. Sci. Rep. 2019, 9, 3460. [Google Scholar] [CrossRef]
- Jiang, Y.; Ramanan, R.; Yoon, S.; Lee, B.M.; Kang, Y.H.; Li, Z. Toxin Production in Bloom-Forming, Harmful Alga Alexandrium pacificum (Group IV) Is Regulated by Cyst Formation-Promoting Bacteria Jannaschia cystaugens NBRC 100362T. Water Res. 2025, 272, 122930. [Google Scholar] [CrossRef] [PubMed]
- Murray, S.A.; Diwan, R.; Orr, R.J.S.; Kohli, G.S.; John, U. Gene Duplication, Loss and Selection in the Evolution of Saxitoxin Biosynthesis in Alveolates. Mol. Phylogenet. Evol. 2015, 92, 165–180. [Google Scholar] [CrossRef]
- Wang, H.; Wu, P.; Xiong, L.; Kim, H.S.; Kim, J.H.; Ki, J.S. Nuclear Genome of Dinoflagellates: Size Variation and Insights into Evolutionary Mechanisms. Eur. J. Protistol. 2024, 93, 126061. [Google Scholar] [CrossRef]
- Lin, S. A Decade of Dinoflagellate Genomics Illuminating an Enigmatic Eukaryote Cell. BMC Genom. 2024, 25, 932. [Google Scholar] [CrossRef]
- Liu, L.; Hastings, J.W. Novel and Rapidly Diverging Intergenic Sequences between Tandem Repeats of the Luciferase Genes in Seven Dinoflagellate Species. J. Phycol. 2006, 42, 96–103. [Google Scholar] [CrossRef]
- Zhang, K.; Xi, M.; Wu, G.; Lu, F.; Wu, G.; Zhou, J.; Zhang, J.; Wang, X.; Li, Y.; Xu, C.; et al. Environmental Drivers and Microbial Interactions in Harmful Dinoflagellate Blooms: Insights from Metagenomics and Machine Learning. Process Saf. Environ. Prot. 2025, 199, 107205. [Google Scholar] [CrossRef]
- Murray, S.; John, U.; Savela, H.; Kremp, A. Alexandrium spp.: Genetic and ecological factors influencing saxitoxin production and proliferation. In Climate Change and Marine and Freshwater Toxins; Walter de Gruyter GmbH & Co KG.: Berlin, Germany, 2015; pp. 123–139. [Google Scholar]
- Savela, H.; Harju, K.; Spoof, L.; Lindehoff, E.; Meriluoto, J.; Vehniäinen, M.; Kremp, A. Quantity of the Dinoflagellate SxtA4 Gene and Cell Density Correlates with Paralytic Shellfish Toxin Production in Alexandrium ostenfeldii Blooms. Harmful Algae 2016, 52, 1–10. [Google Scholar] [CrossRef]
- Wiese, M.; Murray, S.A.; Alvin, A.; Neilan, B.A. Gene Expression and Molecular Evolution of SxtA4 in a Saxitoxin Producing Dinoflagellate Alexandrium catenella. Toxicon 2014, 92, 102–112. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, S.F.; Lin, L.; Wang, D.Z. Whole Transcriptomic Analysis Provides Insights into Molecular Mechanisms for Toxin Biosynthesis in a Toxic Dinoflagellate Alexandrium catenella (ACHK-T). Toxins 2017, 9, 213. [Google Scholar] [CrossRef] [PubMed]
- Akbar, M.A.; Mohd Yusof, N.Y.; Usup, G.; Ahmad, A.; Baharum, S.N.; Bunawan, H. Nutrient Deficiencies Impact on the Cellular and Metabolic Responses of Saxitoxin Producing Alexandrium minutum: A Transcriptomic Perspective. Mar. Drugs 2023, 21, 497. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Kim, H.; Park, H.; Ki, J.S. Temperature Influences the Content and Biosynthesis Gene Expression of Saxitoxins (STXs) in the Toxigenic Dinoflagellate Alexandrium pacificum. Sci. Total Environ. 2022, 802, 149801. [Google Scholar] [CrossRef]
- Hii, K.S.; Lim, P.T.; Kon, N.F.; Takata, Y.; Usup, G.; Leaw, C.P. Physiological and Transcriptional Responses to Inorganic Nutrition in a Tropical Pacific Strain of Alexandrium minutum: Implications for the Saxitoxin Genes and Toxin Production. Harmful Algae 2016, 56, 9–21. [Google Scholar] [CrossRef]
- Lin, Z.R.; Geng, H.X.; Zhang, Q.C.; Chen, Z.F.; Dai, L.; Yu, R.C. Toxin Production of Dinoflagellate Gymnodinium catenatum Isolated from the East China Sea. Harmful Algae 2022, 113, 102188. [Google Scholar] [CrossRef]
- Hernández-Sandoval, F.E.; Bustillos-Guzmán, J.J.; Band-Schmidt, C.J.; Núñez-Vázquez, E.J.; López-Cortés, D.J.; Fernández-Herrera, L.J.; Poot-Delgado, C.A.; Moreno-Legorreta, M. Effect of Different N:P Ratios on the Growth, Toxicity, and Toxin Profile of Gymnodinium catenatum (Dinophyceae) Strains from the Gulf of California. Toxins 2022, 14, 501. [Google Scholar] [CrossRef]
- Sixto, M.; Riobó, P.; Rodríguez, F.; Díaz, P.A.; Figueroa, R.I. Climate Change Stressors, Phosphate Limitation, and High Irradiation Interact to Increase Alexandrium minutum Toxicity and Modulate Encystment Rates. Microorganisms 2024, 12, 1480. [Google Scholar] [CrossRef]
- Cusick, K.D.; Wei, B.; Hall, S.; Brown, N.; Widder, E.A.; Boyer, G.L. Toxin Dynamics among Populations of the Bioluminescent HAB Species Pyrodinium bahamense from the Indian River Lagoon, FL. Mar. Drugs 2024, 22, 311. [Google Scholar] [CrossRef]
- Judd, M.; Place, A.R. A Strategy for Gene Knockdown in Dinoflagellates. Microorganisms 2022, 10, 1131. [Google Scholar] [CrossRef]
- Murray, S.A.; Wiese, M.; Wiese, A.; Brett, S.; Kellmann, R.; Hallegraeff, G.; Neilan, B.A. SxtA-Based Quantitative Molecular Assay to Identify Saxitoxin-Producing Harmful Algal Blooms in Marine Waters. Appl. Environ. Microbiol. 2011, 77, 7050–7057. [Google Scholar] [CrossRef] [PubMed]
- Lin, S. Genomic Understanding of Dinoflagellates. Res. Microbiol. 2011, 162, 551–569. [Google Scholar] [CrossRef]
- Lin, S.; Zhang, H.; Zhuang, Y.; Tran, B.; Gill, J. Spliced Leader-Based Metatranscriptomic Analyses Lead to Recognition of Hidden Genomic Features in Dinoflagellates. Proc. Natl. Acad. Sci. USA 2010, 107, 20033–20038. [Google Scholar] [CrossRef] [PubMed]
- Anderson, D.M.; Kulis, D.M.; Sullivan, J.J.; Hall, S. Toxin Composition Variations in One Isolate of the Dinoflagellate Alexandrium fundyense. Toxicon 1990, 28, 885–893. [Google Scholar] [CrossRef] [PubMed]
- John, E.H.; Flynn, K.J. Growth Dynamics and Toxicity of Alexandrium fundyense (Dinophyceae): The Effect of Changing N[Ratio ]P Supply Ratios on Internal Toxin and Nutrient Levels. Eur. J. Phycol. 2000, 35, 11–23. [Google Scholar] [CrossRef]
- Laureano-Rosario, A.E.; McFarland, M.; Bradshaw, D.J.; Metz, J.; Brewton, R.A.; Pitts, T.; Perricone, C.; Schreiber, S.; Stockley, N.; Wang, G.; et al. Dynamics of Microcystins and Saxitoxin in the Indian River Lagoon, Florida. Harmful Algae 2021, 103, 102012. [Google Scholar] [CrossRef]
- Cusick, K.; Duran, G. SxtA4+ and SxtA4- Genotypes Occur Together within Natural Pyrodinium bahamense Sub-Populations from the Western Atlantic. Microorganisms 2021, 9, 1128. [Google Scholar] [CrossRef]
- Lopes, V.M.; Court, M.; Seco, M.C.; Borges, F.O.; Vicente, B.; Lage, S.; Braga, A.C.; Duarte, B.; Santos, C.F.; Amorim, A.; et al. Gymnodinium catenatum Paralytic Shellfish Toxin Production and Photobiological Responses under Marine Heat Waves. Toxins 2023, 15, 157. [Google Scholar] [CrossRef]
- Alonso-Rodríguez, R.; Pichardo-Velarde, J.G. Effects of Temperature and Nutrients on Growth and Toxicity of Alexandrium Affine from Southeastern Gulf of California. Mar. Pollut. Bull. 2024, 203, 116464. [Google Scholar] [CrossRef]
- Yarimizu, K.; Mardones, J.I.; Paredes-Mella, J.; Norambuena-Subiabre, L.; Carrano, C.J.; Maruyama, F. The Effect of Iron on Chilean Alexandrium catenella Growth and Paralytic Shellfish Toxin Production as Related to Algal Blooms. Biometals 2022, 35, 39–51. [Google Scholar] [CrossRef]
- Cevora, G. The Relationship between Biological and Artificial Intelligence. arXiv 2019, arXiv:1905.00547. [Google Scholar] [CrossRef]
- Kraus, J. louis Artificial Intelligence Applied to the Production of High-Added-Value Dinoflagellates Toxins. AI Soc. 2020, 35, 851–855. [Google Scholar] [CrossRef]
- Marrone, B.L.; Banerjee, S.; Talapatra, A.; Gonzalez-Esquer, C.R.; Pilania, G. Toward a Predictive Understanding of Cyanobacterial Harmful Algal Blooms through AI Integration of Physical, Chemical, and Biological Data. ACS ES&T Water 2023, 4, 844–858. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Niu, J.; Gan, W.; Gou, S.; Zhang, S.; Qiu, H.; Jiang, T. Identification of Paralytic Shellfish Toxin-Producing Microalgae Using Machine Learning and Deep Learning Methods. J. Oceanol. Limnol. 2022, 40, 2202–2217. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, G. Machine Learning Based Toxicity Prediction: From Chemical Structural Description to Transcriptome Analysis. Int. J. Mol. Sci. 2018, 19, 2358. [Google Scholar] [CrossRef]
- Sharifi-Noghabi, H.; Zolotareva, O.; Collins, C.C.; Ester, M. MOLI: Multi-Omics Late Integration with Deep Neural Networks for Drug Response Prediction. Bioinformatics 2019, 35, i501–i509. [Google Scholar] [CrossRef]
- Bui, Q.T.N.; Ki, J.S. Molecular Characterization and Expression Analysis of Saxitoxin Biosynthesis Gene SxtU from Toxigenic Dinoflagellate Alexandrium pacificum. J. Appl. Phycol. 2023, 35, 687–700. [Google Scholar] [CrossRef]
- Mendoza-Flores, A.; Leyva-Valencia, I.; Band-Schmidt, C.J.; Galindo-Sánchez, C.E.; Bustillos-Guzmán, J.J. Identification of the Gene SxtA (Domains SxtA1 and SxtA4) in Mexican Strains of Gymnodinium catenatum (Dinophyceae) and Their Evolution. Front. Mar. Sci. 2018, 5, 390302. [Google Scholar] [CrossRef]
- Gligorijević, V.; Renfrew, P.D.; Kosciolek, T.; Leman, J.K.; Berenberg, D.; Vatanen, T.; Chandler, C.; Taylor, B.C.; Fisk, I.M.; Vlamakis, H.; et al. Structure-Based Protein Function Prediction Using Graph Convolutional Networks. Nat. Commun. 2021, 12, 3168. [Google Scholar] [CrossRef]
- Jha, K.; Saha, S.; Singh, H. Prediction of Protein–Protein Interaction Using Graph Neural Networks. Sci. Rep. 2022, 12, 8360. [Google Scholar] [CrossRef]
- Brandes, N.; Ofer, D.; Peleg, Y.; Rappoport, N.; Linial, M. ProteinBERT: A Universal Deep-Learning Model of Protein Sequence and Function. Bioinformatics 2022, 38, 2102–2110. [Google Scholar] [CrossRef] [PubMed]
- Kulmanov, M.; Khan, M.A.; Hoehndorf, R. DeepGO: Predicting Protein Functions from Sequence and Interactions Using a Deep Ontology-Aware Classifier. Bioinformatics 2018, 34, 660–668. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.T.; Jiang, Z.; Nguyen, V.N.; Le, N.Q.K.; Chua, M.C.H. Integrating ESM-2 and Graph Neural Networks with AlphaFold-2 Structures for Enhanced Protein Function Prediction. ACS Omega 2025, 10, 38103–38111. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Zeng, X.; Zhao, Y.; Chen, R. AlphaFold2 and Its Applications in the Fields of Biology and Medicine. Signal Transduct. Target. Ther. 2023, 8, 115. [Google Scholar] [CrossRef]
- Lai, B.; Xu, J. Accurate protein function prediction via graph attention networks with predicted structure information. Brief. Bioinform. 2022, 23, bbab502. [Google Scholar] [CrossRef]
- Hannigan, G.D.; Prihoda, D.; Palicka, A.; Soukup, J.; Klempir, O.; Rampula, L.; Durcak, J.; Wurst, M.; Kotowski, J.; Chang, D.; et al. A Deep Learning Genome-Mining Strategy for Biosynthetic Gene Cluster Prediction. Nucleic Acids Res. 2019, 47, E110. [Google Scholar] [CrossRef]
- Silberg, J.; Simon, E.; Zou, J. Towards Functional Annotation with Latent Protein Language Model Features. bioRxiv 2025. [Google Scholar] [CrossRef]
- Ghazanfari, S.; Rasteh, A.; Motahari, S.A.; Baghshah, M.S. Isoform Function Prediction Using a Deep Neural Network. arXiv 2022, arXiv:2208.03325. [Google Scholar]
- Ye, M.; Ren, S.; Luo, H.; Wu, X.; Lian, H.; Cai, X.; Ji, Y. Integration of Graph Neural Networks and Transcriptomics Analysis Identify Key Pathways and Gene Signature for Immunotherapy Response and Prognosis of Skin Melanoma. BMC Cancer 2025, 25, 648. [Google Scholar] [CrossRef]
- Piazza, L.; Di Stefano, M.; Poles, C.; Bononi, G.; Poli, G.; Renzi, G.; Galati, S.; Giordano, A.; Macchia, M.; Carta, F.; et al. A Machine Learning Platform for Isoform-Specific Identification and Profiling of Human Carbonic Anhydrase Inhibitors. Pharmaceuticals 2025, 18, 1007. [Google Scholar] [CrossRef]
- Su, J.; Jiao, X. Quartformer: An Accurate Deep Learning Framework for Phylogenetic Tree Construction. bioRxiv 2025. [Google Scholar] [CrossRef]
- Azouri, D.; Abadi, S.; Mansour, Y.; Mayrose, I.; Pupko, T. Harnessing Machine Learning to Guide Phylogenetic-Tree Search Algorithms. Nat. Commun. 2021, 12, 1983. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Sharma, S. Evolutionary Sparse Learning for Phylogenomics. Mol. Biol. Evol. 2021, 38, 4674. [Google Scholar] [CrossRef] [PubMed]
- Xie, T.; Richman, H.; Gao, J.; Matsen, F.A.; Zhang, C. PhyloVAE: Unsupervised Learning of Phylogenetic Trees via Variational Autoencoders. In Proceedings of the 13th International Conference on Learning Representations (ICLR), Singapore, 24–28 April 2025; Volume 2025, pp. 100538–100559. [Google Scholar]
- Smith, M.L.; Hahn, M.W. Phylogenetic Inference Using Generative Adversarial Networks. Bioinformatics 2023, 39, btad543. [Google Scholar] [CrossRef]
- A Guide to Phylotranscriptomic Analysis for Phycologists. 2021. Available online: https://www.e-algae.org/journal/view.php?number=2960 (accessed on 25 November 2025).
- Xu, X.; Bonvin, A.M.J.J. DeepRank-GNN-Esm: A Graph Neural Network for Scoring Protein–Protein Models Using Protein Language Model. Bioinform. Adv. 2024, 4, vbad191. [Google Scholar] [CrossRef]
- Shaw, R.; Love, S.D.; McWhite, C.D. Evaluating Pretrained Protein Language Model Embeddings as Proxies for Functional Similarity. J. Mol. Evol. 2025, 93, 765–776. [Google Scholar] [CrossRef]
- Pantolini, L.; Studer, G.; Pereira, J.; Durairaj, J.; Tauriello, G.; Schwede, T. Embedding-Based Alignment: Combining Protein Language Models with Dynamic Programming Alignment to Detect Structural Similarities in the Twilight-Zone. Bioinformatics 2024, 40, btad786. [Google Scholar] [CrossRef]
- Williams, T.A.; Davin, A.A.; Szánthó, L.L.; Stamatakis, A.; Wahl, N.A.; Woodcroft, B.J.; Soo, R.M.; Eme, L.; Sheridan, P.O.; Gubry-Rangin, C.; et al. Phylogenetic Reconciliation: Making the Most of Genomes to Understand Microbial Ecology and Evolution. ISME J. 2024, 18, wrae129. [Google Scholar] [CrossRef]
- Åkerborg, Ö.; Sennblad, B.; Arvestad, L.; Lagergren, J. Simultaneous Bayesian Gene Tree Reconstruction and Reconciliation Analysis. Proc. Natl. Acad. Sci. USA 2009, 106, 5714–5719. [Google Scholar] [CrossRef]
- Bansal, M.S.; Wu, Y.C.; Alm, E.J.; Kellis, M. Improved Gene Tree Error Correction in the Presence of Horizontal Gene Transfer. Bioinformatics 2015, 31, 1211–1218. [Google Scholar] [CrossRef]
- Bremer, N.; Knopp, M.; Martin, W.F.; Tria, F.D.K. Realistic Gene Transfer to Gene Duplication Ratios Identify Different Roots in the Bacterial Phylogeny Using a Tree Reconciliation Method. Life 2022, 12, 995. [Google Scholar] [CrossRef] [PubMed]
- Mondal, A.; Kordi, M.; Bansal, M.S. A Supervised Ma-Chine Learning Approach for Distinguishing Between Additive and Replacing Horizontal Gene Transfers. In Proceedings of the 11th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics, Online, 21–24 September 2020. [Google Scholar] [CrossRef]
- Ding, X.; Zou, Z.; Brooks, C.L. Deciphering Protein Evolution and Fitness Landscapes with Latent Space Models. Nat. Commun. 2019, 10, 5644. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yao, Y.; Xia, Y.; Tang, M. Searching for Protein Variants with Desired Properties Using Deep Generative Models. BMC Bioinform. 2023, 24, 297. [Google Scholar] [CrossRef] [PubMed]
- Sevgen, E.; Moller, J.; Lange, A.; Parker, J.; Quigley, S.; Mayer, J.; Srivastava, P.; Gayatri, S.; Hosfield, D.; Dilks, C.; et al. ProT-VAE: Protein Transformer Variational AutoEncoder for Functional Protein Design. Proc. Natl. Acad. Sci. USA 2025, 122, e2408737122. [Google Scholar] [CrossRef]
- Mardikoraem, M.; Wang, Z.; Pascual, N.; Woldring, D. Generative Models for Protein Sequence Modeling: Recent Advances and Future Directions. Brief. Bioinform. 2023, 24, bbad358. [Google Scholar] [CrossRef]
- Qiu, J.; Meng, F.; Ding, L.; Che, Y.; McCarron, P.; Beach, D.G.; Li, A. Dynamics of Paralytic Shellfish Toxins and Their Metabolites during Timecourse Exposure of Scallops Chlamys farreri and Mussels Mytilus galloprovincialis to Alexandrium pacificum. Aquat. Toxicol. 2018, 200, 233–240. [Google Scholar] [CrossRef]
- Zhang, S.F.; Zhang, Y.; Lin, L.; Wang, D.Z. ITRAQ-Based Quantitative Proteomic Analysis of a Toxigenic Dinoflagellate Alexandrium catenella and Its Non-Toxigenic Mutant Exposed to a Cell Cycle Inhibitor Colchicine. Front. Microbiol. 2018, 9, 312328. [Google Scholar] [CrossRef]
- Zheng, D.; Zou, L.; Zou, J.; Li, Q.; Lu, S. Multi-Omics Analysis Reveals Potential Mechanisms of Diarrhetic Shellfish Toxin and Fatty Acid Synthesis in Marine Harmful Prorocentrum. J. Hazard. Mater. 2025, 489, 137674. [Google Scholar] [CrossRef]
- Picard, M.; Scott-Boyer, M.P.; Bodein, A.; Périn, O.; Droit, A. Integration Strategies of Multi-Omics Data for Machine Learning Analysis. Comput. Struct. Biotechnol. J. 2021, 19, 3735–3746. [Google Scholar] [CrossRef]
- Zarayeneh, N.; Ko, E.; Oh, J.H.; Suh, S.; Liu, C.; Gao, J.; Kim, D.; Kang, M. Integration of Multi-Omics Data for Integrative Gene Regulatory Network Inference. Int. J. Data Min. Bioinform. 2017, 18, 223–239. [Google Scholar] [CrossRef]
- Ge, J.; Cai, J.; Zhang, G.; Li, D.; Tao, L. Multi-Omics Integration and Machine Learning Uncover Molecular Basal-like Subtype of Pancreatic Cancer and Implicate A2ML1 in Promoting Tumor Epithelial-Mesenchymal Transition. J. Transl. Med. 2025, 23, 741. [Google Scholar] [CrossRef] [PubMed]
- Wekesa, J.S.; Kimwele, M. A Review of Multi-Omics Data Integration through Deep Learning Approaches for Disease Diagnosis, Prognosis, and Treatment. Front. Genet. 2023, 14, 1199087. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhang, H.; Gao, S.; He, K.; Yu, T.; Gao, S.; Wang, J.; Li, H. Unveiling Salt Tolerance Mechanisms in Plants: Integrating the KANMB Machine Learning Model with Metabolomic and Transcriptomic Analysis. Adv. Sci. 2025, 12, 2417560. [Google Scholar] [CrossRef] [PubMed]
- Xin, L.; Huang, C.; Li, H.; Huang, S.; Feng, Y.; Kong, Z.; Liu, Z.; Li, S.; Yu, C.; Shen, F.; et al. Artificial Intelligence for Central Dogma-Centric Multi-Omics: Challenges and Breakthroughs. arXiv 2024, arXiv:2412.12668. [Google Scholar]
- Zhang, H.; Hou, Y.; Miranda, L.; Campbell, D.A.; Sturm, N.R.; Gaasterland, T.; Lin, S. Spliced Leader RNA Trans-Splicing in Dinoflagellates. Proc. Natl. Acad. Sci. USA 2007, 104, 4618–4623. [Google Scholar] [CrossRef]
- Parker, M.T.; Knop, K.; Sherwood, A.V.; Schurch, N.J.; Mackinnon, K.; Gould, P.D.; Hall, A.J.W.; Barton, G.J.; Simpson, G.G. Nanopore Direct RNA Sequencing Maps the Complexity of Arabidopsis MRNA Processing and M6A Modification. eLife 2020, 9, e49658. [Google Scholar] [CrossRef]
- Lee, V.V.; Judd, L.M.; Jex, A.R.; Holt, K.E.; Tonkin, C.J.; Ralph, S.A. Direct Nanopore Sequencing of MRNA Reveals Landscape of Transcript Isoforms in Apicomplexan Parasites. mSystems 2021, 6, 10–1128. [Google Scholar] [CrossRef]
- Zahedi, R.; Ghamsari, R.; Argha, A.; Macphillamy, C.; Beheshti, A.; Alizadehsani, R.; Lovell, N.H.; Lotfollahi, M.; Alinejad-Rokny, H. Deep learning in spatially resolved transcriptomics: A comprehensive technical view. Brief. Bioinform. 2024, 25, bbae082. [Google Scholar] [CrossRef]
- Argelaguet, R.; Arnol, D.; Bredikhin, D.; Deloro, Y.; Velten, B.; Marioni, J.C.; Stegle, O. MOFA+: A Statistical Framework for Comprehensive Integration of Multi-Modal Single-Cell Data. Genome Biol. 2020, 21, 111. [Google Scholar] [CrossRef]
- Chen, L.; Xu, J.; Li, S.C. DeepMF: Deciphering the Latent Patterns in Omics Profiles with a Deep Learning Method. BMC Bioinform. 2019, 20, 648. [Google Scholar] [CrossRef]
- Stein-O’Brien, G.L.; Arora, R.; Culhane, A.C.; Favorov, A.V.; Garmire, L.X.; Greene, C.S.; Goff, L.A.; Li, Y.; Ngom, A.; Ochs, M.F.; et al. Enter the Matrix: Factorization Uncovers Knowledge from Omics. Trends Genet. 2018, 34, 790–805. [Google Scholar] [CrossRef] [PubMed]
- Kang, Z.; Zhang, H.; Liang, C.; Yang, R.; Ye, Y.; Bai, H.; Zhang, Y.; Ning, K. Deciphering Biosynthetic Gene Clusters with a Context-Aware Protein Language Model. bioRxiv 2025. [Google Scholar] [CrossRef]
- Zhu, S.; Xu, H.; Liu, Y.; Hong, Y.; Yang, H.; Zhou, C.; Tao, L. Computational Advances in Biosynthetic Gene Cluster Discovery and Prediction. Biotechnol. Adv. 2025, 79, 108532. [Google Scholar] [CrossRef] [PubMed]
- Almeida, H.; Palys, S.; Tsang, A.; Diallo, A.B. TOUCAN: A Framework for Fungal Biosynthetic Gene Cluster Discovery. NAR Genom. Bioinform. 2020, 2, lqaa098. [Google Scholar] [CrossRef]
- Sztain, T.; Bartholow, T.G.; Lee, D.J.; Casalino, L.; Mitchell, A.; Young, M.A.; Wang, J.; McCammon, J.A.; Burkart, M.D. Decoding Allosteric Regulation by the Acyl Carrier Protein. Proc. Natl. Acad. Sci. USA 2021, 118, e2025597118. [Google Scholar] [CrossRef]
- Dudchenko, O.; Batra, S.S.; Omer, A.D.; Nyquist, S.K.; Hoeger, M.; Durand, N.C.; Shamim, M.S.; Machol, I.; Lander, E.S.; Aiden, A.P.; et al. De Novo Assembly of the Aedes Aegypti Genome Using Hi-C Yields Chromosome-Length Scaffolds. Science 2017, 356, 92–95. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R Package for Weighted Correlation Network Analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Sarkar, J.; Singh, R.; Chandel, S. Understanding LC/MS-Based Metabolomics: A Detailed Reference for Natural Product Analysis. Proteom. Clin. Appl. 2025, 19, e202400048. [Google Scholar] [CrossRef]
- Ji, H.; Du, R.; Dai, Q.; Su, M.; Lyu, Y.; Peng, Y.; Yan, J. DeepMASS: Unknown Compound Annotation Using Semantic Similarity of Mass Spectral Language and Chemical Space Localization. bioRxiv 2024. [Google Scholar] [CrossRef]
- Qiang, H.; Wang, F.; Lu, W.; Xing, X.; Kim, H.; Merette, S.A.; Ayres, L.B.; Oler, E.; AbuSalim, J.E.; Roichman, A.; et al. Language Model-Guided Anticipation and Discovery of Unknown Metabolites. Leonard J. Foster 2024, 10, 22. [Google Scholar] [CrossRef]
- Pang, Y.T.; Kuo, K.M.; Yang, L.; Gumbart, J.C. DeepPath: Overcoming Data Scarcity for Protein Transition Pathway Prediction Using Physics-Based Deep Learning. bioRxiv 2025. [Google Scholar] [CrossRef]
- Hafner, J.; Hatzimanikatis, V. NICEpath: Finding Metabolic Pathways in Large Networks through Atom-Conserving Substrate–Product Pairs. Bioinformatics 2021, 37, 3560–3568. [Google Scholar] [CrossRef] [PubMed]
- Lewis, A.M.; Coates, L.N.; Turner, A.D.; Percy, L.; Lewis, J. A Review of the Global Distribution of Alexandrium minutum (Dinophyceae) and Comments on Ecology and Associated Paralytic Shellfish Toxin Profiles, with a Focus on Northern Europe. J. Phycol. 2018, 54, 581–598. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Yang, S.; Zhao, D.; Hu, C.; Xu, W.; Anderson, D.M.; Li, Y.; Song, X.P.; Boyce, D.G.; Gibson, L.; et al. Coastal Phytoplankton Blooms Expand and Intensify in the 21st Century. Nature 2023, 615, 280–284. [Google Scholar] [CrossRef]
- Ho, J.C.; Michalak, A.M.; Pahlevan, N. Widespread Global Increase in Intense Lake Phytoplankton Blooms since the 1980s. Nature 2019, 574, 667–670. [Google Scholar] [CrossRef]
- Anderson, D.M.; Fachon, E.; Pickart, R.S.; Lin, P.; Fischer, A.D.; Richlen, M.L.; Uva, V.; Brosnahan, M.L.; McRaven, L.; Bahr, F.; et al. Evidence for Massive and Recurrent Toxic Blooms of Alexandrium catenella in the Alaskan Arctic. Proc. Natl. Acad. Sci. USA 2021, 118, e2107387118. [Google Scholar] [CrossRef]
- Kim, D.; Choi, J. AI-Based Toxicity Prediction Models Using ToxCast Data: Current Status and Future Directions for Explainable Models. Toxicology 2025, 517, 154230. [Google Scholar] [CrossRef]
- Mishra, D.R.; Kumar, A.; Ramaswamy, L.; Boddula, V.K.; Das, M.C.; Page, B.P.; Weber, S.J. CyanoTRACKER: A Cloud-Based Integrated Multi-Platform Architecture for Global Observation of Cyanobacterial Harmful Algal Blooms. Harmful Algae 2020, 96, 101828. [Google Scholar] [CrossRef]
- Rayamajhi, V.; Hussain, M.; Shin, H.; Jung, S. Recent advances in the application of artificial intelligence in microalgal cultivation. Processes 2025, 13, 3764. [Google Scholar] [CrossRef]
- Shin, J.; Kim, S.M. Temporal prediction of paralytic shellfish toxins in the mussel Mytilus galloprovincialis using an LSTM neural network model from environmental data. Toxins 2022, 14, 51. [Google Scholar] [CrossRef]
- Wang, W.; Wang, G.; Li, J.; Chen, J.; Gao, Z.; Fang, L.; Ren, S.; Wang, Q. Remote Sensing Identification and Model-Based Prediction of Harmful Algal Blooms in Inland Waters: Current Insights and Future Perspectives. Water Res. X 2025, 28, 100369. [Google Scholar] [CrossRef]
- Bellas Aláez, F.M.; Torres Palenzuela, J.M.; Spyrakos, E.; Vilas, L.G. Machine learning methods applied to the prediction of Pseudo-nitzschia spp. blooms in the Galician Rías Baixas (NW Spain). ISPRS Int. J. Geo-Inf. 2021, 10, 199. [Google Scholar] [CrossRef]
- Shin, J.; Cha, Y. Development of a deep learning–based feature stream network for forecasting riverine harmful algal blooms from a network perspective. Water Res. 2025, 268, 122751. [Google Scholar] [CrossRef] [PubMed]
- Martín-Suazo, S.; Morón-López, J.; Vakaruk, S.; Karamchandani, A.; Aguilar, J.A.P.; Mozo, A.; Gómez-Canaval, S.; Vinyals, M.; Ortiz, J.M. Deep learning methods for multi-horizon long-term forecasting of harmful algal blooms. Knowl.-Based Syst. 2024, 301, 112279. [Google Scholar] [CrossRef]
- Jeon, Y.; Struewing, I.; McIntosh, K.; Tidd, M.; Webb, L.; Ryu, H.; Mash, H.; Lu, J. Spatial and Temporal Variability of Saxitoxin-Producing Cyanobacteria in U.S. Urban Lakes. Toxins 2024, 16, 70. [Google Scholar] [CrossRef]
- Turner, A.D.; Dean, K.J.; Lewis, A.M.; Hartnell, D.M.; Jenkins, Z.; Bear, B.; Hatfield, R.G. ScillyHAB: A multidisciplinary survey of harmful marine phytoplankton and shellfish toxins in the Isles of Scilly: Combining citizen science with state-of-the-art monitoring in an isolated UK island territory. Mar. Drugs 2025, 23, 478. [Google Scholar] [CrossRef]
- Choi, D.H.; Yang, W.; Kim, Y.E.; Park, B.S.; Sung, J.; Choi, J.; Lee, Y. Variability in paralytic shellfish toxin profiles and dinoflagellate diversity in mussels and seawater collected during spring in Korean coastal waters. Toxins 2024, 16, 338. [Google Scholar] [CrossRef]
- Ahn, J.M.; Kim, J.; Kim, K. Ensemble Machine Learning of Gradient Boosting (XGBoost, LightGBM, CatBoost) and Attention-Based CNN-LSTM for Harmful Algal Blooms Forecasting. Toxins 2023, 15, 608. [Google Scholar] [CrossRef]
- Yussof, F.N.; Maan, N.; Reba, M.N.M. LSTM Networks to Improve the Prediction of Harmful Algal Blooms in the West Coast of Sabah. Int. J. Environ. Res. Public Health 2021, 18, 7650. [Google Scholar] [CrossRef]
- dos Santos, R.P.; Matos-Carvalho, J.P.; Leithardt, V.R.Q. Deep Learning in Time Series Forecasting with Transformer Models and RNNs. PeerJ Comput. Sci. 2025, 11, e3001. [Google Scholar] [CrossRef]
- Sharma, A.; Kiciman, E. DoWhy: An End-to-End Library for Causal Inference. arXiv 2020, arXiv:2011.04216. [Google Scholar]
- Fotia, P.; Ferrara, M. A Different Approach for Causal Impact Analysis on Python with Bayesian Structural Time-Series and Bidirectional LSTM Models. Atti Della Accad. Peloritana Dei Pericolanti-Cl. Di Sci. Fis. Mat. E Nat. 2023, 101, 12. [Google Scholar] [CrossRef]
- Shoguchi, E.; Shinzato, C.; Kawashima, T.; Gyoja, F.; Mungpakdee, S.; Koyanagi, R.; Takeuchi, T.; Hisata, K.; Tanaka, M.; Fujiwara, M.; et al. Draft Assembly of the Symbiodinium minutum Nuclear Genome Reveals Dinoflagellate Gene Structure. Curr. Biol. 2013, 23, 1399–1408. [Google Scholar] [CrossRef] [PubMed]
- Aranda, M.; Li, Y.; Liew, Y.J.; Baumgarten, S.; Simakov, O.; Wilson, M.C.; Piel, J.; Ashoor, H.; Bougouffa, S.; Bajic, V.B.; et al. Genomes of Coral Dinoflagellate Symbionts Highlight Evolutionary Adaptations Conducive to a Symbiotic Lifestyle. Sci. Rep. 2016, 6, 39734. [Google Scholar] [CrossRef]
- Ouhmouk, M.; Baichoo, S.; Abik, M. Challenges in AI-Driven Multi-Omics Data Analysis for Oncology: Addressing Dimensionality, Sparsity, Transparency and Ethical Considerations. Inform. Med. Unlocked 2025, 57, 101679. [Google Scholar] [CrossRef]
- Anderson, D.M.; Glibert, P.M.; Burkholder, J.M. Harmful Algal Blooms and Eutrophication: Nutrient Sources, Composition, and Consequences. Estuaries 2002, 25, 704–726. [Google Scholar] [CrossRef]
- Sellner, K.G.; Doucette, G.J.; Kirkpatrick, G.J. Harmful Algal Blooms: Causes, Impacts and Detection. J. Ind. Microbiol. Biotechnol. 2003, 30, 383–406. [Google Scholar] [CrossRef]
- Zainurin, S.N.; Wan Ismail, W.Z.; Mahamud, S.N.I.; Ismail, I.; Jamaludin, J.; Ariffin, K.N.Z.; Wan Ahmad Kamil, W.M. Advancements in Monitoring Water Quality Based on Various Sensing Methods: A Systematic Review. Int. J. Environ. Res. Public Health 2022, 19, 14080. [Google Scholar] [CrossRef]
- Lee, S.; Meister, G.; Franz, B.A. MODIS Aqua reflective solar band calibration for NASA’s R2018 ocean color products. Remote Sens. 2019, 11, 2187. [Google Scholar] [CrossRef]
- National Oceanic and Atmospheric Administration (NOAA). ERDDAP: Environmental Research Division’s Data Access Program. National Centers for Environmental Information. 2025. Available online: https://www.ncei.noaa.gov/erddap/ (accessed on 5 March 2025).
- IOCCG. Phytoplankton Functional Types from Space; Sathyendranath, S., Ed.; Reports of the International Ocean-Colour Coordinating Group, No. 15; IOCCG: Dartmouth, NS, Canada, 2014. [Google Scholar]
- Marx, V. The big challenges of big data. Nature 2013, 498, 255–260. [Google Scholar] [CrossRef]
- Ching, T.; Himmelstein, D.S.; Beaulieu-Jones, B.K.; Kalinin, A.A.; Do, B.T.; Way, G.P.; Greene, C.S. Opportunities and obstacles for deep learning in biology and medicine. J. R. Soc. Interface 2018, 15, 20170387. [Google Scholar] [CrossRef]
- Aliferis, C.; Simon, G. Overfitting, underfitting and general model overconfidence and underperformance pitfalls and best practices in machine learning and AI. In Artificial Intelligence and Machine Learning in Health Care and Medical Sciences: Best Practices and Pitfalls; Elsevier: Amsterdam, The Netherlands, 2024; pp. 477–524. [Google Scholar]
- Altalhan, M.; Algarni, A.; Alouane, M.T.H. Imbalanced data problem in machine learning: A review. IEEE Access 2025, 13, 13686–13699. [Google Scholar] [CrossRef]
- Azodi, C.B.; Tang, J.; Shiu, S.-H. Opening the black box: Interpretable machine learning for geneticists. Trends Genet. 2020, 36, 442–455. [Google Scholar] [CrossRef]
- Hassija, V.; Chamola, V.; Mahapatra, A.; Singal, A.; Goel, D.; Huang, K.; Hussain, A. Interpreting black-box models: A review on explainable artificial intelligence. Cogn. Comput. 2024, 16, 45–74. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.