Abstract
Background: In humans, vitamin D3 synthesis follows a day–night rhythm due to its UV-B-dependent production. Results: As part of the VitDHiD intervention study, we identified 87 in vivo vitamin D target genes with circadian expression patterns in immune cells, forming a regulatory network centered on transcription factors and membrane receptors. These genes exhibit a narrow basal expression range, with 80% downregulated upon vitamin D3 supplementation. Clustering analysis revealed six distinct gene groups, with the two most prominent clusters driven by the transcription factor CSRNP1 (cysteine- and serine-rich nuclear protein 1) and GAS7 (growth arrest-specific 7), a known differentiation inducer. Among the 25 VitDHiD study participants, we identified two subgroups distinguished by significant differences in the responsiveness of 14 in vivo vitamin D target genes. These genes encode transcription factors like CSRNP1, as well as metabolic enzymes and transporters, including NAMPT (nicotinamide phosphoribosyltransferase), PFKFB3 (6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3), and SLC2A3 (solute carrier family 2 member 3). Notably, all 14 genes possess a vitamin D receptor-binding enhancer within a reasonable distance of their transcription start site. Conclusions: These findings highlight a novel link between vitamin D signaling and circadian gene regulation, with potential implications for personalized supplementation strategies.
1. Introduction
Vitamin D3 is traditionally classified as a vitamin, yet humans can endogenously synthesize it through UV-B-induced conversion of 7-dehydrocholesterol in the skin [1,2]. However, approximately 50,000 years ago, the migration of Homo sapiens from the consistently sunlit regions of Africa to areas in Europe and Asia with significantly lower UV exposure transformed this molecule into a vital nutrient for populations in those latitudes [3,4,5,6].
The role of vitamin D in calcium regulation has been critical since vertebrate species transitioned from aquatic to terrestrial environments around 380 million years ago, requiring stable skeletal structures [7]. Consequently, a hallmark of vitamin D deficiency is the development of bone abnormalities [8]. Beyond skeletal health, vitamin D is also integral to the regulation of both innate and adaptive immune systems [9]. Maintaining sufficient vitamin D levels has been associated with reduced risks of various conditions, including musculoskeletal disorders such as osteoporosis [10] and sarcopenia [11], autoimmune diseases like multiple sclerosis [12], and certain cancers, such as colorectal cancer [13]. Nevertheless, large-scale interventional studies, involving thousands of participants over periods of up to five years, have struggled to provide statistically significant evidence for primary health benefits of vitamin D3 supplementation [14,15]. Despite lack of health benefits for the general population, in the DO-HEALTH study, which focused on healthy elderly individuals, participants aged 70–74 years had significantly lower infection rates, suggesting a potential effect of vitamin D3 supplementation on the immune system [16].
The molecular mechanisms of vitamin D are centered on its nuclear receptor, VDR (vitamin D receptor), which exhibits an exceptionally high binding affinity (KD = 0.1 nM) for the active vitamin D metabolite, 1α,25-dihydroxyvitamin D3 (1,25(OH)2D3) [17,18]. In the endocrine system, this active form is primarily synthesized in the kidneys from 25-hydroxyvitamin D3; however, it can also be produced locally in immune and skin cells for autocrine and paracrine signaling [19]. As a transcription factor, VDR governs all genomic actions of vitamin D, with 1,25(OH)2D3 being its sole physiological activator [20]. VDR is widely expressed across most tissues, with the exception of the brain, and regulates hundreds of genes in a tissue- and individual-specific manner [15]. This variability in gene responsiveness forms the basis for the vitamin D response index, which classifies individuals as high, mid, or low responders to vitamin D supplementation [21]. This heterogeneity in response might contribute to the difficulty of finding health benefits for vitamin D3 supplementation in studies targeting the general population.
To explore the molecular effects of vitamin D in vivo under healthy conditions, studies such as VitDbol (NCT02063334) [22,23] and VitDHiD (NCT03537027) [24,25] were conducted with cohorts in Kuopio, Finland, who received in total 80,000 IU of vitamin D3 administered in two equal doses along with breakfast and lunch, which is equivalent to a monthly dose. The PER1 (period circadian regulator 1) gene, a key regulator of circadian rhythms, emerged as a prominent in vivo vitamin D target in the VitDHiD study. Given that vitamin D3 synthesis in the skin naturally follows a day–night rhythm, we sought to explore whether vitamin D also influences the expression of circadian genes. While the impact of vitamin D on immune function is well established, its potential role in modulating circadian gene expression remains largely unexplored. This study seeks to bridge this gap by identifying circadian vitamin D target genes and exploring interindividual variability in responsiveness.
This study presents a secondary analysis of the VitDHiD trial, focusing on 7665 genes with stable expression profiles in peripheral blood mononuclear cells (PBMCs) from 25 study participants. Of the 361 in vivo vitamin D target genes, 87 exhibit a circadian expression pattern. This suggests that fine-tuning the circadian rhythm of gene expression is a previously underexplored physiological function of vitamin D.
2. Materials and Methods
2.1. Transcriptome Data Sources
The VitDHiD trial investigated transcriptome changes in PBMCs from 25 healthy individuals (age 20–54 years, 12 females and 13 males, body mass index 21.4–25.6) supplemented with a single vitamin D3 bolus (80,000 IU) over one day [25]. Our analysis focused on 7665 genes with stable expression levels exceeding 10 counts per million (CPM) across all participants (Table S1). Among these, 361 genes exhibited significant changes in expression (FDR (false discovery rate) ≤ 0.05) when comparing baseline levels to 24 h post-supplementation with vitamin D3. A comprehensive literature and database search for transcriptome datasets describing circadian gene expression in vivo in human blood cells identified only one relevant study. This report analyzed the circadian expression profile of the whole blood transcriptome of 26 participants undergoing a 1-week sleep restriction study controlled with 1-week of sufficient sleep; after each condition, blood was sampled every 3 h for 30 h to detect genes with a circadian rhythm. We used the results from the control group in our analysis [26]. Additionally, seasonally expressed genes were identified based on a 4-year profiling of PBMCs from 105 individuals in California [27]. To ensure consistency and adherence to current nomenclature, gene symbols across all datasets were updated using either the HGNC Multi-Symbol Checker [28] (www.genenames.org/tools/multi-symbol-checker, accessed on 22 November 2024) when only gene symbols were available or g:Profiler’s [29] (https://biit.cs.ut.ee/gprofiler/convert, accessed on 22 November 2024) gene ID conversion for ENSG identifiers. Outdated symbols were replaced with their current approved equivalents, while aliases were excluded to avoid potential errors caused by genes sharing similar aliases, which could introduce false overlap during analysis. If no matching gene symbol was available, or if it was withdrawn, the entry was removed from the list. After this update, the circadian gene list contained 1461 genes, while the seasonal gene list included 757 genes. Within the 7665 commonly expressed genes, 873 and 363 members of these genes were found to be expressed (Table S1).
2.2. Differential Gene Expression Analysis
The raw count data from the VitDHiD study [25] were used in this analysis. Differential gene expression analysis was conducted in R (version 4.4.1) on MacOS 15 (Sequoia, Menlo Park, CA, USA) using EdgeR [30] (version 4.4.2). The analysis focused on 19,824 expressed protein-coding genes to minimize transcriptional noise from non-coding or lowly expressed genes. Read counts were normalized to CPM to account for differences in library size. To remove genes that have insufficient read counts for reliable statistical inference, genes with very low expression were filtered out using the FilterByExpr() function and removing all genes with a CPM ≤ 10. After filtering, library sizes were recomputed, and trimmed mean of M-values (TMM) normalization was applied. Differential expression between clusters was assessed using the generalized linear model (GLM) quasi-likelihood pipeline available in EdgeR, as described by its developer [31].
2.3. Characterization of Target Genes
The STRING database (http://string-db.org, accessed on 22 January 2025, version 12.0) [32] was employed to confirm and visualize the physical and functional interactions among circadian and vitamin D target genes. In the resulting network, each node represents a protein, while edges between nodes indicate associations derived from experimental data, computational predictions, and literature sources. Functional annotations of the genes, with a focus on their primary biological roles, were obtained from resources such as the Human Protein Atlas (www.proteinatlas.org, accessed on 14 December 2024) [33] and GeneCards (www.genecards.org, accessed on 14 December 2024) [34].
To further investigate gene relationships, hierarchical clustering was performed on the Pearson correlation matrix using correlation distance as the metric and complete linkage as the clustering method; the optimal amount of correlation clusters was determined by the silhouette method. For the heatmap visualization of the log2FC (fold change) across individuals, k-means clustering was applied to partition individuals into two groups, while hierarchical clustering was performed on the genes. The resulting correlation matrix and clustering results were visualized as a heatmap using the pheatmap R package (version 1.0.12). GeneMANIA (https://genemania.org, accessed on 2 March 2025) [35] was used to see which genes had previously reported coexpressions.
2.4. Analysis of Genomic Regions of Vitamin D Target Genes
To investigate regulatory elements in key in vivo vitamin D target genes, genomic regions were analyzed to identify VDR-binding enhancers and TSS (transcription start site) regions. This analysis leveraged epigenome-wide data from THP-1 cells stimulated with either 10 nM 1,25(OH)2D3 or a solvent control (0.1% ethanol) for 2 and 24 h. ChIP-seq (chromatin immunoprecipitation sequencing) datasets [36] were used to pinpoint VDR-binding sites, while FAIRE-seq (formaldehyde-assisted isolation of regulatory elements followed by sequencing) data [37] provided insights into chromatin accessibility, aiding in the identification of active regulatory regions. Visualization of these datasets was performed using the IGV browser [38], allowing the examination of VDR-binding enhancers and TSS regions within accessible chromatin that respond to 1,25(OH)2D3. These regulatory elements were assessed within TAD (topologically associating domain) regions of vitamin D target genes. While a broad genomic window of ±1 Mb around each target gene’s TSS was initially screened, only the most relevant regions are reported in this analysis.
2.5. Statistical Tests
To assess the significance of potential overlap among the VitDHiD (361 genes), circadian (873 genes), and seasonal (363 genes) gene sets, a hypergeometric distribution test was performed using R. Additionally, to evaluate statistically significant correlations (p ≤ 0.05) in the expression of circadian and vitamin D target genes, a Pearson correlation analysis was conducted, applying the Benjamini–Hochberg correction for multiple testing. To identify significant differences in gene expression between the two clusters of individuals, a t-test was performed, followed by the Benjamini–Hochberg correction for multiple testing.
3. Results
3.1. Circadian Expression Profile of In Vivo Vitamin D Target Genes
A transcriptomic dataset from the vitamin D3 intervention study VitDHiD, which involved 25 healthy individuals supplemented with 80,000 IU of vitamin D3 over 24 h, was analyzed as a reference for identifying in vivo vitamin D target genes in PBMCs [25]. After filtering for genes with a minimum expression of CPM ≥ 10 across all individuals, 7665 genes were classified as commonly expressed. Of these, 361 genes were significantly responsive to vitamin D3 supplementation (FDR ≤ 0.05) (Table S1).
Among the 7665 commonly expressed genes, 873 were found in the curated and updated list of 1462 circadian genes in the whole blood transcriptome of 26 participants in the control arm of a sleep study [26]. Additionally, 363 of 757 seasonally expressed genes, identified from the curated and updated PBMC transcriptome of 105 individuals [27], overlapped with the 7665 genes (Table S1). Notably, 87 and 18 of the 361 vitamin D target genes exhibited circadian and seasonal expression patterns, respectively (Figure 1A). Furthermore, the list of circadian and seasonal genes overlapped in 37 genes, 6 of which were also vitamin D targets: CREB5 (CAMP responsive element binding protein 5), DYSF (dysferlin), PER1, SIK3 (SIK family kinase 3), SLC8A1, and WDFY3 (WD repeat and FYVE domain containing 3).
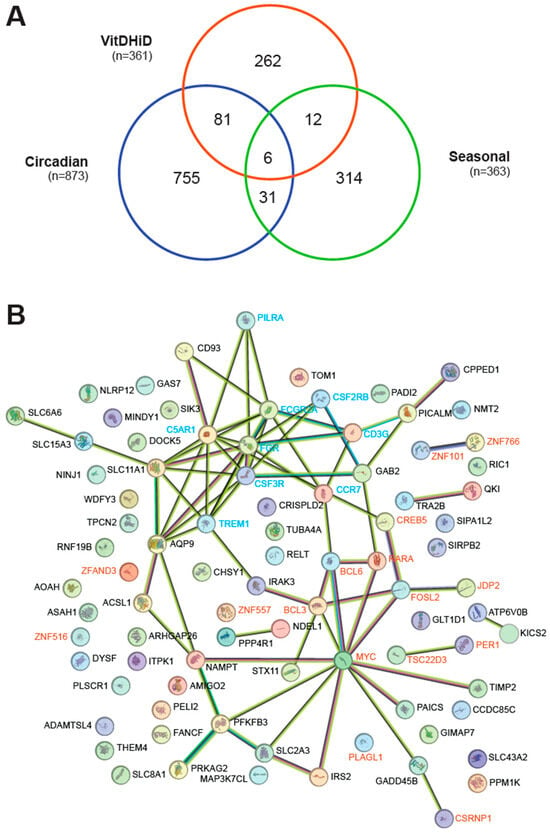
Figure 1.
Circadian expression profile of in vivo vitamin D target genes. (A) A Venn diagram illustrating the overlap between in vivo vitamin D target genes, circadian genes, and seasonal genes. (B) Gene/protein interaction network for the 87 circadian genes identified as in vivo vitamin D targets, based on STRING database analysis. Nodes represent proteins, while edges denote both functional and physical associations. Edge thickness indicates the strength of supporting data. Transcription factors and regulators are highlighted in red, while membrane receptor genes are shown in blue.
Proteins encoded by the 87 in vivo vitamin D target genes exhibiting circadian behavior were analyzed using the STRING database [32]. Approximately half of these proteins are part of a network centered around the transcription factors BCL6 (BCL6 transcription coactivator), CSRNP1, CREB5, FOSL2 (FOS-like 2, AP1 transcription factor subunit), JDP2 (Jun dimerization protein 2), MYC (MYC proto-oncogene, BHLH transcription factor), and RARA (retinoic acid receptor alpha) as well as the transcriptional cofactor BCL3 (Figure 1B). Additionally, the transcriptional cofactors PER1 and TSC22D3 (TSC22 domain family member 3), along with the transcription factors ZNF101 (zinc finger protein 101) and ZNF766, were also found to be connected within this network. Another network centered on the membrane receptor proteins C5AR1 (complement C5a receptor 1), CCR7 (C-C motif chemokine receptor 7), CD3G (CD3 gamma subunit of T-cell receptor complex), CSF2RB (colony stimulating factor 2 receptor subunit beta), CSF3R (colony stimulating factor 3 receptor), FCGR2A (Fc gamma receptor IIa), FGR (FGR proto-oncogene, Src family tyrosine kinase), PILRA (paired immunoglobin-like type 2 receptor alpha), and TREM1 (triggering receptor expressed on myeloid cells 1). In contrast, the analysis of the 18 proteins encoded by vitamin D target genes that exhibit seasonal variation revealed no convincing network using STRING. Only two transcription factor proteins, CREB5 and KLF11 (KLF transcription factor 11), showed a connection (Figure S1). Furthermore, a hypergeometric test indicated that the identification of 18 seasonal genes out of 363 may have occurred by chance (p = 0.4233), whereas the 87 vitamin D target genes among the 873 circadian genes were highly significant (p = 1.1976 × 10−12). Interestingly, 34 of the 87 in vivo vitamin D target genes were also identified in recently published in vitro studies, in which human PBMCs had been stimulated for 24 h with 1,25(OH)2D3 [24,39,40] (Table S1). These genes include ADAMTSL4 (ADAMTS-like 4), AQP9 (aquaporin 9), BCL3, C5AR1, CD93 (CD93 molecule), CPPED1 (calcineurin-like phosphoesterase domain containing 1), CREB5, CRISPLD2 (cysteine-rich secretory protein LCCL domain containing 2), CSF2RB, CSF3R, DYSF, FCGR2A, FOSL2, GAS7, IRAK3 (interleukin 1 receptor-associated kinase 3), JDP2, MAP3K7CL (MAP3K7 C-terminal like), NAMPT, NLRP12 (NLR family pyrin domain containing 12), PADI2 (peptidyl arginine deiminase 2), PILRA, PLAGL1, RNF19B (ring finger protein 19B), SIPA1L2 (signal-induced proliferation-associated 1 like 2), SIRPB2 (signal regulatory protein beta 2), SLC11A1, SLC15A3, SLC43A2, SLC8A1, STX11 (syntaxin 11), TIMP2 (TIMP metallopeptidase inhibitor 2), TREM1, WDFY3, and ZNF516.
In summary, 87 in vivo vitamin D target genes display a circadian rhythm. These target genes encode proteins that form a network, with transcription factors and membrane receptors at its core.
3.2. Characterization of Vitamin D Targets with Circadian Behavior
The 87 in vivo vitamin D target genes exhibiting circadian expression profiles demonstrate a rather narrow range of basal expression levels, with AMIGO2 (adhesion molecule with Ig-like domain 2) being only 35-fold less expressed than TSC22D3 (Figure S2A, Table S1). Of these genes, only 17 are upregulated by vitamin D3 supplementation, with MAP3K7CL showing the highest induction, while 70 are downregulated, with JDP2 being the most repressed (Figure S2B).
Hierarchical clustering of the 87 vitamin D target genes for coexpression identified six distinct gene clusters, each comprising 7 to 16 members with positively correlated expression patterns (Pearson correlation padj ≤ 0.05) (Figure 2). Among these, cluster 1 is the most densely connected. The core regulatory hub of this cluster appears to be the CSRNP1 gene encoding for a transcription factor, whose expression strongly correlates with that of the transcription (co)factors BCL3 and FOSL2, the innate immune receptor C5AR1, the carbohydrate metabolism regulators PFKFB3 and SLC2A3, and the cellular growth and apoptosis regulator GADD45B (growth arrest and DNA damage inducible beta). Cluster 5, in contrast, exhibits a different functional profile. This cluster is primarily driven by the GAS7 gene encoding a differentiation-promoting protein, whose expression correlates with the cytokine receptor CSF3R, the adhesion protein NINJ1 (ninjurin 1), the ion channel TPCN2 (two-pore segment channel 2), the calcium ion sensor DYSF, the cellular trafficking regulator TOM1 (target of Myb1 membrane trafficking protein), and the inflammation-modulating protein NLRP12. The coexpression results for cluster 1, as verified by GeneMANIA, indicate that CSRNP1 is coexpressed with all genes except C5AR1. However, C5AR1 is indirectly linked to CSRNP1 via SLC2A3 and GADD45B. The GAS7 gene does not exhibit any connections with other genes, whereas CSF3R is coexpressed with the genes TOM1, NINJ1, DYSF, and NLRP12. Additionally, the genes in cluster 1 are represented in the STRING network, where each gene has at least one connection, except for C5AR1, in contrast to cluster 5, where no connections are observed.
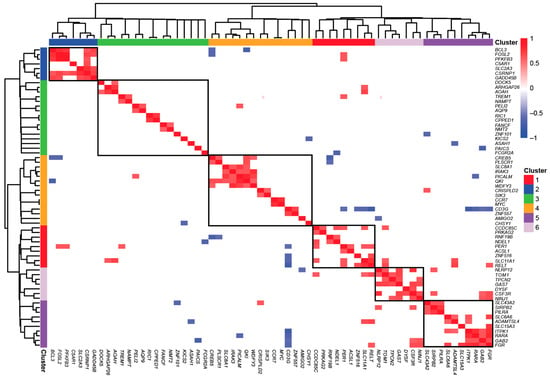
Figure 2.
Characterization of vitamin D targets with circadian behavior. Cluster analysis on the Pearson correlation of the 87 circadian genes identified as in vivo vitamin D targets revealed 6 distinct gene clusters.
Taken together, the 87 circadian genes display a relatively narrow range of basal expression, with 80% being downregulated by vitamin D3 supplementation. Among the 6 identified gene clusters, cluster 1 and cluster 5 stand out, driven respectively by the transcription factor CSRNP1 and the differentiation inducer GAS7, emphasizing their distinct functional roles.
3.3. Individual-Specific Responses of Circadian Vitamin D Target Genes
Expression analysis of 87 in vivo circadian vitamin D target genes before and after vitamin D3 supplementation distinguished the 25 participants of the VitDHiD trial into two groups (Figure 3). Group 1, consisting of 9 individuals, showed significantly higher expression (padj ≤ 0.05) in 14 genes compared to the 16 participants in group 2. These genes were NAMPT, PFKFB3, BCL3, FOSL2, CSRNP2, GADD45B, C5AR1, CREB5, NDEL1 (NudE neurodevelopment protein 1 like 1), RNF19B, SLC2A3, SIRPB2, PILRA, and MAP3K7CL. Differential gene expression analysis revealed no significant differences in gene expression across these clusters prior to vitamin D3 supplementation. However, after supplementation, four genes (CSRNP2, FOSL2, NDEL1, and PFKFB3) were found to be differentially expressed between the two groups (FDR ≤ 0.05, Table S2).
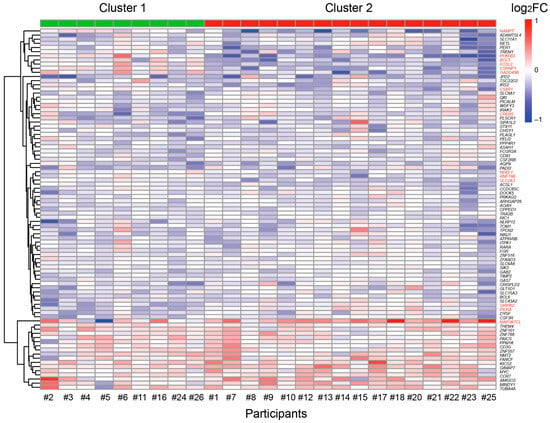
Figure 3.
Individual-specific responses of circadian vitamin D target genes. Hierarchical clustering is based on the log2FC of individual-specific responses of 87 circadian vitamin D target genes. K-means clustering suggested a distinction among the 25 VitDHiD participants, classifying them into two groups (Cluster 1 and Cluster 2). Genes driving the significant differences between these groups are highlighted in red.
The genomic regions within ±1 Mb of the TSS of these in total 14 in vivo vitamin D target genes were screened for experimentally validated VDR binding sites in accessible chromatin. This analysis utilized VDR ChIP-seq and FAIRE-seq datasets from THP-1 cells treated with either solvent or 10 nM 1,25(OH)2D3 for 2 and 24 h. At least one VDR-binding enhancer was identified for each of the 14 genes at varying distances from the TSS: 2 kb (NAMPT), 6 kb (BCL3), 14 kb (C5AR1), 16 kb (RNF19B), 27 kb (CSRNP2), 49 kb (FOSL2), 54 kb (SLC2A3), 55 kb (MAP3K7CL), 69 kb (GADD45B), 76 kb (SIRPB2), 140 kb (NDEL1), 146 kb (PFKFB3), 165 kb (PILRA), and 455 kb (CREB5) (Figure 4 and Figure S3). These enhancers were located within a range that suggests their potential role in regulating gene expression.
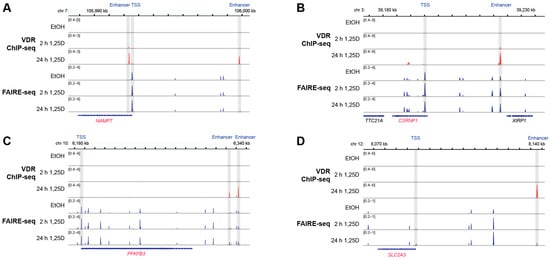
Figure 4.
VDR-binding enhancers in the genomic regions of four vitamin D target genes. The IGV browser was used to visualize ChIP-seq results for VDR (red) and FAIRE-seq data (blue) from THP-1 cells treated with solvent (0 h) or 1,25(OH)2D3 (1,25D) for 2 and 24 h. The genomic region of the vitamin D target genes NAMPT (A), CSRNP1 (B), PFKFB3 (C) and SLC2A3 (D) are displayed. TSS regions and VDR-binding enhancers are shaded in grey. Peak tracks represent merged data from three biological replicates. Gene structures are depicted in blue, with vitamin D target genes highlighted in red. While genomic regions spanning 1 Mb upstream and downstream of each gene’s TSS were analyzed, only areas relevant to 1,25(OH)2D3-dependent regulation are shown.
In summary, based on the expression of 14 out of 87 circadian in vivo vitamin D target genes, the 25 VitDHiD participants can be categorized into two groups of 9 and 16 individuals. Notably, all 14 genes possess VDR-binding enhancers within 2 to 455 kb of their TSS, suggesting a role for VDR-mediated regulation in the observed differential expression patterns.
4. Discussion
This study provides new insights into the circadian expression of vitamin D target genes in vivo, demonstrating a significant overlap between genes responsive to vitamin D3 supplementation and those exhibiting circadian rhythmicity. Our findings emphasize the complexity of vitamin D-mediated transcriptional regulation, particularly within PBMCs of healthy individuals. Using transcriptomic data from the VitDHiD vitamin D intervention study, we identified 87 in vivo vitamin D target genes exhibiting circadian expression patterns. Network analysis revealed that the proteins encoded by these 87 circadian vitamin D target genes are functionally interconnected, with transcription factors (e.g., BCL6, CREB5, FOSL2, JDP2, MYC, RARA) and membrane receptors (e.g., C5AR1, CCR7, CD3G, CSF2RB, CSF3R, FCGR2A, FGR, PILRA, TREM1) forming core regulatory hubs. This suggests that vitamin D signaling influences key transcriptional regulators and immune-related pathways in a time-dependent manner. The rhythmic nature of the immune system has been widely reported [41,42], encompassing cell-type composition, both innate and adaptive immune responses, and immune cell functions such as metabolism, migration, and phagocytic activity. Additionally, previous studies have demonstrated that measuring key clock genes in PBMCs allows individuals to be categorized into two chronotypes like morning types (“larks”) and evening types (“owls”), feeling more alert and productive in the morning or evening, respectively [43]. However, due to the design of the VitDHiD study, it is not possible to determine whether the observed clustering is attributable to chronotype differences. Regardless, the finding that vitamin D3 supplementation can stratify individuals into two distinct groups of 9 and 16 members, respectively, in a circadian manner is a noteworthy observation.
In contrast, seasonally regulated vitamin D target genes did not display a comparable functional network, reinforcing a stronger association between vitamin D and circadian biology rather than seasonal variation. Although seasonal changes, except close to the equator, are typically characterized by fluctuations in mean ambient temperature and sunlight hours [44], which would be expected to influence circadian rhythms, the overlap between circadian and seasonal genes remains surprisingly limited. Given that circadian entrainment primarily depends on light, with contributions from temperature and other factors, this limited overlap is intriguing from an evolutionary perspective. Modern humans (Homo sapiens) evolved in East Africa, where they were consistently exposed to sufficient UV-B radiation year-round, ensuring stable vitamin D3 synthesis and adaptation to a regular day–night cycle for over 200,000 years [45]. In contrast, populations migrating beyond 37° N only began experiencing seasonal variations in sunlight exposure and endogenous vitamin D3 production approximately 45,000 years ago [46]. Thus, there has been significantly less evolutionary time to adapt to seasonal variations compared to circadian rhythms.
Notably, the circadian vitamin D target genes also clustered into six coexpression groups, with two clusters, CSRNP1-driven and GAS7-driven, standing out due to their distinct regulatory functions. The CSRNP1 cluster includes genes involved in transcriptional regulation and metabolism [47]. In this cluster, C5AR1, which is as of now underreported in its relations to the other genes in the cluster, is emerging as a driver of complement-mediated regulation of immunometabolism through mitochondrial activity [48]. Given that CSRNP1 appears to drive this cluster, future research could explore whether CSRNP1 directly influences immunometabolism through transcriptional regulation. The GAS7 cluster is particularly intriguing, as the association among these genes appears underreported. However, when analyzing the functions of its constituents, it emerges as a promising target for further investigation. The GAS7 protein itself regulates actin assembly and membrane outgrowth [49], while TOM1, DYSF, and TPCN2 are linked to lysosomal function [50,51]. CSF3R, the only gene in the cluster with reported coexpression with all other genes, is well known for its central role in granulocyte differentiation. Additionally, NINJ1 plays a key role in membrane rupture during pyroptosis, a form of programmed cell death affecting granulocytes, while NLRP12 is involved in inflammasome formation and may contribute to pyroptosis through the PANosome [52]. Taken together, these findings suggest that the GAS7 cluster and its constituents could serve as valuable targets for further research into immune cell function. More broadly, these results indicate that vitamin D may exert temporally coordinated effects on diverse biological processes.
Interindividual variability in the response to vitamin D3 supplementation was another critical finding of this study. Based on differential gene expression analysis, the 25 VitDHiD participants were classified into two distinct groups, with 14 genes showing significantly higher expression in one group compared to the other. This variability suggests that individual-specific factors, such as genetic background, baseline vitamin D status, or circadian regulatory mechanisms, may influence the responsiveness of vitamin D target genes. Further genomic analysis revealed that all 14 genes distinguishing the two groups contained VDR-binding enhancers within 2 to 455 kb of their TSS, supporting the hypothesis that differential VDR-mediated regulation contributes to interindividual differences in gene expression. However, this aspect needs to be explored in more detail, since the in vitro THP-1 model may not reflect all VDR binding sites used in vivo.
The circadian in vivo vitamin D target genes, AQP9, ASCL1 (acyl-CoA synthetase long chain family member 1), IRS2 (insulin receptor substrate 2), NAMPT, PFKFB3, PRKAG2 (protein kinase AMP-activated non-catalytic subunit gamma 2), SLC2A3, and SLC6A6, encode proteins with crucial roles in energy metabolism and cellular function. ASCL1 contributes to fatty acid metabolism; AQP9 functions as a glycerol and urea transporter; IRS2 mediates the downstream effects of insulin signaling; NAMPT is essential for cellular respiration, as it plays a key role in NAD“ biosynthesis; PFKFB3 regulates glycolysis; PRKAG2 is a subunit of AMPK (AMP-activated protein kinase), the master regulator of metabolism with circadian crosstalk capabilities [53]; while SLC2A3 facilitates glucose transport and SLC6A6 taurine transport. These findings highlight vitamin D’s evolutionarily ancient role in regulating energy metabolism [54], while also emphasizing the significance of key metabolic products, NAD“, glucose, and fatty acids, in circadian biology [55]. In recent years, the circadian aspect of health, particularly its connection to metabolism, has gained increasing attention, notably in the context of the so-called circadian syndrome [56]. This condition links metabolic syndrome to disruptions in the circadian rhythm.
This study has several limitations. Most notably, the relatively small cohort size of the VitDHiD study (25 individuals) limits the statistical power to detect subtle differences between chronotype groups. The limited power may, in fact, suggest that additional, as-yet-undetected differences exist between the clusters studied. Furthermore, the participants were primarily of Finnish origin, representing one of the most genetically homogenous populations worldwide, which may limit the generalizability of the findings. Another limitation is that the VitDHiD study was not originally designed with a circadian focus, and no data on participants’ biorhythms were collected. Finally, although the use of a monthly vitamin D3 bolus appears effective for identifying in vivo vitamin D target genes, the results should be validated using a regimen of daily, moderate vitamin D3 supplementation.
5. Conclusions
Overall, our findings emphasize the circadian nature of vitamin D target gene regulation and highlight interindividual variability in the transcriptional response to vitamin D3 supplementation. These insights have important implications for personalized vitamin D3 supplementation strategies, suggesting that optimal dosing regimens may need to consider both circadian biology and individual responsiveness to vitamin D. For instance, vitamin D has been linked to improved sleep quality [57], indicating that one of its few well-established health benefits may have a circadian component. However, there is currently no consensus on whether vitamin D3 supplementation is best taken at night or during the day, nor definitive evidence that vitamin D acts as a Zeitgeber. A Zeitgeber (German for “time giver”) is any external cue that helps regulate the human body’s circadian rhythms, such as light, mealtime, physical activity, social interactions, noise, or temperature changes. Future studies should further investigate the molecular mechanisms underlying these differences, including potential genetic and epigenetic factors that influence VDR activity and circadian gene expression. Additionally, further research is needed to determine whether vitamin D has Zeitgeber properties and how it may contribute to circadian entrainment. In conclusion, our findings reveal a strong and previously underappreciated link between vitamin D signaling and circadian biology in human PBMCs. The identification of distinct gene expression clusters and interindividual differences in vitamin D response further emphasize the need for a personalized approach to vitamin D3 supplementation, potentially incorporating circadian timing as a factor in optimizing its biological effects.
Supplementary Materials
The following Supporting Information can be downloaded at: https://www.mdpi.com/article/10.3390/nu17071204/s1, Figure S1: Seasonal Vitamin D Target Genes; Figure S2: Basal Expression and Inducibility of Circadian Vitamin D Target Genes; Figure S3: VDR-Binding Enhancers in the Genomic Regions of Ten Vitamin D Target Genes; Table S1: Vitamin D3-Induced Transcriptome of PBMCs In Vivo; Table S2: Differential Gene Regulation Analysis.
Author Contributions
Data analysis was performed by P.M. P.M. provided the first draft of the manuscript text and figures, which was refined by C.C. All authors have read and agreed to the published version of the manuscript.
Funding
This work is part of NUTRIOME, which has received funding from the European Union’s Horizon Europe research and innovation program under grant agreement no. 101119497. In addition, this publication is part of a project that has received funding from the European Union’s Horizon 2020 research and innovation program under grant agreement no. 952601, from the David and Amy Fulton Foundation, Seattle, US, and from the National Science Centre (Poland), project number 2023/49/B/NZ9/00402.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Fastq files of the raw data of the VitDHiD study can be found at Gene Expression Omnibus (GEO) with accession number GSE260981.
Acknowledgments
We would like to thank Ranjini Ghosh Dastidar for providing the raw counts from the VitDHiD study and for conducting analyses related to our study but not included in the final manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| 1,25(OH)2D3 | 1α,25-dihydroxyvitamin D3 |
| AMIGO2 | adhesion molecule with Ig-like domain 2 |
| AMPK | AMP-activated protein kinase |
| AQP9 | aquaporin 9 |
| ASCL1 | acyl-CoA synthetase long chain family member 1 |
| BCL | BCL transcription coactivator |
| C5AR1 | complement C5a receptor 1 |
| CCR7 | C-C motif chemokine receptor 7 |
| CD3G | CD3 gamma subunit of T-cell receptor complex |
| CD93 | CD93 molecule |
| ChIP-seq | chromatin immunoprecipitation sequencing |
| CPM | counts per million |
| CPPED1 | calcineurin-like phosphoesterase domain containing 1 |
| CREB5 | CAMP responsive element binding protein 5 |
| CRISPLD2 | cysteine-rich secretory protein LCCL domain containing 2 |
| CSF2RB | colony stimulating factor 2 receptor subunit beta |
| CSF3R | colony stimulating factor 3 receptor |
| CSRNP1 | cysteine- and serine-rich nuclear protein 1 |
| DYSF | dysferlin |
| FAIRE-seq | formaldehyde-assisted isolation of regulatory elements followed by sequencing |
| FC | fold change |
| FCGR2A | Fc gamma receptor IIa |
| FDR | false discovery rate |
| FGR | FGR proto-oncogene, Src family tyrosine kinase |
| FOSL2 | FOS-like 2, AP1 transcription factor subunit |
| GADD45B | growth arrest and DNA damage inducible beta |
| GAS7 | growth arrest specific 7 |
| IRAK3 | interleukin 1 receptor-associated kinase 3 |
| IRS2 | insulin receptor substrate 2 |
| JDP2 | Jun dimerization protein 2 |
| KLF11 | KLF transcription factor 11 |
| MAP3K7CL | MAP3K7 C-terminal like |
| MYC | MYC proto-oncogene, BHLH transcription factor |
| NAMPT | nicotinamide phosphoribosyltransferase |
| NDEL1 | NudE neurodevelopment protein 1 like 1 |
| NINJ1 | ninjurin 1 |
| NLRP12 | NLR family pyrin domain containing 12 |
| PADI2 | peptidyl arginine deiminase |
| PBMC | peripheral blood mononuclear cell |
| PER1 | period circadian regulator 1 |
| PFKFB3 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| PILRA | paired immunoglobin-like type 2 receptor alpha |
| PRKAG2 | protein kinase AMP-activated non-catalytic subunit gamma 2 |
| RARA | retinoic acid receptor alpha |
| RNA-seq | RNA sequencing |
| RNF19B | ring finger protein 19B |
| SIK3 | SIK family kinase 3 |
| SIPA1L2 | signal-induced proliferation-associated 1 like 2 |
| SIRPB2 | signal regulatory protein beta 2 |
| SLC | solute carrier family |
| STX11 | syntaxin 11 |
| TAD | topologically associating domain |
| TIMP2 | TIMP metallopeptidase inhibitor 2 |
| TOM1 | target of Myb1 membrane trafficking protein |
| TPCN2 | two pore segment channel 2 |
| TREM1 | triggering receptor expressed on myeloid cells 1 |
| TSC22D3 | TSC22 domain family member 3 |
| TSS | transcription start site |
| VDR | vitamin D receptor |
| WDFY3 | WD repeat and FYVE domain containing 3 |
| ZNF | zinc finger protein |
References
- Holick, M.F.; MacLaughlin, J.A.; Doppelt, S.H. Regulation of cutaneous previtamin D3 photosynthesis in man: Skin pigment is not an essential regulator. Science 1981, 211, 590–593. [Google Scholar] [CrossRef] [PubMed]
- Holick, M.F. The cutaneous photosynthesis of previtamin D3: A unique photoendocrine system. J. Investig. Dermatol. 1981, 77, 51–58. [Google Scholar] [CrossRef]
- Hanel, A.; Carlberg, C. Skin color and vitamin D: An update. Exp. Dermatol. 2020, 29, 864–875. [Google Scholar] [CrossRef] [PubMed]
- Pagani, L.; Lawson, D.J.; Jagoda, E.; Morseburg, A.; Eriksson, A.; Mitt, M.; Clemente, F.; Hudjashov, G.; DeGiorgio, M.; Saag, L.; et al. Genomic analyses inform on migration events during the peopling of Eurasia. Nature 2016, 538, 238–242. [Google Scholar] [CrossRef] [PubMed]
- Skoglund, P.; Mathieson, I. Ancient genomics of modern humans: The first decade. Annu. Rev. Genom. Hum. Genet. 2018, 19, 381–404. [Google Scholar] [CrossRef]
- O’Neill, C.M.; Kazantzidis, A.; Ryan, M.J.; Barber, N.; Sempos, C.T.; Durazo-Arvizu, R.A.; Jorde, R.; Grimnes, G.; Eiriksdottir, G.; Gudnason, V.; et al. Seasonal changes in vitamin D-effective UVB availability in Europe and associations with population serum 25-hydroxyvitamin D. Nutrients 2016, 8, 533. [Google Scholar] [CrossRef]
- Bouillon, R.; Suda, T. Vitamin D: Calcium and bone homeostasis during evolution. BoneKEy Rep. 2014, 3, 480. [Google Scholar] [CrossRef]
- Levine, M.A. Diagnosis and management of vitamin D dependent rickets. Front. Pediatr. 2020, 8, 315. [Google Scholar] [CrossRef]
- Bouillon, R.; Carmeliet, G.; Lieben, L.; Watanabe, M.; Perino, A.; Auwerx, J.; Schoonjans, K.; Verstuyf, A. Vitamin D and energy homeostasis-of mice and men. Nat. Rev. Endocrinol. 2014, 10, 79–87. [Google Scholar] [CrossRef]
- Sunyecz, J.A. The use of calcium and vitamin D in the management of osteoporosis. Ther. Clin. Risk Manag. 2008, 4, 827–836. [Google Scholar] [CrossRef]
- Uchitomi, R.; Oyabu, M.; Kamei, Y. Vitamin D and sarcopenia: Potential of vitamin D supplementation in sarcopenia prevention and treatment. Nutrients 2020, 12, 3189. [Google Scholar] [CrossRef]
- Feige, J.; Moser, T.; Bieler, L.; Schwenker, K.; Hauer, L.; Sellner, J. Vitamin D supplementation in multiple sclerosis: A critical analysis of potentials and threats. Nutrients 2020, 12, 783. [Google Scholar] [CrossRef] [PubMed]
- Vaughan-Shaw, P.G.; Buijs, L.F.; Blackmur, J.P.; Theodoratou, E.; Zgaga, L.; Din, F.V.N.; Farrington, S.M.; Dunlop, M.G. The effect of vitamin D supplementation on survival in patients with colorectal cancer: Systematic review and meta-analysis of randomised controlled trials. Br. J. Cancer 2020, 123, 1705–1712. [Google Scholar] [CrossRef] [PubMed]
- Bouillon, R.; Manousaki, D.; Rosen, C.; Trajanoska, K.; Rivadeneira, F.; Richards, J.B. The health effects of vitamin D supplementation: Evidence from human studies. Nat. Rev. Endocrinol. 2022, 18, 96–110. [Google Scholar] [CrossRef] [PubMed]
- Gospodarska, E.; Ghosh Dastidar, R.; Carlberg, C. Intervention approaches in studying the response to vitamin D3 supplementation. Nutrients 2023, 15, 3382. [Google Scholar] [CrossRef]
- Bischoff-Ferrari, H.A.; Vellas, B.; Rizzoli, R.; Kressig, R.W.; da Silva, J.A.P.; Blauth, M.; Felson, D.T.; McCloskey, E.V.; Watzl, B.; Hofbauer, L.C.; et al. Effect of vitamin D supplementation, omega-3 fatty acid supplementation, or a strength-training exercise program on clinical outcomes in older adults: The DO-HEALTH randomized clinical trial. JAMA 2020, 324, 1855–1868. [Google Scholar] [CrossRef]
- Whitfield, G.K.; Dang, H.T.; Schluter, S.F.; Bernstein, R.M.; Bunag, T.; Manzon, L.A.; Hsieh, G.; Dominguez, C.E.; Youson, J.H.; Haussler, M.R.; et al. Cloning of a functional vitamin D receptor from the lamprey (Petromyzon marinus), an ancient vertebrate lacking a calcified skeleton and teeth. Endocrinology 2003, 144, 2704–2716. [Google Scholar] [CrossRef]
- Haussler, M.R.; Haussler, C.A.; Bartik, L.; Whitfield, G.K.; Hsieh, J.C.; Slater, S.; Jurutka, P.W. Vitamin D receptor: Molecular signaling and actions of nutritional ligands in disease prevention. Nutr. Rev. 2008, 66, S98–S112. [Google Scholar] [CrossRef]
- Chun, R.F.; Liu, P.T.; Modlin, R.L.; Adams, J.S.; Hewison, M. Impact of vitamin D on immune function: Lessons learned from genome-wide analysis. Front. Physiol. 2014, 5, 151. [Google Scholar] [CrossRef]
- Pike, J.W.; Meyer, M.B.; Lee, S.M.; Onal, M.; Benkusky, N.A. The vitamin D receptor: Contemporary genomic approaches reveal new basic and translational insights. J. Clin. Investig. 2017, 127, 1146–1154. [Google Scholar] [CrossRef]
- Carlberg, C.; Haq, A. The concept of the personal vitamin D response index. J. Steroid Biochem. Mol. Biol. 2018, 175, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Vukic, M.; Neme, A.; Seuter, S.; Saksa, N.; de Mello, V.D.; Nurmi, T.; Uusitupa, M.; Tuomainen, T.P.; Virtanen, J.K.; Carlberg, C. Relevance of vitamin D receptor target genes for monitoring the vitamin D responsiveness of primary human cells. PLoS ONE 2015, 10, e0124339. [Google Scholar] [CrossRef]
- Seuter, S.; Virtanen, J.K.; Nurmi, T.; Pihlajamäki, J.; Mursu, J.; Voutilainen, S.; Tuomainen, T.P.; Neme, A.; Carlberg, C. Molecular evaluation of vitamin D responsiveness of healthy young adults. J. Steroid Biochem. Mol. Biol. 2017, 174, 314–321. [Google Scholar] [CrossRef]
- Hanel, A.; Neme, A.; Malinen, M.; Hamalainen, E.; Malmberg, H.R.; Etheve, S.; Tuomainen, T.P.; Virtanen, J.K.; Bendik, I.; Carlberg, C. Common and personal target genes of the micronutrient vitamin D in primary immune cells from human peripheral blood. Sci. Rep. 2020, 10, 21051. [Google Scholar] [CrossRef]
- Ghosh Dastidar, R.; Jaroslawska, J.; Malinen, M.; Tuomainen, T.P.; Virtanen, J.K.; Bendik, I.; Carlberg, C. In vivo vitamin D targets reveal the upregulation of focal adhesion-related genes in primary immune cells of healthy individuals. Sci. Rep. 2024, 14, 17552. [Google Scholar] [CrossRef]
- Moller-Levet, C.S.; Archer, S.N.; Bucca, G.; Laing, E.E.; Slak, A.; Kabiljo, R.; Lo, J.C.; Santhi, N.; von Schantz, M.; Smith, C.P.; et al. Effects of insufficient sleep on circadian rhythmicity and expression amplitude of the human blood transcriptome. Proc. Natl. Acad. Sci. USA 2013, 110, E1132–E1141. [Google Scholar] [CrossRef]
- Sailani, M.R.; Metwally, A.A.; Zhou, W.; Rose, S.M.S.; Ahadi, S.; Contrepois, K.; Mishra, T.; Zhang, M.J.; Kidzinski, L.; Chu, T.J.; et al. Deep longitudinal multiomics profiling reveals two biological seasonal patterns in California. Nat. Commun. 2020, 11, 4933. [Google Scholar] [CrossRef]
- Seal, R.L.; Braschi, B.; Gray, K.; Jones, T.E.M.; Tweedie, S.; Haim-Vilmovsky, L.; Bruford, E.A. Genenames.org: The HGNC resources in 2023. Nucleic Acids Res. 2023, 51, D1003–D1009. [Google Scholar] [CrossRef]
- Kolberg, L.; Raudvere, U.; Kuzmin, I.; Adler, P.; Vilo, J.; Peterson, H. g:Profiler-interoperable web service for functional enrichment analysis and gene identifier mapping (2023 update). Nucleic Acids Res. 2023, 51, W207–W212. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Chen, Y.; Lun, A.T.; Smyth, G.K. From reads to genes to pathways: Differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000Research 2016, 5, 1438. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef] [PubMed]
- Sjostedt, E.; Zhong, W.; Fagerberg, L.; Karlsson, M.; Mitsios, N.; Adori, C.; Oksvold, P.; Edfors, F.; Limiszewska, A.; Hikmet, F.; et al. An atlas of the protein-coding genes in the human, pig, and mouse brain. Science 2020, 367, eaay5947. [Google Scholar] [CrossRef] [PubMed]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinform. 2016, 54, 1.30.1–1.30.33. [Google Scholar] [CrossRef]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010, 38, W214–W220. [Google Scholar] [CrossRef]
- Neme, A.; Seuter, S.; Carlberg, C. Selective regulation of biological processes by vitamin D based on the spatio-temporal cistrome of its receptor. Biochim. Biophys. Acta 2017, 1860, 952–961. [Google Scholar] [CrossRef]
- Seuter, S.; Neme, A.; Carlberg, C. Epigenome-wide effects of vitamin D and their impact on the transcriptome of human monocytes involve CTCF. Nucleic Acids Res. 2016, 44, 4090–4104. [Google Scholar] [CrossRef]
- Thorvaldsdottir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef]
- Hanel, A.; Carlberg, C. Time-resolved gene expression analysis monitors the regulation of inflammatory mediators and attenuation of adaptive immune response by vitamin D. Int. J. Mol. Sci. 2022, 23, 911. [Google Scholar] [CrossRef]
- Malmberg, H.R.; Hanel, A.; Taipale, M.; Heikkinen, S.; Carlberg, C. Vitamin D treatment sequence is critical for transcriptome modulation of immune challenged primary human cells. Front. Immunol. 2021, 12, 754056. [Google Scholar] [CrossRef]
- Scheiermann, C.; Kunisaki, Y.; Frenette, P.S. Circadian control of the immune system. Nat. Rev. Immunol. 2013, 13, 190–198. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Chen, P.; Qi, C. Circadian rhythm regulation in the immune system. Immunology 2024, 171, 525–533. [Google Scholar] [CrossRef] [PubMed]
- Teboul, M.; Barrat-Petit, M.A.; Li, X.M.; Claustrat, B.; Formento, J.L.; Delaunay, F.; Levi, F.; Milano, G. Atypical patterns of circadian clock gene expression in human peripheral blood mononuclear cells. J. Mol. Med. 2005, 83, 693–699. [Google Scholar] [CrossRef]
- Tahara, Y.; Aoyama, S.; Shibata, S. The mammalian circadian clock and its entrainment by stress and exercise. J. Physiol. Sci. 2017, 67, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Luxwolda, M.F.; Kuipers, R.S.; Kema, I.P.; Dijck-Brouwer, D.A.; Muskiet, F.A. Traditionally living populations in East Africa have a mean serum 25-hydroxyvitamin D concentration of 115 nmol/l. Br. J. Nutr. 2012, 108, 1557–1561. [Google Scholar] [CrossRef]
- Mylopotamitaki, D.; Weiss, M.; Fewlass, H.; Zavala, E.I.; Rougier, H.; Sumer, A.P.; Hajdinjak, M.; Smith, G.M.; Ruebens, K.; Sinet-Mathiot, V.; et al. Homo sapiens reached the higher latitudes of Europe by 45,000 years ago. Nature 2024, 626, 341–346. [Google Scholar] [CrossRef]
- Macdonald, C.D.; Falconer, A.M.D.; Chan, C.M.; Wilkinson, D.J.; Skelton, A.; Reynard, L.; Litherland, G.J.; Europe-Finner, G.N.; Rowan, A.D. Cytokine-induced cysteine-serine-rich nuclear protein-1 (CSRNP1) selectively contributes to MMP1 expression in human chondrocytes. PLoS ONE 2018, 13, e0207240. [Google Scholar] [CrossRef]
- Niyonzima, N.; Rahman, J.; Kunz, N.; West, E.E.; Freiwald, T.; Desai, J.V.; Merle, N.S.; Gidon, A.; Sporsheim, B.; Lionakis, M.S.; et al. Mitochondrial C5aR1 activity in macrophages controls IL-1beta production underlying sterile inflammation. Sci. Immunol. 2021, 6, eabf2489. [Google Scholar] [CrossRef]
- She, B.R.; Liou, G.G.; Lin-Chao, S. Association of the growth-arrest-specific protein Gas7 with F-actin induces reorganization of microfilaments and promotes membrane outgrowth. Exp. Cell Res. 2002, 273, 34–44. [Google Scholar] [CrossRef]
- Roach, T.G.; Lang, H.K.M.; Xiong, W.; Ryhanen, S.J.; Capelluto, D.G.S. Protein trafficking or cell signaling: A dilemma for the adaptor protein TOM1. Front. Cell Dev. Biol. 2021, 9, 643769. [Google Scholar] [CrossRef]
- Zong, X.; Schieder, M.; Cuny, H.; Fenske, S.; Gruner, C.; Rotzer, K.; Griesbeck, O.; Harz, H.; Biel, M.; Wahl-Schott, C. The two-pore channel TPCN2 mediates NAADP-dependent Ca(2+)-release from lysosomal stores. Pflug. Arch. Eur. J. Physiol. 2009, 458, 891–899. [Google Scholar] [CrossRef]
- Sundaram, B.; Pandian, N.; Mall, R.; Wang, Y.; Sarkar, R.; Kim, H.J.; Malireddi, R.K.S.; Karki, R.; Janke, L.J.; Vogel, P.; et al. NLRP12-PANoptosome activates PANoptosis and pathology in response to heme and PAMPs. Cell 2023, 186, 2783–2801.e20. [Google Scholar] [CrossRef] [PubMed]
- Garcia, D.; Shaw, R.J. AMPK: Mechanisms of cellular energy sensing and restoration of metabolic balance. Mol. Cell 2017, 66, 789–800. [Google Scholar] [CrossRef] [PubMed]
- Carlberg, C. Vitamin D in the context of evolution. Nutrients 2022, 14, 3018. [Google Scholar] [CrossRef]
- Lowrey, P.L.; Takahashi, J.S. Mammalian circadian biology: Elucidating genome-wide levels of temporal organization. Annu. Rev. Genom. Hum. Genet. 2004, 5, 407–441. [Google Scholar] [CrossRef]
- Zimmet, P.; Alberti, K.; Stern, N.; Bilu, C.; El-Osta, A.; Einat, H.; Kronfeld-Schor, N. The circadian syndrome: Is the metabolic syndrome and much more! J. Intern. Med. 2019, 286, 181–191. [Google Scholar] [CrossRef]
- Abboud, M. Vitamin D supplementation and sleep: A systematic review and meta-analysis of intervention studies. Nutrients 2022, 14, 1076. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).