Abstract
This paper assesses the effects of exposure to toxic concentrations (1200 to 6000 cells/mL) of the dinoflagellates Prorocentrum lima, Prorocentrum minimum, and Prorocentrum rhathymum and several concentrations of aqueous and organic extracts obtained from the same species (0 to 20 parts per thousand) on the Crassostrea gigas (5–7 mm) proteomic profile. Through comparative proteomic map analyses, several protein spots were detected with different expression levels, of which eight were selected to be identified by liquid chromatography–mass spectrometry (LC–MS/MS) analyses. The proteomic response suggests that, after 72 h of exposure to whole cells, the biological functions of C. gigas affected proteins in the immune system, stress response, contractile systems and cytoskeletal activities. The exposure to organic and aqueous extracts mainly showed effects on protein expressions in muscle contraction and cytoskeleton morphology. These results enrich the knowledge on early bivalve developmental stages. Therefore, they may be considered a solid base for new bioassays and/or generation of specific analytical tools that allow for some of the main effects of algal proliferation phenomena on bivalve mollusk development to be monitored, characterized and elucidated.
Key Contribution:
Dinoflagellates of the genus Prorocentrum produce toxic compounds capable of causing mortalities and affectations at the proteome level in C. gigas, modifying the expression of proteins related to the immune system, stress response, muscle contraction, and cytoskeleton activities. The kind of exposure (to whole cells, aqueous, or organic extracts) reveals differential responses on C. gigas, which are more complex and lethal with whole cells. These results indicate that the susceptibility of young C. gigas is not only due to the toxins present on the Prorocentrum species tested.
1. Introduction
Phycotoxins are natural metabolites produced by micro-algae or seaweeds, which are mass molecules of around 300–3500 Da that belong to diverse groups of chemical compounds [1,2]. In the marine environment, these organic compounds are produced by diatoms, dinoflagellates, cyanobacteria, and other flagellated phytoplankton [1,3]. Of the 5000 phytoplankton species known, about 300 of them are involved in proliferation events [4,5]. These events are natural phenomena characterized by an exponential increase in cell density, as a result of changes in several environmental factors (temperature, salinity, nutrients, ocean acidification, precipitation, etc.). Nevertheless, how the integration of these climate drivers might have driven proliferation is still unclear [6].
Among the 300 phytoplankton species mentioned above, only about 100 produce toxins that can cause intoxication or even death in humans and animals [5]. The phytoplankton species mostly involved in these toxic events are dinoflagellates and diatoms. Furthermore, species of the genus Prorocentrum have been recurrently reported from tropical and temperate waters. The genus Prorocentrum is a group of dinoflagellates distributed worldwide in planktonic and benthic marine ecosystems, with 78 species accepted taxonomically hitherto [7]. At least ten species of Prorocentrum (P. lima, P. cordatum (as P. minimum), P. borbonicum, P. concavum, P. leve, P. rhathymum, P. hoffmannianum (as P. maculosum), P. caipirignum, P. belizeanum, and P. faustiae) have been confirmed to produce a suite of toxins called okadaic acid (OA) and its analogues, also referred to as dinophysistoxins (DTXs). Additionally, borbotoxins and other unidentified toxins have been associated with diarrheic shellfish poisoning [8,9]. These compounds are complex lipid- and water-soluble polyether molecules and are specific inhibitors of serine/threonine phosphatases at the molecular level. However, at cellular levels, they cause alterations in DNA and on cellular components as well as effects on immune and nervous systems and on embryonic development [10].
Phycotoxins are acquired by fish and shellfish—mainly bivalve mollusks (oysters, mussels, scallops, or clams)—by direct filtration of dinoflagellates from the water column and via feeding on resuspended benthic material. Subsequently, they can be consumed by humans, causing alimentary intoxication. Studies on the effects of the genus Prorocentrum on bivalve mollusks have been performed, but most of them have addressed biological responses, such as particle selection, survival, clearance rate, depuration, growth, and toxin accumulation [11,12,13,14,15,16,17]. Moreover, studies at the molecular level are scarce despite the wide interest in knowing the cellular response mechanisms caused by organisms of the genus Prorocentrum [18,19,20,21,22,23].
The Pacific oyster C. gigas is an ideal marine invertebrate model for the studies previously mentioned because of its ecological and economic significance [24]. In addition to its biology, genetics, and innate immunity being extensively studied [25,26], it is the first marine sessile bivalve for which the genome has been completely sequenced [27]. C. gigas inhabits estuarine and intertidal zones, where it is exposed to dramatic environmental fluctuations, including phytoplankton proliferations. Thus, vast interest exists in understanding several cellular response mechanisms to alterations in the physical-chemical environment at the organism level. Proteome rearrangements are common responses to environmental stress, so they must be distinctive of certain types of toxic exposure [28]. Currently, proteomic studies conducted on bivalves have been focused primarily on protein expression profile analyses after exposure to a variety of environmental contaminants [29,30,31,32,33,34,35,36,37]. Others have investigated the response to ocean acidification [38,39,40,41] and evaluated the effect of toxic phytoplankton [42]. Some studies have revealed that early stages of development are the most susceptible to stress and that >7% of the oyster larval proteome is altered, affecting growth, development, and fitness and causing shell morphological abnormalities in these juvenile oysters. Therefore, this study investigates the effects of three toxic Prorocentrum species (P. lima, P. minimum, and P. rhathymum) and their aqueous (AE) and organic (OE) extracts on C. gigas spat proteomic response.
2. Results
2.1. Mortality of C. gigas Exposed to Prorocentrum Complete Cells or Extracts (Time, Number, and Concentration)
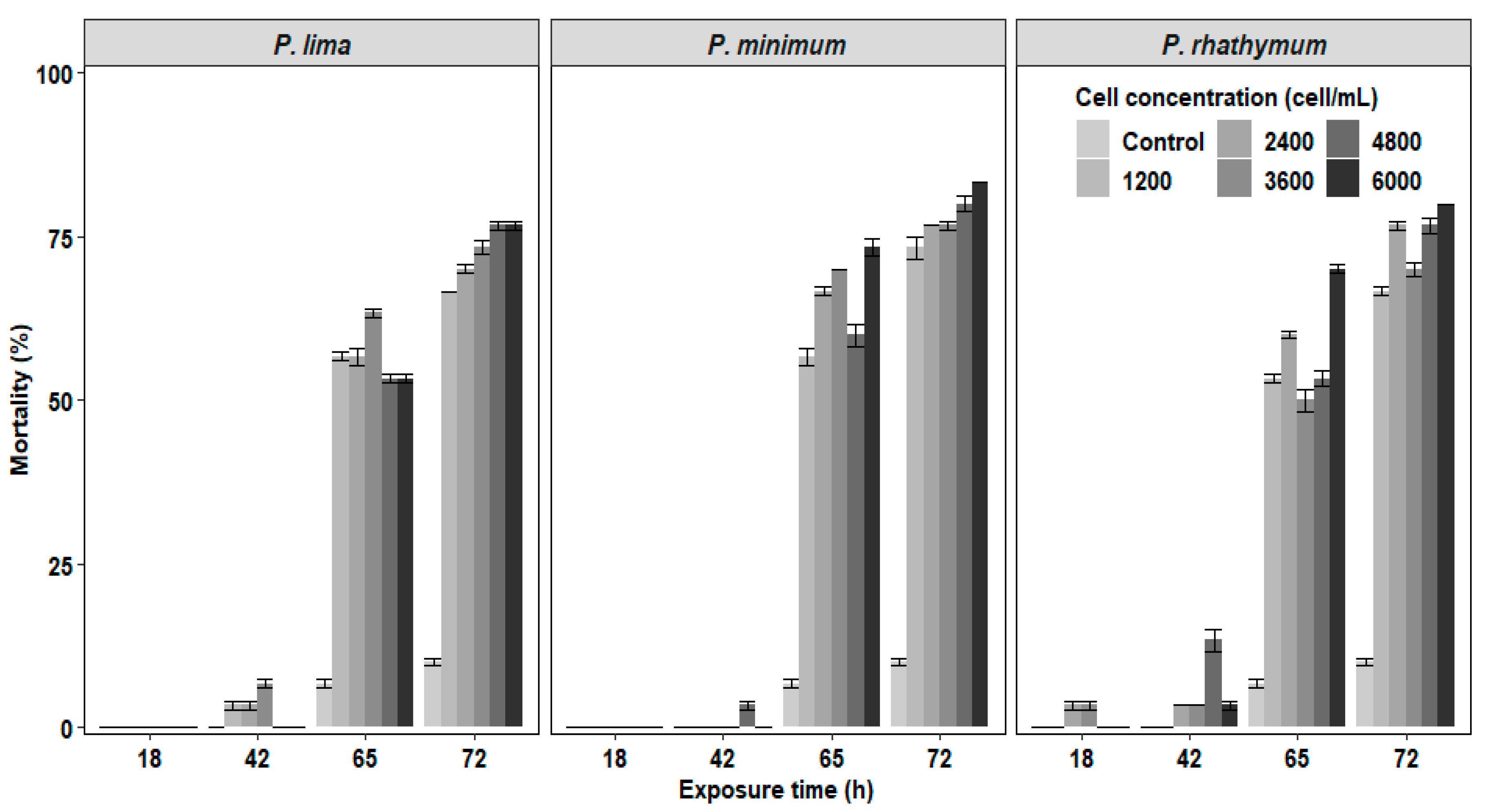
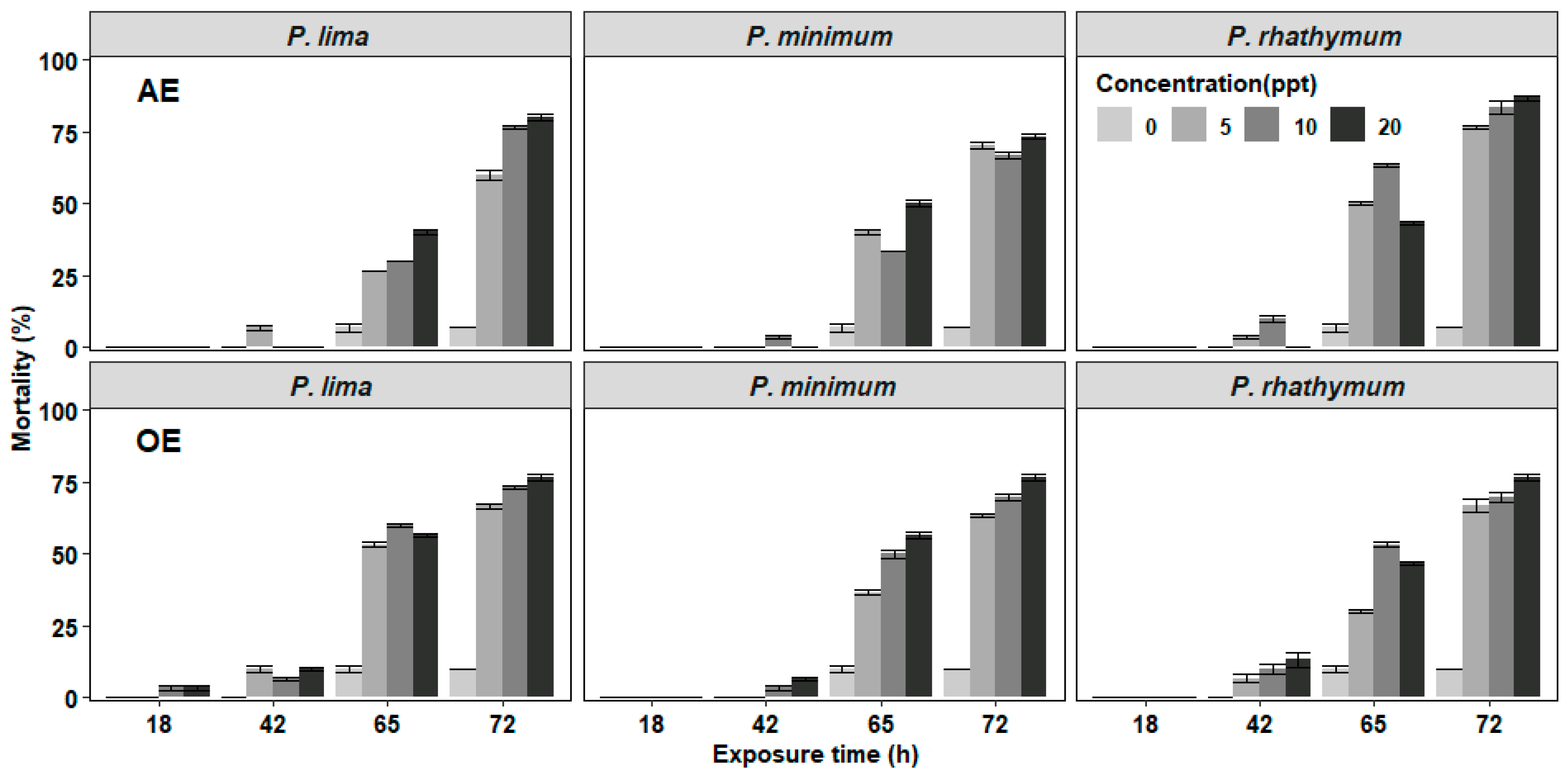
In the experiments of C. gigas with exposure to complete cells or extracts of Prorocentrum species, the mortality of juveniles was observed after 42 h exposure with the highest number of deaths at 72 h without reaching 100% at the end of the bioassay (Figure 1 and Figure 2). In organisms exposed to whole cells (Figure 1), the highest cumulative mortality was observed with treatments of 6000 cells/mL; P. minimun caused the highest mortality (83.3%), followed by P. rhathymum (80%) and P. lima (76.6%). Similarly, organisms exposed to different extracts showed massive mortality after 42 h exposure (Figure 2). The highest cumulative mortality occurred with extract concentrations of 20 parts per thousand (ppt). As in the case of exposure to whole cells, the cumulative mortality using AE varied according to the Prorocentrum species. The highest cumulative mortality was observed with P. rhathymum (86.6%), followed by P. lima (80%) and P. minimum (73.3%). With OE, the three Prorocentrum species promoted similar cumulative mortality percentages in C. gigas (76.6%), while in the control group, mortality reached only 6.67%.
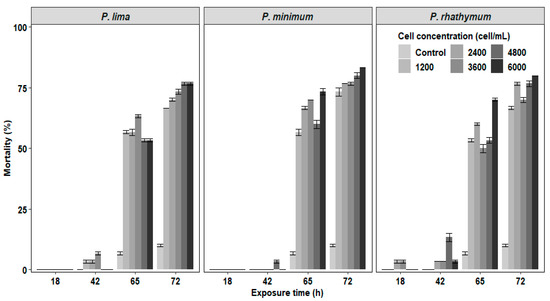
Figure 1.
Percentage of cumulative mortality of C. gigas oyster spat exposed throughout time to several concentrations of whole cells (1200 2400 3600 4800 and 6000 cells/mL) of three different Prorocentrum species. Note: Isochrysis galbana at 6000 cells/mL−1 was used as the control group.
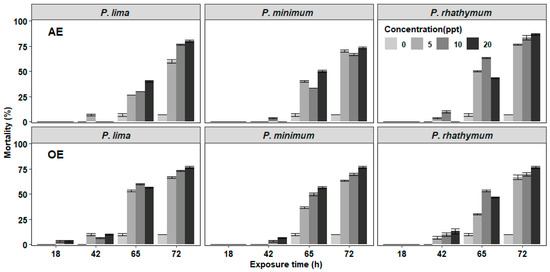
Figure 2.
Percentage of cumulative mortality of C. gigas oyster spat exposed throughout time to several concentrations of aqueous or organic extracts (AE and OE, respectively) of three different Prorocentrum species. Note: Extract concentrations used: 0, 5, 10, and 20 parts per thousand (ppt). As controls, distilled water and 1% Tween 60 were used on AE and OE experiments (respectively).
When the effects caused by each considered factor (number of cells/mL−1, species, exposure time) on C. gigas mortality were compared in each experiment (Table 1), the statistical analyses showed: (1) whole cell exposure (Table S1), in which (i) cell concentration of Prorocentrum species did not influence spat mortality significantly (p < 0.05) and (ii) significant differences were observed on mortality percentage between P. minimum with respect to P. lima and P. rhathymum after 65 h of exposure (p < 0.05); (2) aqueous (AE) or organic (OE) extract cell exposure. Similar results were observed when any Prorocentrum species was used: (i) No significant differences (p < 0.05) were observed between the tested concentrations of both extracts (AE or OE); thus, the tested extract concentration is not a factor that affects mortality. However, significant differences on C. gigas mortality were observed (p < 0.05) only after 65 and 72 h exposure to Prorocentrum species extracts. (ii) When AE was used, statistically significant differences were observed only on C. gigas mortality exposed to Prorocentrum species with regard to those of the control group (p < 0.05) (Table S2). (iii) When OE was tested, no significant differences were observed in oyster mortality between organisms exposed to P. lima, P. minimum and P. rhathymum with regard to oysters exposed to the control treatment (p < 0.05) (Table S3).

Table 1.
Mortality of C. gigas oyster spat exposed to whole cells (WC: 1200 2400 3600 4800 and 6000 cells/mL) and several concentrations of aqueous (AE) or organic (OE) P. lima, P. minimum, or P. rhathymum extracts.
2.2. Proteomic Response
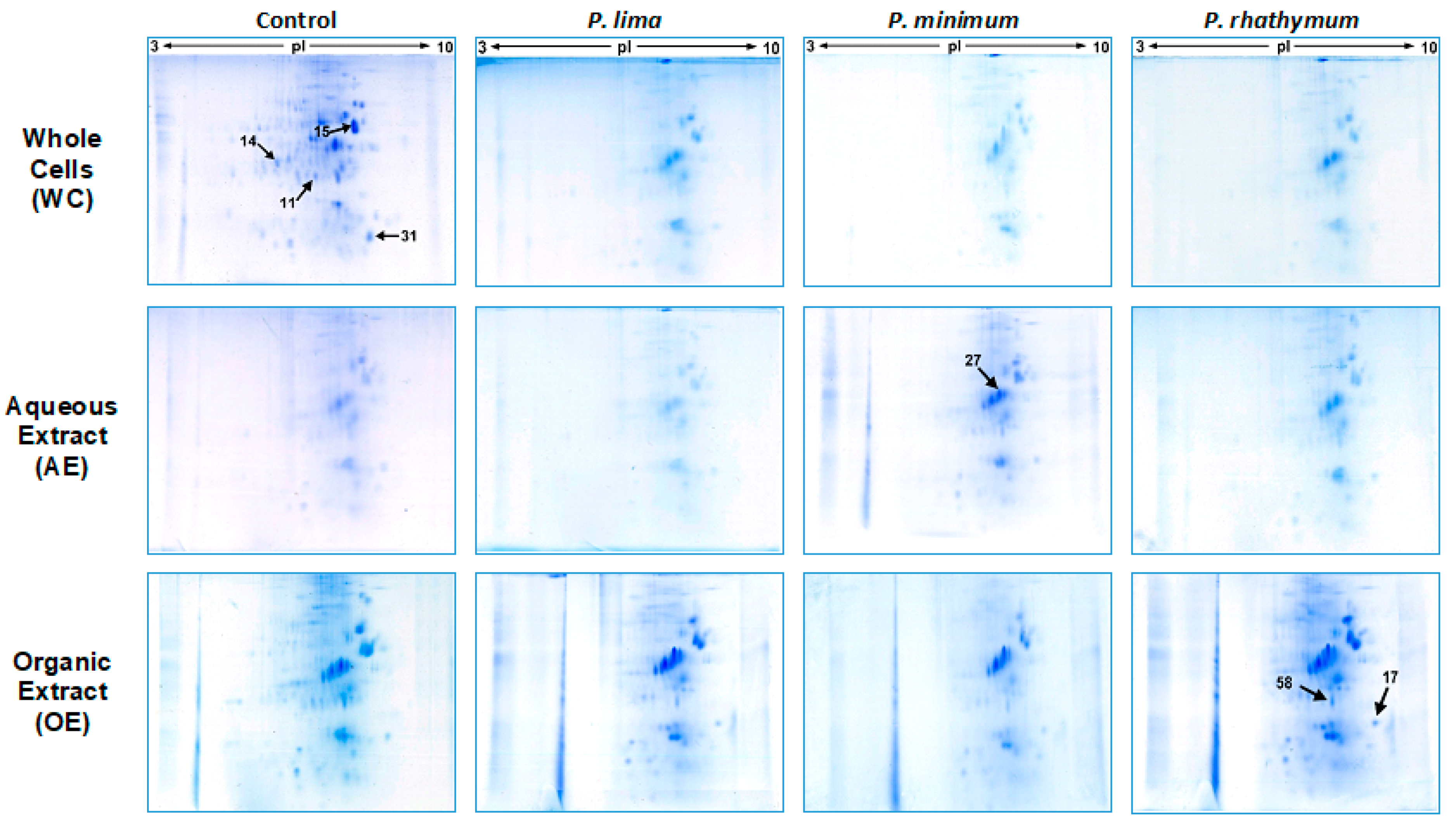
Considering the results obtained in the mortality assay, organisms exposed to 6000 cells/mL and extract concentrations of 20 ppt from each dinoflagellate species were used for the proteomic analyses. In this manner, 12 protein extracts were prepared by homogenization of 20 spat for each treatment (in triplicate), obtaining samples with around 3 mg/mL of total soluble proteins (in average); proteomic maps (2D) of soluble proteins from all assays are shown in Figure 3. In all cases, 2D maps were similar among triplicates showing a high reproducibility of two-dimensional gel electrophoresis (2D-GE) technique in this study.
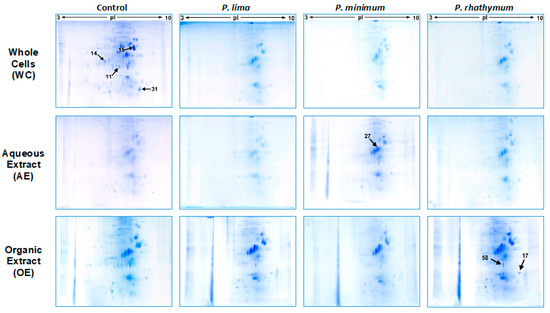
Figure 3.
Representative changes on 2D-GE protein patterns of C. gigas spat in response to exposure to whole cells (WC), aqueous (AE), and organic (OE) extracts of P. lima, P. minimum, or P. rhathymum; I. galbana was used as the control. For whole cell assays, 6000 cells/mL of each species were used. For exposure to extract assays, 20 parts per thousand (ppt) were used on each case, and AE = Distilled water and OE = 1% Tween 60 were used as controls.
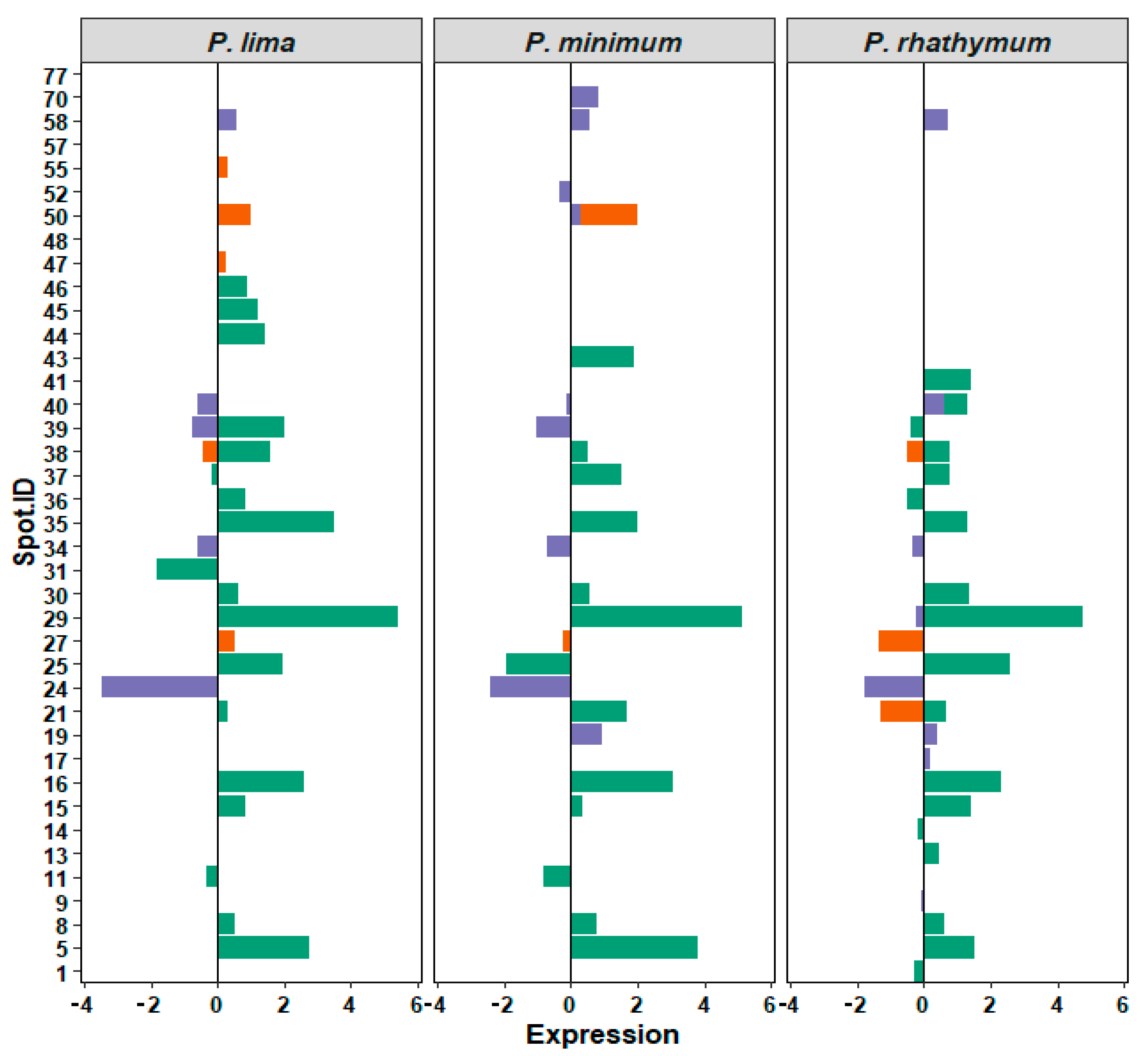
Analyzing 2D-GE images of the treatment groups with respect to the control, a pattern of 434 protein spots were observed in treatments with whole cells, distributed in a pH range from 5 to 8 with mass (Mr) values from 200 to 20 kDa. From these spots (Table 2), the statistical analysis revealed significant differences (p < 0.05) in intensity levels of 26 protein spots when the control was compared with each Prorocentrum species (Figure 3, Table S4). When oysters were exposed to P. lima, upregulation of 14 protein spots as well as downregulation, suppression, and induction of three protein spots for each case were observed. P. minimum cells promoted the upregulation of ten protein spots, the downregulation and suppression of five protein spots, and the induction of one protein spot. Finally, C. gigas exposure to P. rhathymum cells promoted the upregulation of 13 protein spots, the downregulation of four spots, the suppression of three spots, and the induction of two (Table 2 and Figure 4).

Table 2.
Number of protein spots by treatment detected on total protein extracts obtained from C. gigas exposed to whole cells (WC), aqueous extract (AE), or organic extract (OE) of three Prorocentrum species. Resolution technique 2D-GE; software Melanie 7.0.
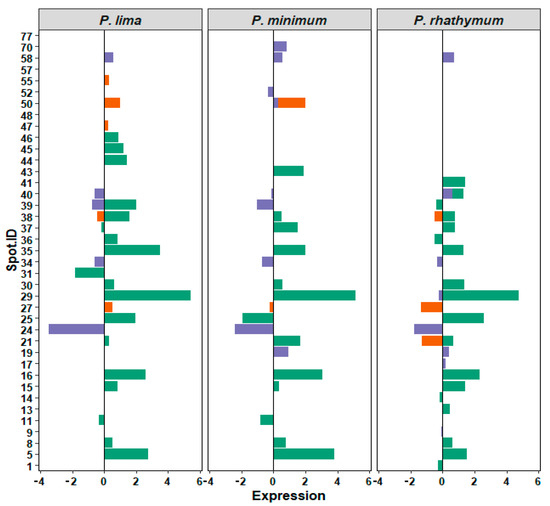
Figure 4.
Relative expression of 39 regulated protein spots identified by 2D-GE image analyses. Bars corresponds to treatments (green WC, whole cells; orange AE, aqueous extract; and violet OE, organic extract). X axis: relative expression on arbitrary units. Zero means no changes with respect to control, and signs, positive or negative, denotes overexpression or subexpression, respectively. Y axis, ID (identification number) of each spot. Panels correspond to species: P. lima, P. minimum, or P. rhathymum.
In the case of oysters exposed to AE (20 ppt), the analyses of 2D-GE images showed a pattern of 476 protein spots distributed in a pH range from 6 to 8 (Figure 3). From the 476 spots, the statistical analysis revealed significant differences (p < 0.05) in seven spots (Figure 4 and Table S4). Under these exposure conditions, the suppressed number of protein spots was higher than any other exposure condition, mainly in extract presence of P. minimum (4 spots), followed by P. rhathymum and P. lima (two proteins spots, respectively). Downregulation was observed on the protein 2D-GE patterns of organisms exposed to P. rhathymum and P. lima extracts (three and one protein spots, respectively), while upregulated proteins were observed with P. lima (two) and P. minimum (one). Finally, induced proteins appeared only in extracts from oysters exposed to P. lima (two) and P. minimum (one) (Table 2 and Figure 4).
In the case of OE (20 ppt), the comparison between 2D-GE protein patterns showed that C. gigas obtained from the control and extract treatments revealed a pattern of 623 protein spots, distributed in a pH range from 6 to 8 (Figure 3). The statistical analysis revealed significant differences (p < 0.05) in 13 of the 623 protein spots (Figure 4 and Table S4). In this experiment, the highest number of downregulated protein spots was observed in the presence of P. minimum extracts (six), followed by P. rhathymum and P. lima (four for each one). The upregulation of three protein spots was observed when extracts of P. rhathymum or P. minimum were used, and only one with P. lima. The suppressed proteins were six with the P. lima extract and three with P. rhathymum and P. minimum. Finally, the induced proteins were two with P. minimum, and one each with P. lima or P. rhathymum species (Table 2 and Figure 4).
2.3. Protein Identification
A total of seven isolated protein spots were successfully identified, four from assays of C. gigas exposure to whole cells of three Prorocentrum species, one from exposure to AE, and two from assays of exposure to OE (Table 3 and Table S5). Multiple proteins were identified in some spots: three in spots 14 and 58 and two in spots 11 and 27.

Table 3.
Identification of differentially expressed proteins in response to exposure to whole cells (6000 cells/mL) and aqueous (AE) or organic (OE) extracts, 20 parts per thousand (ppt) of P. lima, P. minimum, and P. rhathymum.
3. Discussion
Currently, a wide variety of chemical, biological, and molecular techniques and methods have been developed that can be used efficiently for the study of marine toxins; nevertheless, their toxic potential can only be adequately assessed by bioassays with living organisms, which provide a measure of the total toxicity based on the organisms’ biological response to toxins [43]. Bioassays with bivalves have been commonly used in recent decades to determine the physiological and behavioral exposure responses to organisms producing algal proliferation [11,12]. The direct exposure of bivalves to whole cells of algal proliferating organisms is one of the most widely used bioassays to assess toxicity effect. As cells enter the mollusks through a natural feeding process, they represent the natural exposure scenario, and the observations obtained reflect the effects caused by an algal proliferation scenario more reliably.
In regard to oyster mortality, no significant differences among treatments and controls were observed. In the bioassay with whole cells, the results suggest that cell number does not influence mortality on each species. However, when the results were compared among Prorocentrum species, significant differences were observed between P. minimum regarding P. lima and P. rhathymum (Figure 1 and Figure 2). These results could be explained by the ability of mollusks to classify and select their food, where the main characteristics involved are particle size and shape. In addition to their chemical and/or biochemical characteristics, size and shape played a main role in this research study because C. gigas spat fed mainly small cells of phytoplankton, while larger cells had lower or moderate ingestion rates [44]. The cell size used in this research was 18–20 μm long and 15–16 μm wide in P. minimum, 32–50 μm long and 20–28 μm wide in P. lima [45], and 31–36 µm long and 17–21 µm wide in P. rhathymum [46].
Likewise, this ability to classify and select food could be one of the causes of pseudo-feces production observed, especially in organisms fed with P. lima and P. rhathymum where larger cells were rejected by the gills and lip palps, or covered with mucus to later be removed from the inhalation chamber in the form of pseudo-feces. These results were consistent with previous studies carried out in juveniles of C. gigas where exposure to P. lima cells [47,48] as well as to a mixture of P. lima and G. catenatum cells [49,50] modified the feeding behavior of C. gigas, showing a low filtration rate and the formation of pseudo-feces, mainly made up of intact P. lima cells, favoring the intake of G. catenatum.
On the other hand, laboratory studies have demonstrated that bivalves were capable of feeding on toxic phytoplankton, generating various effects. Pearce et al. [51] fed C. gigas (4 mm) with P. rhathymum under conditions that simulated harmful algal blooms (104 cells/mL) for 21 days without recording mortality. Their results differ from those obtained in this investigation, where the death of 80% of the organisms exposed to 6 × 103 cells/mL of P. rhathymum was observed with a direct relationship between the number of dinoflagellate cells and mortality. This relationship agrees with the results obtained by de las Heras [52] when C. gigas (3–5 mm) was exposed to this species. However, de las Heras [52] recorded 100% mortality in organisms exposed to 4.8 × 103 and 6 × 103 cells/mL and in the most concentrated extracts (20) of the three dinoflagellate species after 65 h exposure. The differences in mortality between de las Heras’ work [52] and the results in this study could be explained through the analysis of the organisms used. In this research, C. gigas spat in the bioassays were slightly larger (5–7 mm), which indirectly provides them with greater resistance to this type of exposure by decreasing the mass/mass ratio (dinoflagellate/oyster) or volume/mass (extract/oyster), which finally has a dilution effect. In addition to the aforementioned, the potential effects of different species of harmful algae and their toxins can vary in bivalves depending on a series of species-specific or individual responses [53].
The impact of phycotoxins in bivalves can be grouped into behavioral, pathological, genetic, genomic, or proteomic effects [54]. The study of proteomics effects in the early stages of development plays an important role because the proteomic changes that can occur in the early life stages of organisms emphasize the importance of their development and survival in changing environments, such as coastal environments [28].
The proteins involved in immune system were Toll-interacting protein isoform X4 (Tollip) and Cathepsin L1, identified in C. gigas exposed to whole cells. Tollip negatively controlled the Toll-like receptor (TLR) signaling pathways with which TLR-mediated activation of innate immunity controls not only host defense against pathogens but also immune disorders [55]. Based on this fact, the organisms exposed to P. lima and P. minimum by decreasing Tollip expression should cease to control the TLR receptors, experiencing an overactivation of the immune system—a mechanism reported by de las Heras [52] when an increase in hemocyte concentration was observed. On the other hand, the organisms exposed to P. rhathymum inhibit the signaling cascades activated by TLR when they showed Tollip upregulation. Thus, the immune system defenses do not work, making them prone to attacks by pathogens and causing organism death, whereas the presence of cathepsin L1 on bivalve mollusk has been demonstrated in previous studies and their expression pattern exhibited an increase after they were exposed to various treatments [56,57,58,59,60]. In this study, Ctsl1 was suppressed in the presence of P. lima and P. minimum, showing downregulation in the presence of P. rhathymum. This result indicates that Ctsl1 found in this study could have been involved in some defense mechanisms against pathogens or in response to non-own particle recognition. The suppression and downregulation of this protein indicates that exposure to organisms of the genus Prorocentrum could affect the first line of immune defense, leaving organisms vulnerable to pathogen attack, thereby causing death.
The proteins associated with the stress response identified in this study have been investigated very little in mollusks. Corporeau et al. [61] reported the downregulation of Dihydropteridine reductase in C. gigas caused by stress due to herpesvirus OsHV-1 infection, suggesting a possible effect on endothelial cells. This effect could occur in organisms challenged by whole cells of P. lima and P. minimum, where they did not counteract stress when downregulation was recorded, showing higher mortality (83.33%). The main studies carried out in periostin have reported that this protein is differentially expressed in various tissues. Luo et al. [62] reported downregulation in the gonads of male oysters Crassostrea angulata exposed to Bisphenol-A. Huan et al. [63] proposed that this protein was related to development and growth because the periostin sequences showed a different expression pattern among different larval stages of the clam Meretrix meretrix. Payton et al. [64] described an upregulation in the periostin genes when C. gigas was exposed to Alexandrium minutum—a species known to produce paralytic shellfish toxin and harmful algal blooms. The opposite effect was recorded in this study, where the proteomic analysis revealed a suppression when C. gigas was challenged by P. lima and P. minimum and a downregulation of this protein after exposure to P. rhathymum. Despite these studies, no direct evidence has been provided on the role of periostin in mollusks. However, these results suggest that it could be involved in some regulatory mechanism for environmental stress.
During C. gigas challenge by organisms and extracts of the genus Prorocentrum, the differentially expressed proteins identified and involved in muscle contraction and cytoskeleton morphology were Actin cytoplasmic, Myosin essential light chain, Tropomyosin isoform 2, and Troponin C. The expressions of these proteins suggest that biological processes, such as cytoskeletal activities, contractile cell functions, and muscle contraction, in oysters is affected, mainly in the abductor muscle where valve closure is affected. Similar results have been observed in previous studies in some bivalve species when challenged to different contaminants, where proteomic studies have reported the upregulation of actin, isoforms of tropomyosin, and light chain myosin on Chamelea gallina when exposed to Aroclor 1254 and copper(II) and the downregulation of actin in the presence of tributyltin and arsenic (III) exposure [32]. In the same way, Thompson et al. [65,66] found that proteins associated with cytoskeleton activity (actin, actin-2, tropomyosin, and myosin) were affected in Saccostrea glomerata by exposure to copper (Cu) and lead (Pb), and with the latter, tropomyosin concentration increased fourfold, affecting motility and cellular plasticity of hemocytes. Troponin C, the calcium receptor subunit [67], is still unclear in mollusks; some studies on TnC have been performed on Patinopecten yessoensis [68,69], Ruditapes philippinarum [70], and C. gigas [71], associating it with physiological functions, mainly in muscle tissues as an important part in muscle formation and contraction. However, studies carried out by Funabara et al. [72,73] in Pinctada fucata indicate that TnC genes are mainly expressed in adductor phasic muscles and rarely in adductor catch muscles, gills, mantles, and feet. The previous suggests that TnC may not have a role in catching muscle contraction; thus, the physiological roles of Tn in mollusks somehow may have undergone some divergence from vertebrates.
4. Conclusions
The dinoflagellate species P. lima, P. minimum, and P. rhathymum produce toxic compounds capable of causing mortalities and affectations at the proteome level in C. gigas spat, where exposure time was the factor that influenced mortality the most.
After 72 h of exposure, the biological functions of proteins affected by exposure to whole cells in C. gigas were the immune system, stress response, contractile systems, and cytoskeletal activities, while exposure to organic and aqueous extracts mainly affected proteins involved in muscle contraction and cytoskeleton morphology.
In general, this research demonstrates the applicability of proteomics in the study of oysters exposed to toxic organisms of Prorocentrum genus. Thus, the way these toxins act is mainly by affecting the expression of proteins that are responsible for the contractile systems.
The information generated in this study enriches the knowledge on early bivalve developmental stages. It also provides an approach for new bioassays and the generation of new, more specific analytical tools that allow for the main effects of algal proliferation on the early stages of bivalve mollusks to be monitored, characterized, and elucidated.
5. Materials and Methods
5.1. Biological Material
The Japanese oyster C. gigas, 5–7 mm in shell length, were purchased from the “Acuacultura Robles SPR de RI” farm; upon arrival to the laboratory, the oysters were placed in an acclimation tank with natural filtered seawater to a temperature of 21 °C, salinity 34 UPS, and a light regime (12 h/12 h L/D) for five days. The oysters were fed with I. galbana microalgae (6 × 103 cells/oyster/day).
The Prorocentrum species used in this study were obtained from the collection of marine dinoflagellates of CIBNOR S.C. (CODIMAR [74]). P. lima (PLHV-1) was grown in f/2 medium [75], while P. rhathymum (PXPV-1) and P. minimum (PIPV-1; currently regarded as a synonym of Prorocentrum cordatum (Ostenfeld) J.D. Dodge [7]) were grown in modified GSe medium [76]. The cultures were grown up to the exponential phase in 2L Erlenmeyer flasks under controlled conditions of temperature and illumination (24 ± 1 °C; photoperiod 12:12 h light–dark with cold white fluorescent lights (irradiance 150 µE m−2 s−1). Cell density was determined by Sedgewick-Rafter camera counts in a microscope (OLYMPUS BX43 optical, Olympus Corp., Tokyo, Japan) using 20× and 40× magnification.
5.2. Extract Preparation
The toxin extracts were prepared using a modification of the Yasumoto method [77]. In brief, 50 mL of each dinoflagellate culture was centrifuged at 1500× g at 4 °C for 10 min. The total cell concentrations of each species were P. lima 2.95 × 106 cells, P. rhathymum 1.35 × 106 cells, and P. minimum 9.4 × 106 cells. The cell pellet was extracted with 10 volumes of 80% methanol; subsequently, a volume of dichloromethane was added. The obtained phases (aqueous, upper; organic, lower), were collected separately. All fractions were dried by vacuum distillation. The organic phase (OE) was dissolved in 5 mL of 1% Tween 60 (Sigma Chem. Co., St. Louis, MO, USA) and the aqueous phase (AE) in 5 mL of distilled water.
5.3. Exposure Experiments
All bioassays were performed in triplicate in 6-well plastic culture plates (BD Falcon Multi-well flat bottom sterile plates, Cat. 62406-155). Ten organisms of C. gigas were placed in each well, with 5 mL of diluted extracts or whole cells (depending on the case). On each cell, a complete treatment refill was carried out daily under identical conditions. Observations were made at 18, 42, 65, and 72 h of exposure; dead organisms were collected daily and stored at −80 °C until processing.
5.3.1. Exposure of C. gigas to Organic (OE) and Aqueous (AE) Extracts
C. gigas 5–7 mm in length were exposed separately to organic (OE) and aqueous (AE) extracts of each dinoflagellate species at concentrations of 20, 10, and 5 ppt. The controls consisted of a 1:50 dilution of 1% Tween 60 for OE and a 1:50 dilution of distilled water for AE, both on sterile seawater.
5.3.2. Exposure of C. gigas to Whole Live Cells of Prorocentrum spp.
The second experiment consisted of exposing/feeding oysters with whole cells of each tested species at densities of 1200 2400 3600 4800 and 6000 cells/mL. The control treatments consisted of 6000 cells/mL of fresh I. galbana culture.
5.4. Protein Extraction and Quantification
For protein extraction, ten individuals of C. gigas from each treatment were macerated (on ice bath) in 1.5 mL Eppendorf tubes with a glass pistil in 250 mL of cold phosphate buffer (4 °C; pH 7.8) containing 0.01% of a cocktail of protease inhibitors (PMSF SIGMA -ALDRICH, St. Louis, MO, USA) (in three replicates). After grinding for 5 min, samples were centrifuged at 8900 × g for 10 min at 4 °C. The supernatants obtained were recovered in new tubes, and total protein content was quantified by Lowry’s method [78]. Samples were kept at –80 °C until use for further analysis.
5.5. Proteomic Analysis
5.5.1. Isoelectric Focusing (IEF, First Dimension)
For the proteomic analysis, first-dimensional electrophoresis was performed on immobilized pH gradient (IPG) strips with a pH gradient of 3–10 (ReadyStrip®IPG 7 cm; #163 2000, Bio-Rad Laboratories, Hercules, CA, USA) following the manufacturer’s instructions. First, 500 µg of protein was dissolved in 600 µL of rehydration buffer (urea 8M, CHAPS 2%, DTT 50 mM, Bio-Lyte® Ampholites (Atlanta, GA, USA) 3–10 0.2%, and bromophenol blue 0.005% (w/v)). Then, the IPG strips were rehydrated at 20 °C for 14 h with 300 µL of the previous buffer–protein mixture. After rehydration, the strips were removed from the focus tray and the unabsorbed proteins were carefully removed. The IEF was performed using a PROTEAN® IEF cell (Cat. 165-4000, Bio-Rad Laboratories, Hercules, CA, USA) according to the following IEF parameters: 20 min on a gradient up to 250 V-h, 120 min on a gradient up to 4000 V-h, and 160 min on a gradient up to 10,000 V-h until a total of 14,000 V-h was reached at 20 °C in 5 h. Along the complete run, the current was limited to 25 µA per strip, and temperature was 20 °C to avoid overheating and protein degradation. Finally, the gel strips were removed and stored at 80 °C until use for the second electrophoretic dimension.
5.5.2. Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis (SDS-PAGE, Second Dimension)
After the first dimension, the IPG strips were equilibrated for 10 min (with gentle vertical shaking) on the equilibration buffer (urea 6 M, SDS 2% (w/v), Tris-HCl pH 8.8 0.375 M, glycerol 20% (v/v), and DTT 2% (w/v)). Then, a second buffer-equilibration was performed, but in this case, the DTT was replaced with iodoacetamide 2.5% (w/v) in the equilibrium buffer. Then, the IPG strip were placed on the polyacrylamide gel with an agarose overlay (12.5%). The electrophoresis was run on the Mini-protean II Cell (Cat. 165-2941, Bio-Rad Laboratories, Hercules, CA, USA) at a constant voltage of 100 V until the blue dye reached the bottom of the gel. The SDS-gels were stained for 3 h in a solution containing hydrated aluminum sulfate 5%, Coomassie Brilliant Blue G-250 0.2%, methanol 10%, and ortho-phosphoric acid 2% and de-stained using a solution containing 10% ethanol and 2% orthophosphoric acid.
5.5.3. Image Analysis and Spot Detection
The two-dimensional gels were digitized by using a transparency scanner (UMAX PowerLook 2100XL, UMAX Technologies, Taiwan) with 16-bit depth and a resolution of 600 dpi. The images were aligned, and spots were detected and quantified with the Melanie7.0 software using the automated algorithm. All detected spots were carefully checked manually, removing artifact spots; thus, the analyses were limited to points that showed statistically significant variations in expression among the groups (ANOVA < 0.05) and absolute fold change (FC > 1.5).
5.5.4. Identification of Differentially Expressed Proteins
After the differentially expressed proteins were detected (in quantitative terms), the selected proteins were manually picked from the gels and sent to the Laboratory of Biochemical and Instrumental Analysis of the Centro de Investigación y de Estudios Avanzados (CINVESTAV) del Instituto Politécnico Nacional (IPN) for MS analysis. The protein sequences obtained were identified by a BLAST-P search against the NCBInr sequence database, with an e-value cutoff of 1 × 10−3. The Kyoto encyclopedia of genes and genomes pathway database was used to identify in which metabolic pathways the differential expressed proteins were involved.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/toxins13070504/s1, Table S1: Summary of three-way ANOVA of C. gigas exposure to whole live cells of Prorocentrum spp., Table S2: Summary of three-way ANOVA of C. gigas exposure to the aqueous extract (OA) of Prorocentrum spp., Table S3: Summary of three-way ANOVA of C. gigas exposure to the organic extract (OE) of Prorocentrum spp., Table S4: Relative expression values of regulated protein spots that showed statistical differences (p < 0.005). Note: The processed data were obtained by subtracting the expression value of each spot with respect to the control value, Table S5: Peptide sequences of protein spots expressed differently and selected for identification by mass spectrometry.
Author Contributions
Conceptualization, N.Y.H.S.; methodology, M.A.M.H. and N.Y.H.S.; validation, M.A.M.H.; formal analysis, M.A.M.H. and N.Y.H.S.; investigation, M.A.M.H.; resources, N.Y.H.S.; data curation, M.A.M.H.; writing—original draft preparation, M.A.M.H.; writing—review and editing, N.Y.H.S.; visualization, M.A.M.H. and N.Y.H.S.; supervision, N.Y.H.S.; project administration, N.Y.H.S.; funding acquisition, N.Y.H.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by CONACyT, grant number CB08 83442, Molecular identification and study of the regulation of differential gene expression in the Pacific oyster Crassostrea gigas, in response to exposure to marine toxins and the APC was funded by the Fisheries Ecology Program of the CIBNOR.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sharma, A.; Gautam, S.; Kumar, S. Phycotoxins. In Encyclopedia of Food Microbiology, 2nd ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2014; pp. 25–29. ISBN 978-0-12384-733-1. [Google Scholar]
- Rossini, G.P.; Hess, P. Phycotoxins: Chemistry, mechanisms of action and shellfish poisoning. In Molecular, Clinical and Environmental Toxicology; Luch, A., Ed.; Birkhäuser: Basel, Switzerland, 2010; Volume 100, pp. 65–122. ISBN 978-3-7643-8338-1. [Google Scholar]
- Philip, F.S.; Val, R. Beasley Phycotoxins. In Handbook of Toxicologic Pathology; Wanda, M.H., Colin, G.R., Matthew, A.W., Eds.; Academic Press: Cambridge, MA, USA, 2002; Volume 1, pp. 631–643. ISBN 978-0-12-330215-1. [Google Scholar]
- Gerssen, A.; Pol-Hofstad, I.E.; Poelman, M.; Mulder, P.P.J.; Van Den Top, H.J.; De Boer, J. Marine Toxins: Chemistry, Toxicity, Occurrence and Detection, with Special Reference to the Dutch Situation. Toxins 2010, 2, 878–904. [Google Scholar] [CrossRef]
- Visciano, P.; Schirone, M.; Berti, M.; Milandri, A.; Tofalo, R.; Suzzi, G. Marine Biotoxins: Occurrence, Toxicity, Regulatory Limits and Reference Methods. Front. Microbiol. 2016, 7, 1051. [Google Scholar] [CrossRef] [PubMed]
- Wells, M.L.; Karlson, B.; Wulff, A.; Kudela, R.; Trick, C.; Asnaghi, V.; Berdalet, E.; Cochlan, W.; Davidson, K.; De Rijcke, M.; et al. Future HAB science: Directions and challenges in a changing climate. Harmful Algae 2020, 91. [Google Scholar] [CrossRef] [PubMed]
- Guiry, M.D.; Guiry, G.M. Algaebase. Available online: https://www.algaebase.org (accessed on 19 July 2020).
- Hoppenrath, M.; Chomérat, N.; Horiguchi, T.; Schweikert, M.; Nagahama, Y.; Murray, S. Taxonomy and phylogeny of the benthic Prorocentrum species (Dinophyceae)—A proposal and review. Harmful Algae 2013, 27, 1–28. [Google Scholar] [CrossRef]
- Verma, A.; Kazandjian, A.; Sarowar, C.; Harwood, D.T.; Murray, J.S.; Pargmann, I.; Hoppenrath, M.; Murray, S.A. Morphology and Phylogenetics of Benthic Prorocentrum Species (Dinophyceae) from Tropical Northwestern Australia. Toxins 2019, 11, 571. [Google Scholar] [CrossRef]
- Valdiglesias, V.; Prego-Faraldo, M.V.; Paśaro, E.; Meńdez, J.; Laffon, B. Okadaic Acid: More than a diarrheic toxin. Mar. Drugs 2013, 11, 4328–4349. [Google Scholar] [CrossRef] [PubMed]
- Hégaret, H.; Wikfors, G.H.; Shumway, S.E. Diverse Feeding Responses of Five Species of Bivalve Mollusc When Exposed to Three Species of Harmful Algae. J. Shellfish Res. 2007, 26, 549–559. [Google Scholar] [CrossRef]
- Wikfors, G.H.; Smolowitz, R.M. Experimental and Histological Studies of Four Life- History Stages of the Eastern Oyster, Crassostrea virginica, Exposed to a Cultured Strain of the Dinoflagellate Prorocentrum minimum. Biol. Bull. 1995, 188, 313–328. [Google Scholar] [CrossRef]
- Galimany, E.; Sunila, I.; Hégaret, H.; Ramón, M.; Wikfors, G.H. Pathology and immune response of the blue mussel (Mytilus edulis L.) after an exposure to the harmful dinoflagellate Prorocentrum minimum. Harmful Algae 2008, 7, 630–638. [Google Scholar] [CrossRef]
- Li, Y.; Sunila, I.; Wikfors, G.H. Bioactive effects of Prorocentrum minimum on juvenile bay scallops (Argopecten irradians irradians) are dependent upon algal physiological status. Bot. Mar. 2012, 55, 19–29. [Google Scholar] [CrossRef]
- Bauder, A.; Cembella, A.; Bricelj, V.; Quilliam, M. Uptake and fate of diarrhetic shellfish poisoning toxins from the dinoflagellate Prorocentrum lima in the bay scallop Argopecten irradians. Mar. Ecol. Prog. Ser. 2001, 213, 39–52. [Google Scholar] [CrossRef]
- Kameneva, P.A.; Krasheninina, E.A.; Slobodskova, V.V.; Kukla, S.P.; Orlova, T.Y. Accumulation and tissue distribution of dinophysitoxin-1 and dinophysitoxin-3 in the mussel crenomytilus grayanus feeding on the benthic dinoflagellate prorocentrum foraminosum. Mar. Drugs 2017, 15, 330. [Google Scholar] [CrossRef]
- Prado-Alvarez, M.; Flórez-Barrós, F.; Méndez, J.; Fernandez-Tajes, J. Effect of okadaic acid on carpet shell clam (Ruditapes decussatus) haemocytes by in vitro exposure and harmful algal bloom simulation assays. Cell Biol. Toxicol. 2013, 29, 189–197. [Google Scholar] [CrossRef]
- Huang, L.; Liu, S.L.; Zheng, J.W.; Li, H.Y.; Liu, J.S.; Yang, W.D. P-glycoprotein and its inducible expression in three bivalve species after exposure to Prorocentrum lima. Aquat. Toxicol. 2015, 169, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Dou, M.; Jiao, Y.H.; Zheng, J.W.; Zhang, G.; Li, H.Y.; Liu, J.S.; Yang, W.D. De novo transcriptome analysis of the mussel Perna viridis after exposure to the toxic dinoflagellate Prorocentrum lima. Ecotoxicol. Environ. Saf. 2020, 192, 110265. [Google Scholar] [CrossRef]
- Romero-Geraldo, R.d.J.; Hernández-Saavedra, N.Y. Stress Gene Expression in Crassostrea gigas (Thunberg, 1793) in response to experimental exposure to the toxic dinoflagellate Prorocentrum lima (Ehrenberg) Dodge, 1975. Aquac. Res. 2014, 45, 1512–1522. [Google Scholar] [CrossRef]
- Prego-Faraldo, M.V.; Martínez, L.; Méndez, J. RNA-Seq Analysis for Assessing the Early Response to DSP Toxins in Mytilus galloprovincialis Digestive Gland and Gill. Toxins 2018, 10, 417. [Google Scholar] [CrossRef]
- Prego-Faraldo, M.V.; Valdiglesias, V.; Laffon, B.; Mendez, J.; Eirin Lopez, J.M. Early genotoxic and cytotoxic effects of the toxic dinoflagellate Prorocentrum lima in the Mussel Mytilus galloprovincialis. Toxins 2016, 8, 159. [Google Scholar] [CrossRef] [PubMed]
- Svensson, S.; Förlin, L. Intracellular effects of okadaic acid in the blue mussel Mytilus edulis, and rainbow trout Oncorhynchus mykiss. Mar. Environ. Res. 1998, 46, 449–452. [Google Scholar] [CrossRef]
- Li, A.; Li, L.; Wang, W.; Zhang, G. Acetylome Analysis Reveals Population Differentiation of the Pacific Oyster Crassostrea gigas in Response to Heat Stress. Mar. Biotechnol. 2020, 22, 233–245. [Google Scholar] [CrossRef]
- De Lorgeril, J.; Zenagui, R.; Rosa, R.D.; Piquemal, D.; Bachère, E. Whole transcriptome profiling of successful immune response to vibrio infections in the oyster crassostrea gigas by digital gene expression analysis. PLoS ONE 2011, 6, e23142. [Google Scholar] [CrossRef]
- De Lorgeril, J.; Petton, B.; Lucasson, A.; Perez, V.; Stenger, P.L.; Dégremont, L.; Montagnani, C.; Escoubas, J.M.; Haffner, P.; Allienne, J.F.; et al. Differential basal expression of immune genes confers Crassostrea gigas resistance to Pacific oyster mortality syndrome. BMC Genom. 2020, 21, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, G.; Fang, X.; Guo, X.; Li, L.; Luo, R.; Xu, F.; Yang, P.; Zhang, L.; Wang, X.; et al. The oyster genome reveals stress adaptation and complexity of shell formation. Nature 2012, 490, 49–54. [Google Scholar] [CrossRef]
- Tomanek, L. Environmental Proteomics: Changes in the Proteome of Marine Organisms in Response to Environmental Stress, Pollutants, Infection, Symbiosis, and Development. Ann. Rev. Mar. Sci. 2011, 3, 373–399. [Google Scholar] [CrossRef]
- Olsson, B.; Bradley, B.P.; Gilek, M.; Reimer, O.; Shepard, J.L.; Tedengren, M. Physiological and proteomic responses in Mytilus edulis exposed to PCBs and PAHs extracted from Baltic Sea sediments. Hydrobiologia 2004, 514, 15–27. [Google Scholar] [CrossRef]
- Shepard, J.L.; Bradley, B.P. Protein expression signatures and lysosomal stability in Mytilus edulis exposed to graded copper concentrations. Mar. Environ. Res. 2000, 50, 457–463. [Google Scholar] [CrossRef]
- Shepard, J.L.; Olsson, B.; Tedengren, M.; Bradley, B.P. Protein expression signatures identified in Mytilus edulis exposed to PCBs, copper and salinity stress. Mar. Environ. Res. 2000, 50, 337–340. [Google Scholar] [CrossRef]
- Rodríguez-Ortega, M.J.; Grøsvik, B.E.; Rodríguez-Ariza, A.; Goksøyr, A.; López-Barea, J. Changes in protein expression profiles in bivalve molluscs (Chamaelea gallina) exposed to four model environmental pollutants. Proteomics 2003, 3, 1535–1543. [Google Scholar] [CrossRef]
- Wei, L.; Wang, Q.; Wu, H.; Ji, C.; Zhao, J. Proteomic and metabolomic responses of Pacific oyster Crassostrea gigas to elevated pCO2 exposure. J. Proteom. 2015, 112, 83–94. [Google Scholar] [CrossRef]
- Campos, A.; Tedesco, S.; Vasconcelos, V.; Cristobal, S. Proteomic research in bivalves: Towards the identification of molecular markers of aquatic pollution. J. Proteom. 2012, 75, 4346–4359. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Wang, D.-Z.; Wang, W.-X. Cadmium-induced changes in trace element bioaccumulation and proteomics perspective in four marine bivalves. Environ. Toxicol. Chem. 2012, 31, 1292–1300. [Google Scholar] [CrossRef]
- Meng, J.; Wang, W.; Li, L.; Yin, Q.; Zhang, G. Cadmium effects on DNA and protein metabolism in oyster (Crassostrea gigas) revealed by proteomic analyses. Sci. Rep. 2017, 7, 1–16. [Google Scholar] [CrossRef]
- Miserazzi, A.; Perrigault, M.; Sow, M.; Gelber, C.; Ciret, P.; Lomenech, A.M.; Dalens, J.M.; Weber, C.; Le Floch, S.; Lacroix, C.; et al. Proteome changes in muscles, ganglia, and gills in Corbicula fluminea clams exposed to crude oil: Relationship with behavioural disturbances. Aquat. Toxicol. 2020, 223, 105482. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, M.; Wang, W.; Liu, Z.; Xu, J.; Jia, Z.; Chen, H.; Qiu, L.; Lv, Z.; Wang, L.; et al. Transcriptional changes of Pacific oyster Crassostrea gigas reveal essential role of calcium signal pathway in response to CO2-driven acidification. Sci. Total Environ. 2020, 741, 140177. [Google Scholar] [CrossRef]
- Dineshram, R.; Quan, Q.; Sharma, R.; Chandramouli, K.; Yalamanchili, H.K.; Chu, I.; Thiyagarajan, V. Comparative and quantitative proteomics reveal the adaptive strategies of oyster larvae to ocean acidification. Proteomics 2015, 15, 4120–4134. [Google Scholar] [CrossRef] [PubMed]
- Cao, R.; Wang, Q.; Yang, D.; Liu, Y.; Ran, W.; Qu, Y.; Wu, H.; Cong, M.; Li, F.; Ji, C.; et al. CO2-induced ocean acidification impairs the immune function of the Pacific oyster against Vibrio splendidus challenge: An integrated study from a cellular and proteomic perspective. Sci. Total Environ. 2018, 625, 1574–1583. [Google Scholar] [CrossRef] [PubMed]
- Dineshram, R.; Wong, K.K.W.W.; Xiao, S.; Yu, Z.; Qian, P.Y.; Thiyagarajan, V. Analysis of Pacific oyster larval proteome and its response to high-CO2. Mar. Pollut. Bull. 2012, 64, 2160–2167. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Zou, Y.; Weng, H.W.; Li, H.Y.; Liu, J.S.; Yang, W.D. Proteomic profile in Perna viridis after exposed to Prorocentrum lima, a dinoflagellate producing DSP toxins. Environ. Pollut. 2015, 196, 350–357. [Google Scholar] [CrossRef]
- Fernández, M.L.; Richard, D.J.A.; Cembella, A.D. In vivo assays for phycotoxins. In Manual on Harmful Marine Microalgae; Hallegraeff, G.M., Anderson, D.M., Cembella, A.D., Enevoldsen, H.O., Eds.; UNESCO: Paris, France, 2003; pp. 297–346. ISBN 978-9-23103-871-6. [Google Scholar]
- Blanchard, M.; Pechenik, J.A.; Giudicelli, E.; Connan, J.P.; Robert, R. Competition for food in the larvae of two marine molluscs, Crepidula fornicata and Crassostrea gigas. Aquat. Living Resour. 2008, 21, 197–205. [Google Scholar] [CrossRef][Green Version]
- Dodge, J.D. Marine Dinoflagellates of the British Isles; Dodge, J.D., Ed.; Govt. Bookshops: London, UK, 1982; Volume 13, ISBN 011-2411-967. [Google Scholar]
- Hernández-Becerril, D.U.; Cortés Altamirano, R.; Alonso, R.R. The dinoflagellate genus Prorocentrum along the coasts of the Mexican Pacific. Hydrobiologia 2000, 418, 111–121. [Google Scholar] [CrossRef]
- De Jesuś Romero-Geraldo, R.; Garciá-Lagunas, N.; Hernańdez-Saavedra, N.Y. Effects of in vitro exposure to diarrheic toxin producer Prorocentrum lima on gene expressions related to cell cycle regulation and immune response in Crassostrea gigas. PLoS ONE 2014, 9, e97181. [Google Scholar] [CrossRef]
- Romero-Geraldo, R.d.J.; García-Lagunas, N.; Hernández-Saavedra, N.Y. Crassostrea gigas exposure to the dinoflagellate Prorocentrum lima: Histological and gene expression effects on the digestive gland. Mar. Environ. Res. 2016, 120, 93–102. [Google Scholar] [CrossRef]
- García-Lagunas, N.; Romero-Geraldo, R.; Kao-Godinez, A.K.; Hernández-Saavedra, N.Y. Differential expression of immune response genes in Pacific oyster, Crassostrea gigas spat, fed with dinoflagellates Gymnodinium catenatum and Prorocentrum lima. Lat. Am. J. Aquat. Res. 2019, 47, 699–705. [Google Scholar] [CrossRef]
- Cuevas, A.L. Efecto de la Exposición de Juveniles del Ostión Japonés Crassostrea Gigas (Thunberg, 1793) a Dinoflagelados Tóxicos Productores de Toxinas Dsp y Psp; Centro de Investigaciones Biológicas del Noroeste, S.C.: La Paz, Mexico, 2013. [Google Scholar]
- Pearce, I.; Handlinger, J.H.; Hallegraeff, G.M. Histopathology in Pacific oyster (Crassostrea gigas) spat caused by the dinoflagellate Prorocentrum rhathymum. Harmful Algae 2005, 4, 61–74. [Google Scholar] [CrossRef]
- De las Heras, S.S. Efecto de Dinoflagelados Tóxicos del Género Prorocentrum (Dinophyceae) en el Ostión Japonés Crassostrea Gigas (Thunberg, 1793); Universidad Autónoma De Baja California Sur: La Paz, Mexico, 2009. [Google Scholar]
- Landsberg, J.H. The Effects of Harmful Algal Blooms on Aquatic Organisms. Rev. Fish. Sci. 2002, 10, 113–390. [Google Scholar] [CrossRef]
- Fernández Robledo, J.A.; Yadavalli, R.; Allam, B.; Pales Espinosa, E.; Gerdol, M.; Greco, S.; Stevick, R.J.; Gómez-Chiarri, M.; Zhang, Y.; Heil, C.A.; et al. From the raw bar to the bench: Bivalves as models for human health. Dev. Comp. Immunol. 2019, 92, 260–282. [Google Scholar] [CrossRef] [PubMed]
- Takeda, K.; Akira, S. Toll-like receptors in innate immunity. Int. Immunol. 2005, 17, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Roberts, S.; Goetz, G.; White, S.; Goetz, F. Analysis of genes isolated from plated hemocytes of the Pacific oyster, Crassostreas gigas. Mar. Biotechnol. 2009, 11, 24–44. [Google Scholar] [CrossRef] [PubMed]
- Gueguen, Y.; Cadoret, J.P.; Flament, D.; Barreau-Roumiguière, C.; Girardot, A.L.; Garnier, J.; Hoareau, A.; Bachère, E.; Escoubas, J.M. Immune gene discovery by expressed sequence tags generated from hemocytes of the bacteria-challenged oyster, Crassostrea gigas. Gene 2003, 303, 139–145. [Google Scholar] [CrossRef]
- Ma, J.; Zhang, D.; Jiang, J.; Cui, S.; Pu, H.; Jiang, S. Molecular characterization and expression analysis of cathepsin L1 cysteine protease from pearl oyster Pinctada fucata. Fish Shellfish Immunol. 2010, 29, 501–507. [Google Scholar] [CrossRef] [PubMed]
- Lv, Z.; Qiu, L.; Liu, Z.; Wang, W.; Chen, H.; Jia, Y.; Jia, Z.; Jiang, S.; Wang, L.; Song, L. Molecular characterization of a cathepsin L1 highly expressed in phagocytes of pacific oyster Crassostrea gigas. Dev. Comp. Immunol. 2018, 89, 152–162. [Google Scholar] [CrossRef] [PubMed]
- Venier, P.; De Pittà, C.; Pallavicini, A.; Marsano, F.; Varotto, L.; Romualdi, C.; Dondero, F.; Viarengo, A.; Lanfranchi, G. Development of mussel mRNA profiling: Can gene expression trends reveal coastal water pollution? Mutat. Res. Fundam. Mol. Mech. Mutagen. 2006, 602, 121–134. [Google Scholar] [CrossRef] [PubMed]
- Corporeau, C.; Tamayo, D.; Pernet, F.; Quéré, C.; Madec, S. Proteomic signatures of the oyster metabolic response to herpesvirus OsHV-1 μVar infection. J. Proteom. 2014, 109, 176–187. [Google Scholar] [CrossRef]
- Luo, L.; Zhang, Q.; Kong, X.; Huang, H.; Ke, C. Differential effects of bisphenol A toxicity on oyster (Crassostrea angulata) gonads as revealed by label-free quantitative proteomics. Chemosphere 2017, 176, 305–314. [Google Scholar] [CrossRef]
- Huan, P.; Wang, H.; Liu, B. Transcriptomic Analysis of the Clam Meretrix meretrix on Different Larval Stages. Mar. Biotechnol. 2012, 14, 69–78. [Google Scholar] [CrossRef]
- Payton, L.; Perrigault, M.; Hoede, C.; Massabuau, J.C.; Sow, M.; Huvet, A.; Boullot, F.; Fabioux, C.; Hegaret, H.; Tran, D. Remodeling of the cycling transcriptome of the oyster Crassostrea gigas by the harmful algae Alexandrium minutum. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef]
- Thompson, E.L.; Taylor, D.A.; Nair, S.V.; Birch, G.; Haynes, P.A.; Raftos, D.A. A proteomic analysis of the effects of metal contamination on Sydney Rock Oyster (Saccostrea glomerata) haemolymph. Aquat. Toxicol. 2011, 103, 241–249. [Google Scholar] [CrossRef]
- Thompson, E.L.; Taylor, D.A.; Nair, S.V.; Birch, G.; Haynes, P.A.; Raftos, D.A. Proteomic discovery of biomarkers of metal contamination in Sydney Rock oysters (Saccostrea glomerata). Aquat. Toxicol. 2012, 109, 202–212. [Google Scholar] [CrossRef]
- Cao, T.; Thongam, U.; Jin, J.P. Invertebrate troponin: Insights into the evolution and regulation of striated muscle contraction. Arch. Biochem. Biophys. 2019, 666, 40–45. [Google Scholar] [CrossRef]
- Nishita, K.; Tanaka, H.; Ojima, T. Amino Acid Sequence of Troponin C from Scallop Striated Adductor Muscle. J. Biol. Chem. 1994, 269, 3464–3468. [Google Scholar] [CrossRef]
- Yuasa, H.J.; Takagi, T. The genomic structure of the scallop, Patinopecten yessoensis, troponin C gene: A hypothesis for the evolution of troponin C. Gene 2000, 245, 275–281. [Google Scholar] [CrossRef]
- Umasuthan, N.; Elvitigala, D.A.S.; Revathy, K.S.; Lee, Y.; Whang, I.; Park, M.-A.; Lee, J. Identification and in silico analysis of a novel troponin C like gene from Ruditapes philippinarum (Bivalvia: Veneridae) and its transcriptional response for calcium challenge. Gene 2012. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.Y.; Choi, Y.H. Regulation of adductor muscle growth by the IGF-1/AKT pathway in the triploid Pacific oyster, Crassostrea gigas. Fish. Aquat. Sci. 2019, 22, 19. [Google Scholar] [CrossRef]
- Funabara, D.; Ishikawa, D.; Urakawa, Y.; Kanoh, S. Ca2+-Induced Conformational Change of Troponin C from the Japanese Pearl Oyster, Pinctada fucata. Am. J. Mol. Biol. 2018, 8, 205–214. [Google Scholar] [CrossRef][Green Version]
- Funabara, D.; Urakawa, Y.; Kanoh, S. Molecular Cloning and Tissue Distribution of Troponin C from the Japanese Pearl Oyster, Pinctada fucata. Am. J. Mol. Biol. 2018, 8, 166–177. [Google Scholar] [CrossRef][Green Version]
- Morquecho, L. Listado de Cepas. Available online: https://www.cibnor.gob.mx/investigacion/colecciones-biologicas/codimar/listado-de-cepas (accessed on 30 June 2021).
- Guillard, R.R.; Ryther, J.H. Studies of marine planktonic diatoms. I. Cyclotella nana Hustedt, and Detonula confervacea (cleve) Gran. Can. J. Microbiol. 1962, 8, 229–239. [Google Scholar] [CrossRef]
- Arredondo, V.B.O.; Band, C.J.S. Aislamiento Mantenimiento y Cultivo de Microalgas Nocivas. Manual de Curso de Cultivos; Centro de Investigaciones Biologicas de Noroeste: La Paz, Mexico, 2000. [Google Scholar]
- Lagos, N. Principales toxinas de origenfitoplanctónico: Identificación y cuantificaciónmediante cromatografía líquida de alta resolucion (HPLC). In Floraciones Algales Nocivas en el Cono Sur Américano; Sar, E.A., Ferrario, M.E., Reguera, B., Eds.; Instituto Español de Oceanografía: Madrid, Spain, 2002; Volume 1, pp. 57–72. ISBN 84-95877-01-5. [Google Scholar]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).