Genetic Characterization of Staphylococcus aureus Isolates Associated with Toxic Shock Syndrome Toxin Production: An Epidemiological and Bioinformatics Approach
Abstract
1. Introduction
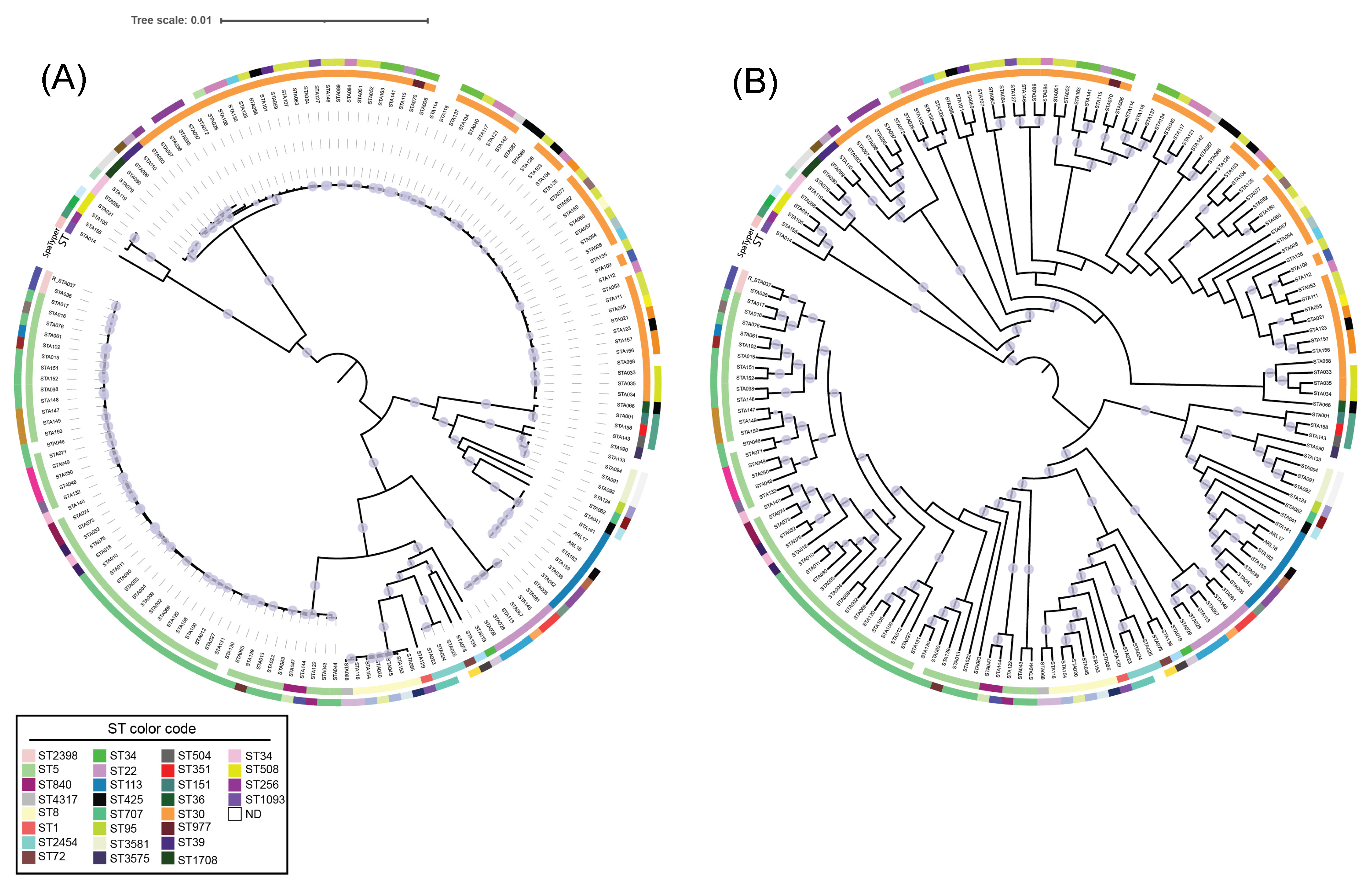
2. Results
3. Discussion
4. Conclusions
5. Methodology
5.1. DNA Extraction and Sequencing
5.2. Genomic Characterization by a Bioinformatics Approach
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wertheim, H.F.L.; Melles, D.C.; Vos, M.C.; Van Leeuwen, W.; Van Belkum, A.; Verbrugh, H.A.; Nouwen, J.L. The Role of Nasal Carriage in Staphylococcus aureus Infections. Lancet Infect. Dis. 2005, 5, 751–762. [Google Scholar] [CrossRef] [PubMed]
- Pendleton, J.N.; Gorman, S.P.; Gilmore, B.F. Clinical Relevance of the ESKAPE Pathogens. Expert Rev. Anti Infect. Ther. 2013, 11, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Howden, B.P.; Giulieri, S.G.; Wong Fok Lung, T.; Baines, S.L.; Sharkey, L.K.; Lee, J.Y.H.; Hachani, A.; Monk, I.R.; Stinear, T.P. Staphylococcus aureus Host Interactions and Adaptation. Nat. Rev. Microbiol. 2023, 21, 380–395. [Google Scholar] [CrossRef] [PubMed]
- Tong, S.Y.C.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G., Jr. Staphylococcus aureus Infections: Epidemiology, Pathophysiology, Clinical Manifestations, and Management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef]
- Léguillier, V.; Pinamonti, D.; Chang, C.-M.; Mukherjee, R.; Kumar, H.; Cossetini, A.; Manzano, M.; Anba-Mondoloni, J.; Malet-Villemagne, J.; Vidic, J. A Review and Meta-Analysis of Staphylococcus aureus Prevalence in Foods. Microbe 2024, 4, 100131. [Google Scholar] [CrossRef]
- Otto, M. Staphylococcus aureus Toxins. Curr. Opin. Microbiol. 2014, 17, 32–37. [Google Scholar] [CrossRef]
- Humphreys, H. Staphylococcus: Skin Infections; Osteomyelitis; Bloodstream Infection; Food Poisoning; Foreign Body Infections; MRSA. In Medical Microbiology; Elsevier: Amsterdam, The Netherlands, 2012; pp. 176–182. [Google Scholar]
- Liao, F.; Gu, W.; Fu, X.; Yuan, B.; Zhang, Y. Community-Acquired Methicillin-Resistant Staphylococcus aureus Provoked Cytokine Storm Causing Severe Infection on BALB/c Mice. Mol. Immunol. 2021, 140, 167–174. [Google Scholar] [CrossRef]
- Goda, K.; Kenzaka, T.; Hoshijima, M.; Yachie, A.; Akita, H. Toxic Shock Syndrome with a Cytokine Storm Caused by Staphylococcus Simulans: A Case Report. BMC Infect. Dis. 2021, 21, 19. [Google Scholar] [CrossRef]
- Samia, N.I.; Robicsek, A.; Heesterbeek, H.; Peterson, L.R. Methicillin-Resistant Staphylococcus aureus Nosocomial Infection Has a Distinct Epidemiological Position and Acts as a Marker for Overall Hospital-Acquired Infection Trends. Sci. Rep. 2022, 12, 17007. [Google Scholar] [CrossRef]
- Fetsch, A.; Contzen, M.; Hartelt, K.; Kleiser, A.; Maassen, S.; Rau, J.; Kraushaar, B.; Layer, F.; Strommenger, B. Staphylococcus aureus Food-Poisoning Outbreak Associated with the Consumption of Ice-Cream. Int. J. Food Microbiol. 2014, 187, 1–6. [Google Scholar] [CrossRef]
- Hennekinne, J.-A.; De Buyser, M.-L.; Dragacci, S. Staphylococcus aureus and Its Food Poisoning Toxins: Characterization and Outbreak Investigation. FEMS Microbiol. Rev. 2012, 36, 815–836. [Google Scholar] [CrossRef] [PubMed]
- Sospedra, I.; Mañes, J.; Soriano, J.M. Report of Toxic Shock Syndrome Toxin 1 (TSST-1) from Staphylococcus aureus Isolated in Food Handlers and Surfaces from Foodservice Establishments. Ecotoxicol. Environ. Saf. 2012, 80, 288–290. [Google Scholar] [CrossRef] [PubMed]
- Goudsmit, A.; Markowicz, S.; Lali, S.E.; Cherifi, S. Food Poisoning Due to a TSST1-Producing Staphylococcus aureus. IDCases 2021, 26, e01272. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.N.; Han, S.G. Bovine Mastitis: Risk Factors, Therapeutic Strategies, and Alternative Treatments—A Review. Asian-Australas. J. Anim. Sci. 2020, 33, 1699–1713. [Google Scholar] [CrossRef]
- SENASICA. Agriculture, Livestock Farmers and Industry Promote Consumption of Domestically Produced Milk. Available online: https://www.gob.mx/senasica/documentos/agriculture-livestock-farmers-and-industry-promote-consumption-of-domestically-produced-milk (accessed on 23 May 2025).
- Mason, A.; Foster, D.; Bradley, P.; Golubchik, T.; Doumith, M.; Gordon, N.C.; Pichon, B.; Iqbal, Z.; Staves, P.; Crook, D.; et al. Accuracy of Different Bioinformatics Methods in Detecting Antibiotic Resistance and Virulence Factors from Staphylococcus aureus Whole-Genome Sequences. J. Clin. Microbiol. 2018, 56, e01815-17. [Google Scholar] [CrossRef]
- Tuan, V.P.; Narith, D.; Tshibangu-Kabamba, E.; Dung, H.D.Q.; Viet, P.T.; Sokomoth, S.; Binh, T.T.; Sokhem, S.; Tri, T.D.; Ngov, S.; et al. A Next-Generation Sequencing-Based Approach to Identify Genetic Determinants of Antibiotic Resistance in Cambodian Helicobacter Pylori Clinical Isolates. J. Clin. Med. 2019, 8, 858. [Google Scholar] [CrossRef]
- Gahlawat, A.; Varma, T.; Kamble, P.; Banerjee, A.; Sandhu, H.; Garg, P. Bioinformatics: Theory and Applications. In The Quintessence of Basic and Clinical Research and Scientific Publishing; Springer: Berlin/Heidelberg, Germany, 2023; pp. 539–555. [Google Scholar]
- Cronin, M.T.D.; Yoon, M. Computational Methods to Predict Toxicity. In The History of Alternative Test Methods in Toxicology; Elsevier: Amsterdam, The Netherlands, 2019; pp. 287–300. [Google Scholar]
- Katsila, T.; Spyroulias, G.A.; Patrinos, G.P.; Matsoukas, M.-T. Computational Approaches in Target Identification and Drug Discovery. Comput. Struct. Biotechnol. J. 2016, 14, 177–184. [Google Scholar] [CrossRef]
- Sharma-Kuinkel, B.K.; Mongodin, E.F.; Myers, J.R.; Vore, K.L.; Canfield, G.S.; Fraser, C.M.; Rude, T.H.; Fowler, V.G., Jr.; Gill, S.R. Potential Influence of Staphylococcus aureus Clonal Complex 30 Genotype and Transcriptome on Hematogenous Infections. Open Forum Infect. Dis. 2015, 2, ofv093. [Google Scholar] [CrossRef]
- Fowler, V.G., Jr.; Nelson, C.L.; McIntyre, L.M.; Kreiswirth, B.N.; Monk, A.; Archer, G.L.; Federspiel, J.; Naidich, S.; Remortel, B.; Rude, T. Potential Associations between Hematogenous Complications and Bacterial Genotype in Staphylococcus aureus Infection. J. Infect. Dis. 2007, 196, 738–747. [Google Scholar] [CrossRef]
- Gill, S.R.; McIntyre, L.M.; Nelson, C.L.; Remortel, B.; Rude, T.; Reller, L.B.; Fowler Jr, V.G. Potential Associations between Severity of Infection and the Presence of Virulence-Associated Genes in Clinical Strains of Staphylococcus aureus. PLoS ONE 2011, 6, e18673. [Google Scholar] [CrossRef]
- Guillén, R.; Salinas, C.; Mendoza-Álvarez, A.; Rubio Rodríguez, L.A.; Díaz-de Usera, A.; Lorenzo-Salazar, J.M.; González-Montelongo, R.; Flores, C.; Rodríguez, F. Genomic Epidemiology of the Primary Methicillin-Resistant Staphylococcus aureus Clones Causing Invasive Infections in Paraguayan Children. Microbiol. Spectr. 2024, 12, e03012-23. [Google Scholar] [CrossRef] [PubMed]
- Souza, S.S.R.; Smith, J.T.; Marcovici, M.M.; Eckhardt, E.M.; Hansel, N.B.; Martin, I.W.; Andam, C.P. Demographic Fluctuations in Bloodstream Staphylococcus aureus Lineages Configure the Mobile Gene Pool and Antimicrobial Resistance. npj Antimicrob. Resist. 2024, 2, 14. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Sánchez, D.A.; Grillo, S.; Carrera-Salinas, A.; González-Díaz, A.; Cuervo, G.; Grau, I.; Camoez, M.; Martí, S.; Berbel, D.; Tubau, F. Molecular Epidemiology, Antimicrobial Susceptibility, and Clinical Features of Methicillin-Resistant Staphylococcus aureus Bloodstream Infections over 30 Years in Barcelona, Spain (1990–2019). Microorganisms 2022, 10, 2401. [Google Scholar] [CrossRef]
- Di Gregorio, S.; Vielma, J.; Haim, M.S.; Rago, L.; Campos, J.; Kekre, M.; Abrudan, M.; Famiglietti, Á.; Canigia, L.F.; Rubinstein, G.; et al. Genomic Epidemiology of Staphylococcus aureus Isolated from Bloodstream Infections in South America during 2019 Supports Regional Surveillance. Microb. Genom. 2023, 9, 001020. [Google Scholar] [CrossRef]
- Deurenberg, R.H.; Nulens, E.; Valvatne, H.; Sebastian, S.; Driessen, C.; Craeghs, J.; De Brauwer, E.; Heising, B.; Kraat, Y.J.; Riebe, J. Cross-Border Dissemination of Methicillin-Resistant Staphylococcus aureus, Euregio Meuse-Rhin Region. Emerg. Infect. Dis. 2009, 15, 727. [Google Scholar] [CrossRef]
- Monecke, S.; Gavier-Widen, D.; Hotzel, H.; Peters, M.; Guenther, S.; Lazaris, A.; Loncaric, I.; Müller, E.; Reissig, A.; Ruppelt-Lorz, A.; et al. Diversity of Staphylococcus aureus Isolates in European Wildlife. PLoS ONE 2016, 11, e0168433. [Google Scholar] [CrossRef]
- Chen, F.; Yin, Y.; Chen, H.; Wang, R.; Wang, S.; Wang, H. Global Genetic Diversity and Asian Clades Evolution: A Phylogeographic Study of Staphylococcus aureus Sequence Type 5. Antimicrob. Agents Chemother. 2024, 68, e01175-23. [Google Scholar] [CrossRef]
- Planet, P.J.; Narechania, A.; Chen, L.; Mathema, B.; Boundy, S.; Archer, G.; Kreiswirth, B. Architecture of a Species: Phylogenomics of Staphylococcus aureus. Trends Microbiol. 2017, 25, 153–166. [Google Scholar] [CrossRef]
- Di Gregorio, S.; Haim, M.S.; Vielma Vallenilla, J.; Cohen, V.; Rago, L.; Gulone, L.; Aanensen, D.M.; Argimón, S.; Mollerach, M. Genomic Epidemiology of CC30 Methicillin-Resistant Staphylococcus aureus Strains from Argentina Reveals Four Major Clades with Distinctive Genetic Features. mSphere 2021, 6, e01297-20. [Google Scholar] [CrossRef]
- Peng, Q.; Tang, X.; Dong, W.; Sun, N.; Yuan, W. A Review of Biofilm Formation of Staphylococcus aureus and Its Regulation Mechanism. Antibiotics 2022, 12, 12. [Google Scholar] [CrossRef]
- Romanova, Y.M.; Mulabaev, N.S.; Tolordava, E.R.; Seregin, A.V.; Seregin, I.V.; Alexeeva, N.V.; Stepanova, T.V.; Levina, G.A.; Barkhatova, O.I.; Gamova, N.A.; et al. Microbial Communities on Kidney Stones. Mol. Genet. Microbiol. Virol. 2015, 30, 78–84. [Google Scholar] [CrossRef]
- Swidsinski, A.; Mendling, W.; Loening-Baucke, V.; Ladhoff, A.; Swidsinski, S.; Hale, L.P.; Lochs, H. Adherent Biofilms in Bacterial Vaginosis. Obstet. Gynecol. 2005, 106, 1013–1023. [Google Scholar] [CrossRef] [PubMed]
- Perry, E.K.; Tan, M.-W. Bacterial Biofilms in the Human Body: Prevalence and Impacts on Health and Disease. Front. Cell Infect. Microbiol. 2023, 13, 1237164. [Google Scholar] [CrossRef]
- Carron, M.A.; Tran, V.R.; Sugawa, C.; Coticchia, J.M. Identification of Helicobacter Pylori Biofilms in Human Gastric Mucosa. J. Gastrointest. Surg. 2006, 10, 712–717. [Google Scholar] [CrossRef]
- Ahmad-Mansour, N.; Loubet, P.; Pouget, C.; Dunyach-Remy, C.; Sotto, A.; Lavigne, J.-P.; Molle, V. Staphylococcus aureus Toxins: An Update on Their Pathogenic Properties and Potential Treatments. Toxins 2021, 13, 677. [Google Scholar] [CrossRef]
- Moraveji, Z.; Tabatabaei, M.; Aski, H.S.; Khoshbakht, R. Characterization of Hemolysins of Staphylococcus Strains Isolated from Human and Bovine, Southern Iran. Iran. J. Vet. Res. 2014, 15, 326. [Google Scholar]
- Burnside, K.; Lembo, A.; de Los Reyes, M.; Iliuk, A.; BinhTran, N.-T.; Connelly, J.E.; Lin, W.-J.; Schmidt, B.Z.; Richardson, A.R.; Fang, F.C.; et al. Regulation of Hemolysin Expression and Virulence of Staphylococcus aureus by a Serine/Threonine Kinase and Phosphatase. PLoS ONE 2010, 5, e11071. [Google Scholar] [CrossRef]
- Atchade, E.; De Tymowski, C.; Grall, N.; Tanaka, S.; Montravers, P. Toxic Shock Syndrome: A Literature Review. Antibiotics 2024, 13, 96. [Google Scholar] [CrossRef]
- Klompas, M. Toxic Shock Syndromes. Decis. Mak. Med. 2010, 336, 727–746. [Google Scholar]
- Thomas, S.; Liu, W.; Arora, S.; Ganesh, V.; Ko, Y.-P.; Höök, M. The Complex Fibrinogen Interactions of the Staphylococcus aureus Coagulases. Front. Cell Infect. Microbiol. 2019, 9, 106. [Google Scholar] [CrossRef]
- Panizzi, P.; Friedrich, R.; Fuentes-Prior, P.; Kroh, H.K.; Briggs, J.; Tans, G.; Bode, W.; Bock, P.E. Novel Fluorescent Prothrombin Analogs as Probes of Staphylocoagulase-Prothrombin Interactions. J. Biol. Chem. 2006, 281, 1169–1178. [Google Scholar] [CrossRef] [PubMed]
- Dubin, G. Extracellular Proteases of Staphylococcus spp. Biol. Chem. 2002, 383, 1075–1086. [Google Scholar]
- Hait, J.; Tallent, S.; Melka, D.; Keys, C.; Bennett, R. S Taphylococcus Aureus Outbreak Investigation of an I Llinois Bakery. J. Food Saf. 2012, 32, 435–444. [Google Scholar] [CrossRef]
- Hait, J.M.; Cao, G.; Kastanis, G.; Yin, L.; Pettengill, J.B.; Tallent, S.M. Evaluation of Virulence Determinants Using Whole-Genome Sequencing and Phenotypic Biofilm Analysis of Outbreak-Linked Staphylococcus aureus Isolates. Front. Microbiol. 2021, 12, 687625. [Google Scholar] [CrossRef]
- Rhem, M.N.; Lech, E.M.; Patti, J.M.; McDevitt, D.; Höök, M.; Jones, D.B.; Wilhelmus, K.R. The Collagen-Binding Adhesin Is a Virulence Factor in Staphylococcus aureus Keratitis. Infect. Immun. 2000, 68, 3776–3779. [Google Scholar] [CrossRef]
- Zong, Y.; Xu, Y.; Liang, X.; Keene, D.R.; Höök, A.; Gurusiddappa, S.; Höök, M.; Narayana, S.V.L. A ‘Collagen Hug’ Model for Staphylococcus aureus CNA Binding to Collagen. EMBO J. 2005, 24, 4224–4236. [Google Scholar] [CrossRef]
- Arumugam, P.; Kielian, T. Metabolism Shapes Immune Responses to Staphylococcus aureus. J. Innate Immun. 2024, 16, 12–30. [Google Scholar] [CrossRef]
- Thammavongsa, V.; Kim, H.K.; Missiakas, D.; Schneewind, O. Staphylococcal Manipulation of Host Immune Responses. Nat. Rev. Microbiol. 2015, 13, 529–543. [Google Scholar] [CrossRef]
- Bowman, L.; Palmer, T. The Type VII Secretion System of Staphylococcus. Annu. Rev. Microbiol. 2021, 75, 471–494. [Google Scholar] [CrossRef]
- Truong-Bolduc, Q.C.; Wang, Y.; Ferrer-Espada, R.; Reedy, J.L.; Martens, A.T.; Goulev, Y.; Paulsson, J.; Vyas, J.M.; Hooper, D.C. Staphylococcus aureus AbcA Transporter Enhances Persister Formation under β-Lactam Exposure. Antimicrob. Agents Chemother. 2024, 68, e01340-23. [Google Scholar] [CrossRef]
- Hiron, A.; Posteraro, B.; Carrière, M.; Remy, L.; Delporte, C.; La Sorda, M.; Sanguinetti, M.; Juillard, V.; Borezée-Durant, E. A Nickel ABC-transporter of Staphylococcus aureus Is Involved in Urinary Tract Infection. Mol. Microbiol. 2010, 77, 1246–1260. [Google Scholar] [CrossRef] [PubMed]
- Shalem, O.; Sanjana, N.E.; Hartenian, E.; Shi, X.; Scott, D.A.; Mikkelsen, T.S.; Heckl, D.; Ebert, B.L.; Root, D.E.; Doench, J.G.; et al. Genome-Scale CRISPR-Cas9 Knockout Screening in Human Cells. Science 2014, 343, 84–87. [Google Scholar] [CrossRef] [PubMed]
- Alpern, D.; Gardeux, V.; Russeil, J.; Mangeat, B.; Meireles-Filho, A.C.A.; Breysse, R.; Hacker, D.; Deplancke, B. BRB-Seq: Ultra-Affordable High-Throughput Transcriptomics Enabled by Bulk RNA Barcoding and Sequencing. Genome Biol. 2019, 20, 71. [Google Scholar] [CrossRef]
- Quince, C.; Walker, A.W.; Simpson, J.T.; Loman, N.J.; Segata, N. Shotgun Metagenomics, from Sampling to Analysis. Nat. Biotechnol. 2017, 35, 833–844. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 3 June 2025).
- Krueger, F. Trim Galore. A wrapper tool around Cutadapt and FastQC to consistently apply quality and adapter trimming to FastQ files. Babraham Bioinform. 2015, 516, 517. [Google Scholar]
- Coil, D.; Jospin, G.; Darling, A.E. A5-Miseq: An Updated Pipeline to Assemble Microbial Genomes from Illumina MiSeq Data. Bioinformatics 2015, 31, 587–589. [Google Scholar] [CrossRef]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-Access Bacterial Population Genomics: BIGSdb Software, the PubMLST. Org Website and Their Applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef]
- Francisco, A.P.; Vaz, C.; Monteiro, P.T.; Melo-Cristino, J.; Ramirez, M.; Carriço, J.A. PHYLOViZ: Phylogenetic Inference and Data Visualization for Sequence Based Typing Methods. BMC Bioinform. 2012, 13, 87. [Google Scholar] [CrossRef]
- Seemann, T. ABRicate: Mass Screening of Contigs for Antimicrobial and Virulence Genes. Department of Microbiology and Immunology, The University of Melbourne, Melbourne, Australia. 2018. Available online: https://github.com/tseemann/abricate (accessed on 28 February 2019).
- Liu, B.; Zheng, D.; Jin, Q.; Chen, L.; Yang, J. VFDB 2019: A Comparative Pathogenomic Platform with an Interactive Web Interface. Nucleic Acids Res. 2019, 47, D687–D692. [Google Scholar] [CrossRef]
- Sanchez-Herrero, J.F.; Sullivan, M. SpaTyper: Staphylococcal Protein A (Spa) Characterization Pipeline. Zenodo 2020. [Google Scholar] [CrossRef]
- Treangen, T.J.; Ondov, B.D.; Koren, S.; Phillippy, A.M. The Harvest Suite for Rapid Core-Genome Alignment and Visualization of Thousands of Intraspecific Microbial Genomes. Genome Biol. 2014, 15, 524. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (ITOL) v6: Recent Updates to the Phylogenetic Tree Display and Annotation Tool. Nucleic Acids Res. 2024, 52, gkae268. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aguirre-Sánchez, J.R.; Chaidez-Quiroz, C.; Castro-del Campo, N.; Castro-del Campo, N. Genetic Characterization of Staphylococcus aureus Isolates Associated with Toxic Shock Syndrome Toxin Production: An Epidemiological and Bioinformatics Approach. Toxins 2025, 17, 440. https://doi.org/10.3390/toxins17090440
Aguirre-Sánchez JR, Chaidez-Quiroz C, Castro-del Campo N, Castro-del Campo N. Genetic Characterization of Staphylococcus aureus Isolates Associated with Toxic Shock Syndrome Toxin Production: An Epidemiological and Bioinformatics Approach. Toxins. 2025; 17(9):440. https://doi.org/10.3390/toxins17090440
Chicago/Turabian StyleAguirre-Sánchez, J. R., C. Chaidez-Quiroz, Nohemi Castro-del Campo, and Nohelia Castro-del Campo. 2025. "Genetic Characterization of Staphylococcus aureus Isolates Associated with Toxic Shock Syndrome Toxin Production: An Epidemiological and Bioinformatics Approach" Toxins 17, no. 9: 440. https://doi.org/10.3390/toxins17090440
APA StyleAguirre-Sánchez, J. R., Chaidez-Quiroz, C., Castro-del Campo, N., & Castro-del Campo, N. (2025). Genetic Characterization of Staphylococcus aureus Isolates Associated with Toxic Shock Syndrome Toxin Production: An Epidemiological and Bioinformatics Approach. Toxins, 17(9), 440. https://doi.org/10.3390/toxins17090440





