Lactiplantibacillus plantarum LM1001 Supplementation Attenuates Muscle Atrophy and Function Decline in Aged Mice
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
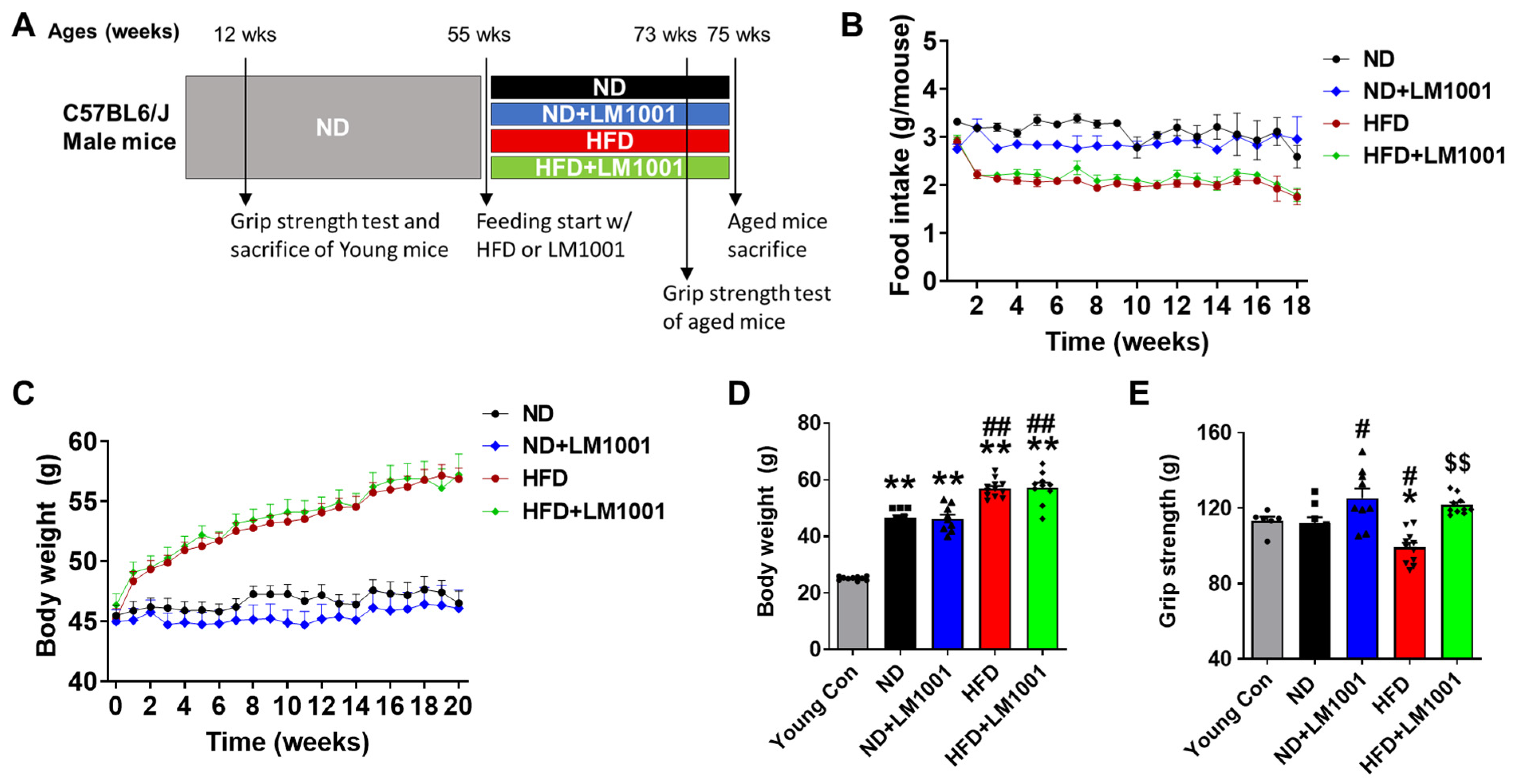
2.2. Probiotic Supplementation and Experimental Groups
2.3. Grip Strength Test
2.4. Hematoxylin & Eosin Analysis and Injury Scoring
2.5. Immunofluorescence (IF) Staining
2.6. Western Blot Analysis of Muscle Tissues
2.7. Stool Sample Collection and DNA Extraction
2.8. Amplification of the 16S rRNA Genes and Library Preparation
2.9. Bioinformatic Data Processing
2.10. In Vitro Cell Culture and Differentiation
2.11. Western Blot Analysis of C2C12 Cells
2.12. Creatine Kinase Activity of C2C12 Cells
2.13. Statistical Analysis
3. Results
3.1. LM1001 Supplementation Improves the Muscle Strength in Aged Mice
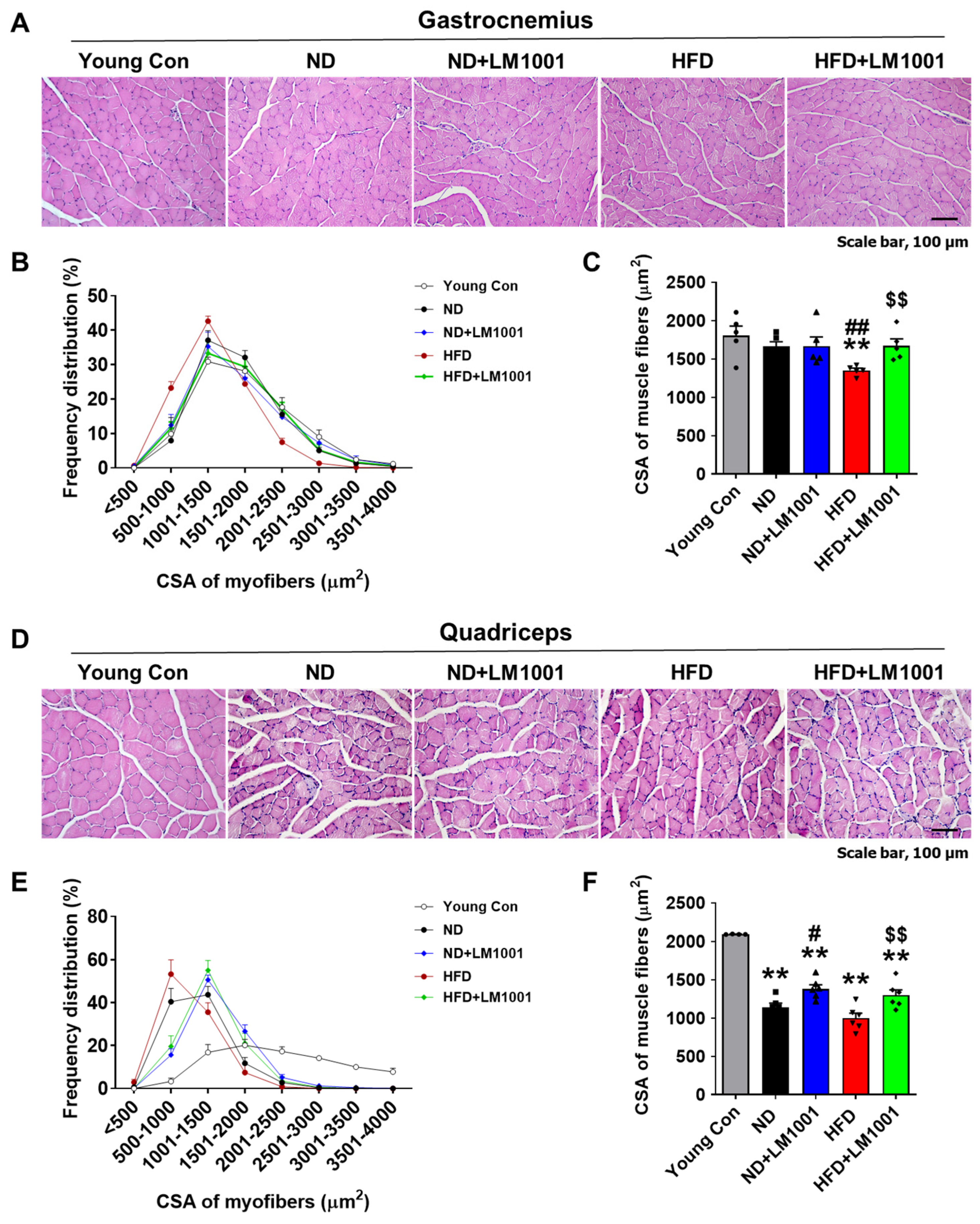
3.2. LM1001 Supplementation Restores the Myofiber Size That Is Altered in Aged Mice
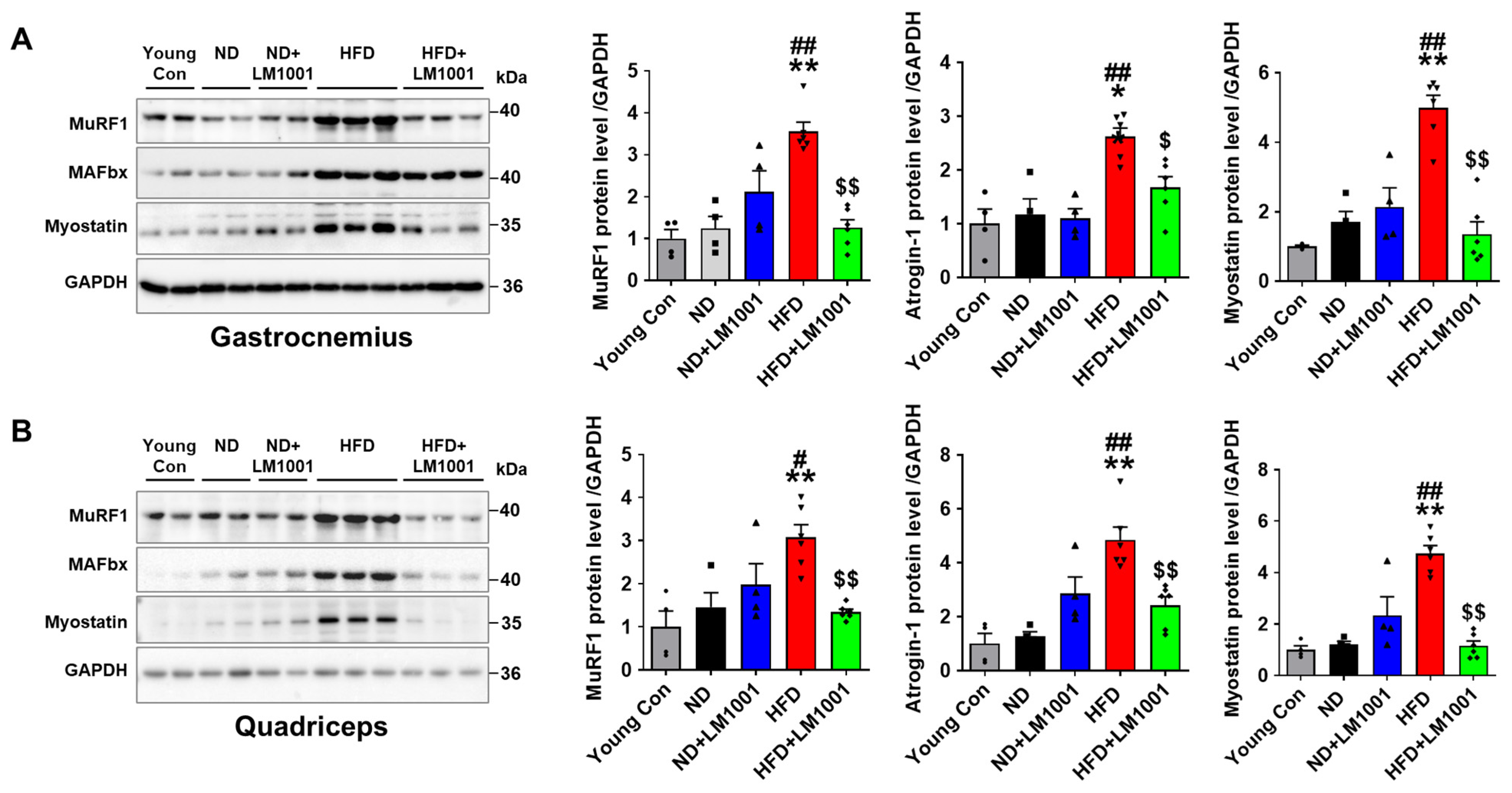
3.3. LM1001 Supplementation Reduces Muscle Atrophy Induced by HFD in Aged Mice
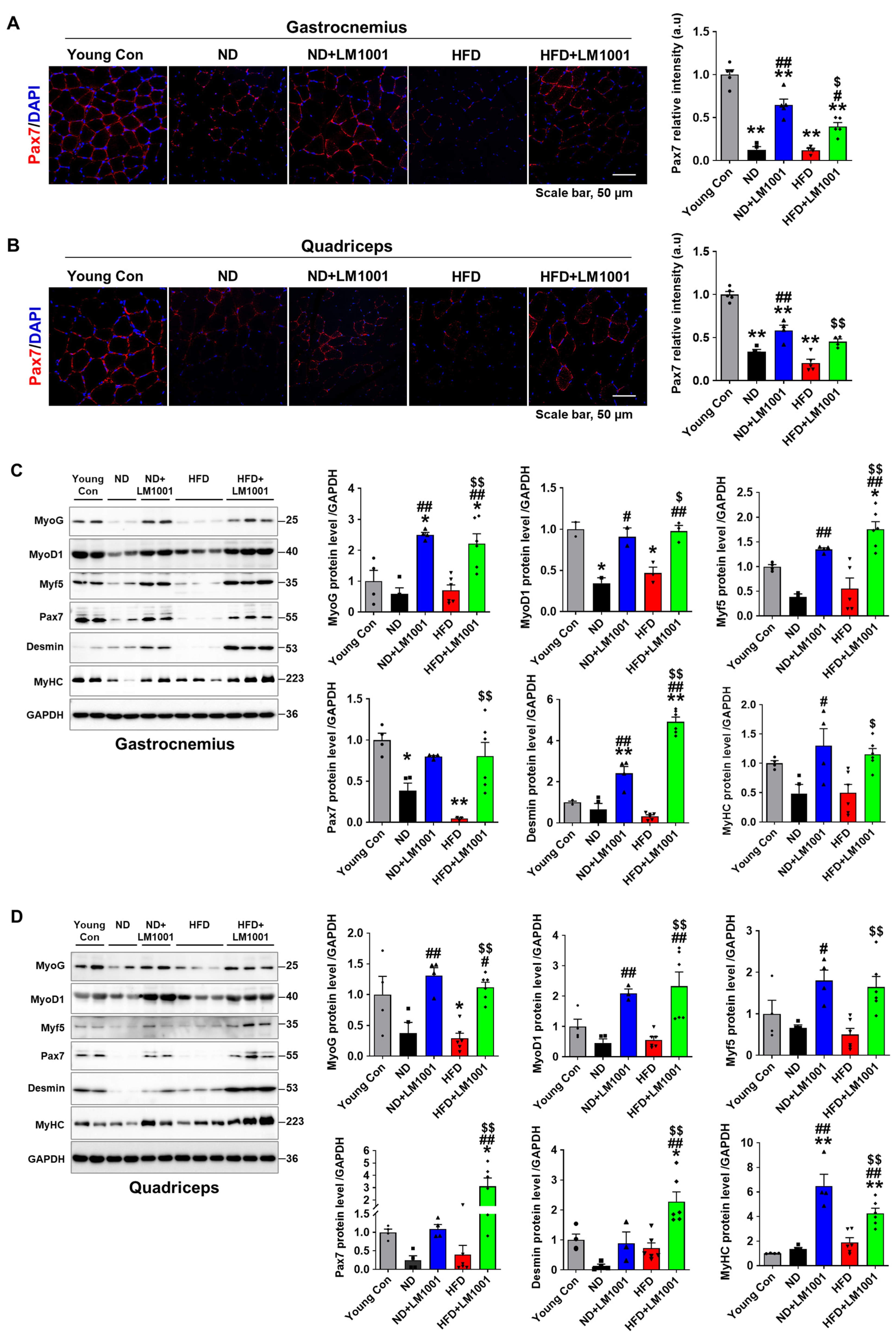
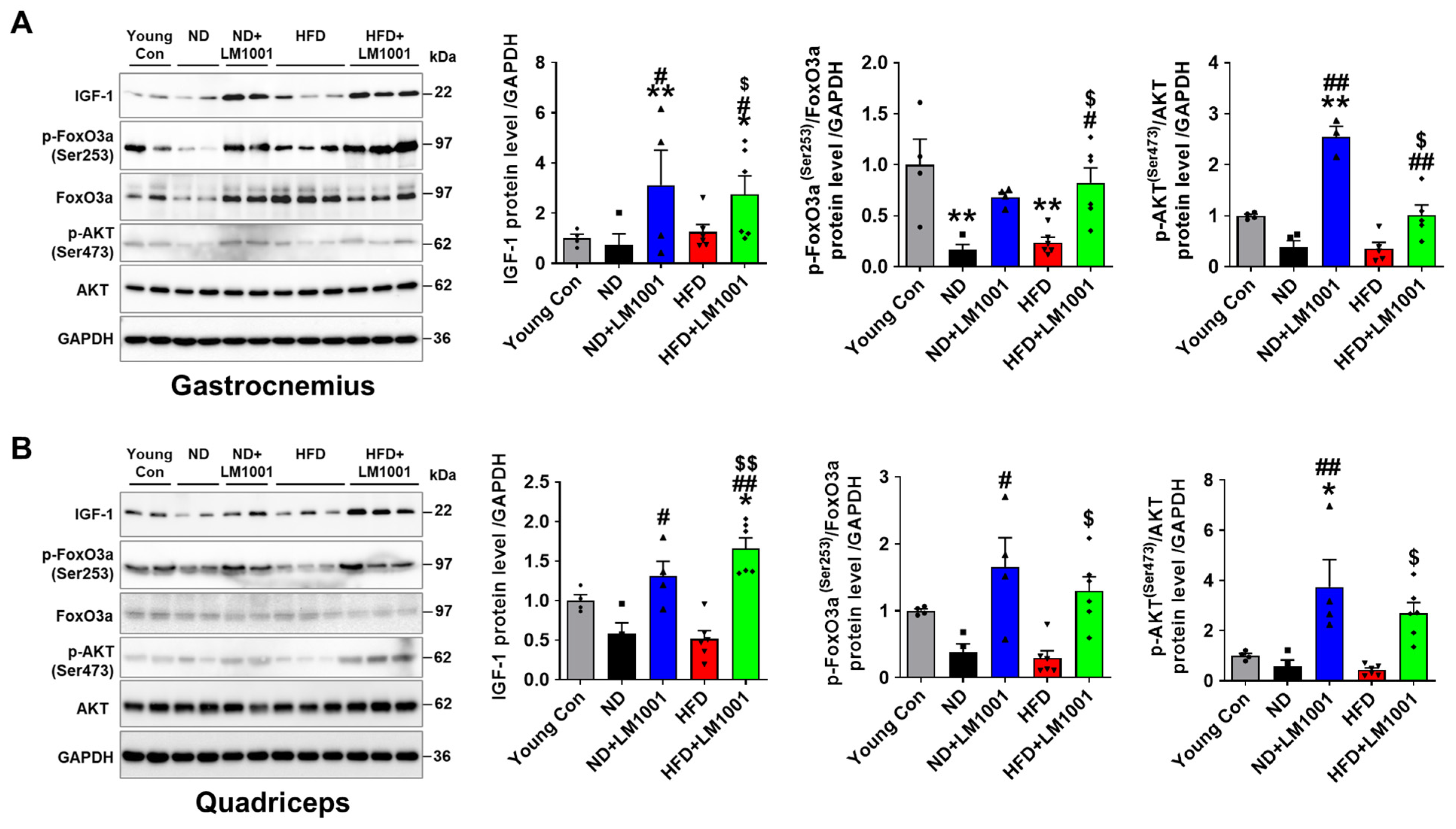
3.4. LM1001 Supplementation Promotes Myogenesis in the Skeletal Muscle of Aged Mice
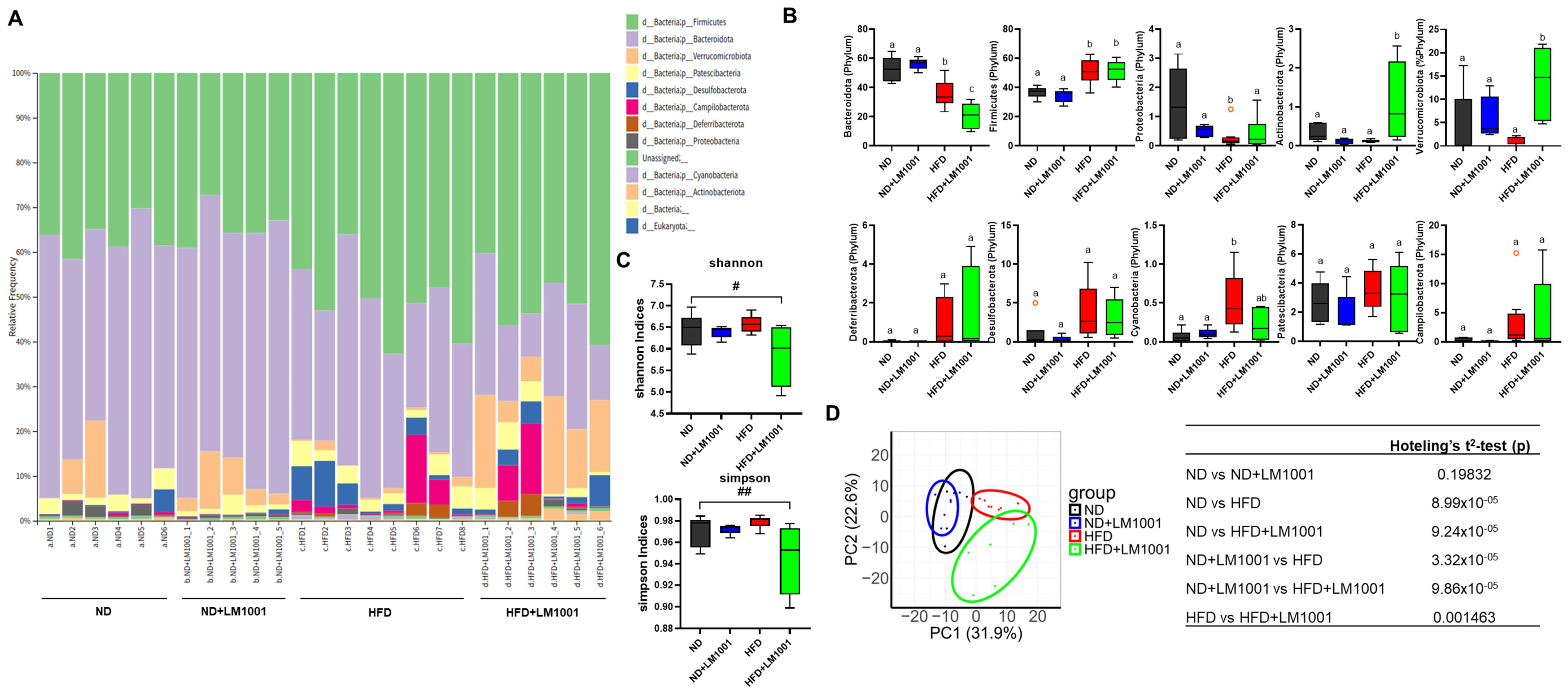
3.5. LM1001 Supplementation Improves the Gut Microbiota Altered in Aged HFD-Fed Mice
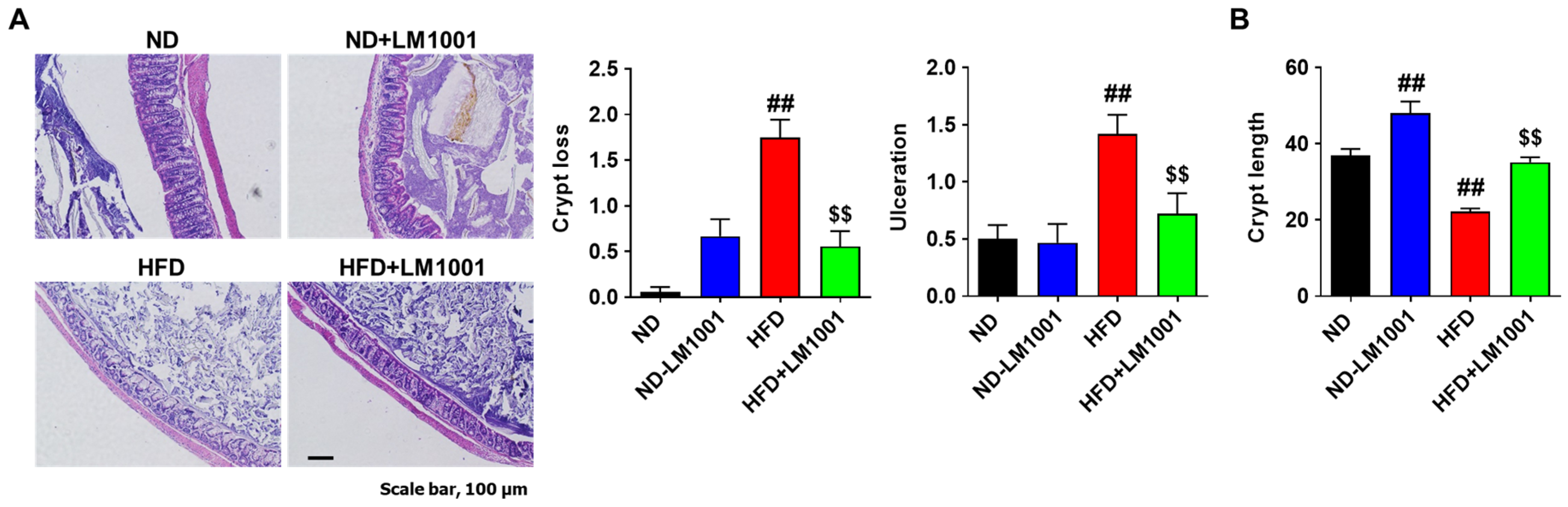
3.6. LM1001 Supplementation Reduces the Large Intestinal Injury Induced by HFD in Aged Mice
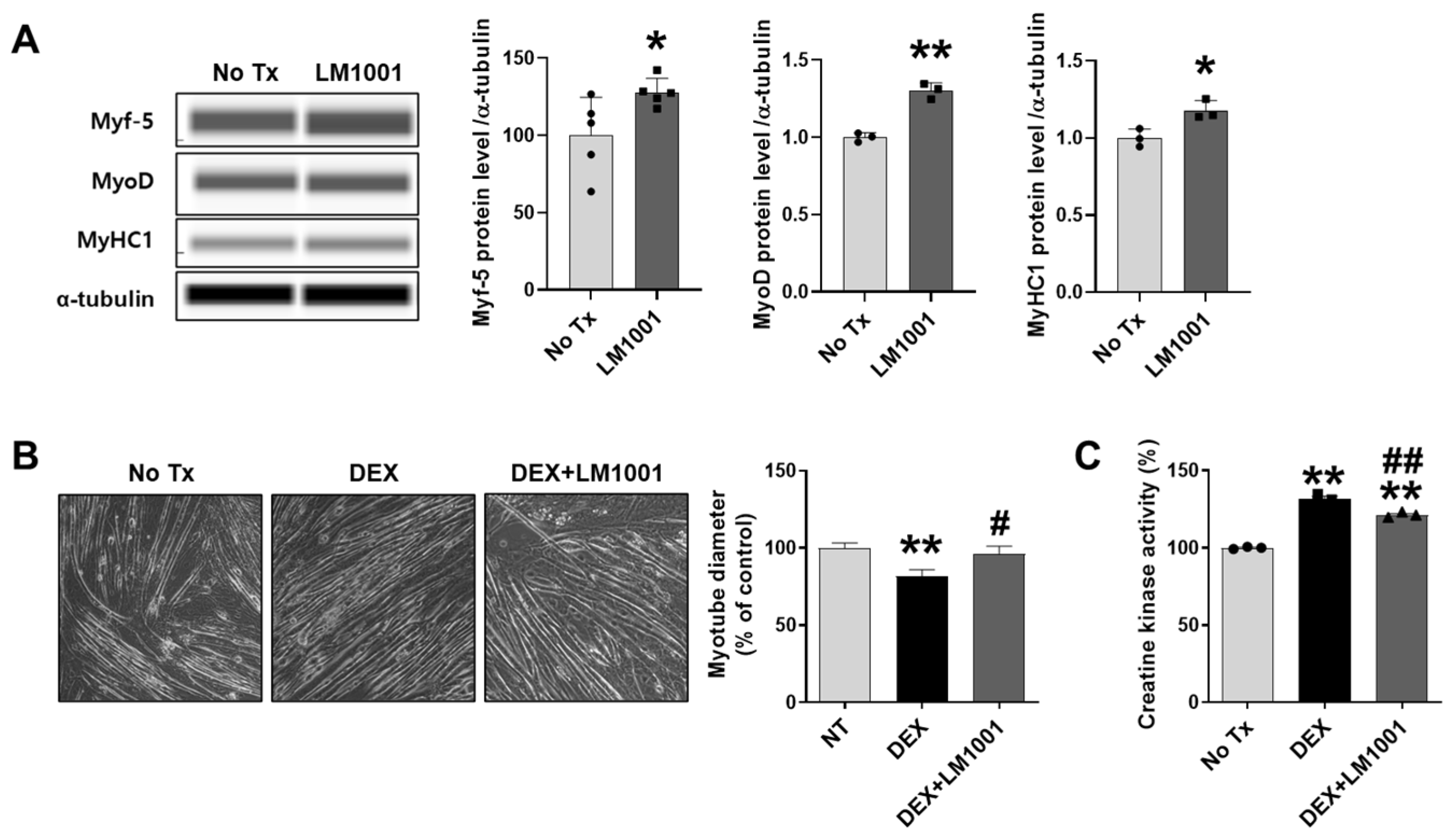
3.7. LM1001 Promotes Myogenic Differentiation and Reduces Dexamethasone-Induced Muscle Atrophy in C2C12 Cells
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Larsson, L.; Degens, H.; Li, M.; Salviati, L.; Lee, Y.I.; Thompson, W.; Kirkland, J.L.; Sandri, M. Sarcopenia: Aging-Related Loss of Muscle Mass and Function. Physiol. Rev. 2019, 99, 427–511. [Google Scholar] [CrossRef]
- Volpi, E.; Nazemi, R.; Fujita, S. Muscle tissue changes with aging. Curr. Opin. Clin. Nutr. Metab. Care 2004, 7, 405–410. [Google Scholar] [CrossRef] [PubMed]
- Anker, S.D.; Morley, J.E.; von Haehling, S. Welcome to the ICD-10 code for sarcopenia. J. Cachexia Sarcopenia Muscle 2016, 7, 512–514. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.K.; Woo, J.; Arai, H. Asian Working Group for Sarcopenia Response to the Emphasis on Anterior Thigh Muscle Mass in Sarcopenia Diagnosis. J. Am. Med. Dir. Assoc. 2020, 21, 1174–1175. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Jentoft, A.J.; Bahat, G.; Bauer, J.; Boirie, Y.; Bruyere, O.; Cederholm, T.; Cooper, C.; Landi, F.; Rolland, Y.; Sayer, A.A.; et al. Sarcopenia: Revised European consensus on definition and diagnosis. Age Ageing 2019, 48, 16–31. [Google Scholar] [CrossRef]
- Fernandes, L.V.; Paiva, A.E.G.; Silva, A.C.B.; de Castro, I.C.; Santiago, A.F.; de Oliveira, E.P.; Porto, L.C.J. Prevalence of sarcopenia according to EWGSOP1 and EWGSOP2 in older adults and their associations with unfavorable health outcomes: A systematic review. Aging Clin. Exp. Res. 2022, 34, 505–514. [Google Scholar] [CrossRef]
- Yuan, S.; Larsson, S.C. Epidemiology of sarcopenia: Prevalence, risk factors, and consequences. Metabolism 2023, 144, 155533. [Google Scholar] [CrossRef]
- Axelrod, C.L.; Dantas, W.S.; Kirwan, J.P. Sarcopenic obesity: Emerging mechanisms and therapeutic potential. Metabolism 2023, 146, 155639. [Google Scholar] [CrossRef]
- Chen, Y.; Wu, J. Aging-Related Sarcopenia: Metabolic Characteristics and Therapeutic Strategies. Aging Dis. 2024, 16, 1003–1022. [Google Scholar] [CrossRef]
- Martinez-Arnau, F.M.; Fonfria-Vivas, R.; Buigues, C.; Castillo, Y.; Molina, P.; Hoogland, A.J.; van Doesburg, F.; Pruimboom, L.; Fernandez-Garrido, J.; Cauli, O. Effects of Leucine Administration in Sarcopenia: A Randomized and Placebo-controlled Clinical Trial. Nutrients 2020, 12, 932. [Google Scholar] [CrossRef]
- Kwak, J.Y.; Kwon, K.S. Pharmacological Interventions for Treatment of Sarcopenia: Current Status of Drug Development for Sarcopenia. Ann. Geriatr. Med. Res. 2019, 23, 98–104. [Google Scholar] [CrossRef]
- Gentile, G.; De Stefano, F.; Sorrentino, C.; D’Angiolo, R.; Lauretta, C.; Giovannelli, P.; Migliaccio, A.; Castoria, G.; Di Donato, M. Androgens as the “old age stick” in skeletal muscle. Cell Commun. Signal 2025, 23, 167. [Google Scholar] [CrossRef]
- Dalton, J.T.; Barnette, K.G.; Bohl, C.E.; Hancock, M.L.; Rodriguez, D.; Dodson, S.T.; Morton, R.A.; Steiner, M.S. The selective androgen receptor modulator GTx-024 (enobosarm) improves lean body mass and physical function in healthy elderly men and postmenopausal women: Results of a double-blind, placebo-controlled phase II trial. J. Cachexia Sarcopenia Muscle 2011, 2, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Giron, M.; Thomas, M.; Dardevet, D.; Chassard, C.; Savary-Auzeloux, I. Gut microbes and muscle function: Can probiotics make our muscles stronger? J. Cachexia Sarcopenia Muscle 2022, 13, 1460–1476. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.H.; Chang, S.S.; Chang, H.Y.; Wu, C.H.; Pan, C.H.; Chang, C.C.; Chan, C.H.; Huang, H.Y. Probiotic supplementation attenuates age-related sarcopenia via the gut-muscle axis in SAMP8 mice. J. Cachexia Sarcopenia Muscle 2022, 13, 515–531. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; So, Y.J.; Jung, W.H.; Kim, T.R.; Sohn, M.; Jeong, Y.J.; Imm, J.Y. Lactiplantibacillus plantarum LM1001 Improves Digestibility of Branched-Chain Amino Acids in Whey Proteins and Promotes Myogenesis in C2C12 Myotubes. Food Sci. Anim. Resour. 2024, 44, 951–965. [Google Scholar] [CrossRef]
- Ray, N.; Jeong, H.; Kwon, D.; Kim, J.; Moon, Y. Antibiotic Exposure Aggravates Bacteroides-Linked Uremic Toxicity in the Gut-Kidney Axis. Front. Immunol. 2022, 13, 737536. [Google Scholar] [CrossRef]
- Sun, J.; Kim, J.; Jeong, H.; Kwon, D.; Moon, Y. Xenobiotic-induced ribosomal stress compromises dysbiotic gut barrier aging: A one health perspective. Redox Biol. 2023, 59, 102565. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Agnetti, G.; Herrmann, H.; Cohen, S. New roles for desmin in the maintenance of muscle homeostasis. FEBS J. 2022, 289, 2755–2770. [Google Scholar] [CrossRef]
- Plotkin, D.L.; Mattingly, M.L.; Anglin, D.A.; Michel, J.M.; Godwin, J.S.; McIntosh, M.C.; Kontos, N.J.; Bergamasco, J.G.A.; Scarpelli, M.C.; Angleri, V.; et al. Skeletal muscle myosin heavy chain fragmentation as a potential marker of protein degradation in response to resistance training and disuse atrophy. Exp. Physiol. 2024, 109, 1739–1754. [Google Scholar] [CrossRef]
- Zhang, W.B.; Milman, S. Looking at IGF-1 through the hourglass. Aging 2022, 14, 6379–6380. [Google Scholar] [CrossRef]
- Rodrigues, V.F.; Elias-Oliveira, J.; Pereira, I.S.; Pereira, J.A.; Barbosa, S.C.; Machado, M.S.G.; Carlos, D. Akkermansia muciniphila and Gut Immune System: A Good Friendship That Attenuates Inflammatory Bowel Disease, Obesity, and Diabetes. Front. Immunol. 2022, 13, 934695. [Google Scholar] [CrossRef] [PubMed]
- Wade, H.; Pan, K.; Duan, Q.; Kaluzny, S.; Pandey, E.; Fatumoju, L.; Saraswathi, V.; Wu, R.; Harris, E.N.; Su, Q. Akkermansia muciniphila and its membrane protein ameliorates intestinal inflammatory stress and promotes epithelial wound healing via CREBH and miR-143/145. J. Biomed. Sci. 2023, 30, 38. [Google Scholar] [CrossRef] [PubMed]
- Bedu-Ferrari, C.; Biscarrat, P.; Langella, P.; Cherbuy, C. Prebiotics and the Human Gut Microbiota: From Breakdown Mechanisms to the Impact on Metabolic Health. Nutrients 2022, 14, 2096. [Google Scholar] [CrossRef] [PubMed]
- Myhrstad, M.C.W.; Tunsjo, H.; Charnock, C.; Telle-Hansen, V.H. Dietary Fiber, Gut Microbiota, and Metabolic Regulation-Current Status in Human Randomized Trials. Nutrients 2020, 12, 859. [Google Scholar] [CrossRef] [PubMed]
- Cronin, P.; Joyce, S.A.; O’Toole, P.W.; O’Connor, E.M. Dietary Fibre Modulates the Gut Microbiota. Nutrients 2021, 13, 1655. [Google Scholar] [CrossRef]
- Mankhong, S.; Kim, S.; Moon, S.; Kwak, H.B.; Park, D.H.; Kang, J.H. Experimental Models of Sarcopenia: Bridging Molecular Mechanism and Therapeutic Strategy. Cells 2020, 9, 1385. [Google Scholar] [CrossRef]
- Romagnoli, C.; Iantomasi, T.; Brandi, M.L. Available In Vitro Models for Human Satellite Cells from Skeletal Muscle. Int. J. Mol. Sci. 2021, 22, 13221. [Google Scholar] [CrossRef]
- Bentzinger, C.F.; Wang, Y.X.; von Maltzahn, J.; Rudnicki, M.A. The emerging biology of muscle stem cells: Implications for cell-based therapies. Bioessays 2013, 35, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Schiaffino, S.; Mammucari, C. Regulation of skeletal muscle growth by the IGF1-Akt/PKB pathway: Insights from genetic models. Skelet. Muscle 2011, 1, 4. [Google Scholar] [CrossRef] [PubMed]
- Stitt, T.N.; Drujan, D.; Clarke, B.A.; Panaro, F.; Timofeyva, Y.; Kline, W.O.; Gonzalez, M.; Yancopoulos, G.D.; Glass, D.J. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol. Cell 2004, 14, 395–403. [Google Scholar] [CrossRef] [PubMed]
- Gellhaus, B.; Boker, K.O.; Schilling, A.F.; Saul, D. Therapeutic Consequences of Targeting the IGF-1/PI3K/AKT/FOXO3 Axis in Sarcopenia: A Narrative Review. Cells 2023, 12, 2787. [Google Scholar] [CrossRef] [PubMed]
- Bodine, S.C.; Baehr, L.M. Skeletal muscle atrophy and the E3 ubiquitin ligases MuRF1 and MAFbx/atrogin-1. Am. J. Physiol. Endocrinol. Metab. 2014, 307, E469–E484. [Google Scholar] [CrossRef]
- Yoshida, T.; Delafontaine, P. Mechanisms of IGF-1-Mediated Regulation of Skeletal Muscle Hypertrophy and Atrophy. Cells 2020, 9, 1970. [Google Scholar] [CrossRef]
- Manning, B.D.; Toker, A. AKT/PKB Signaling: Navigating the Network. Cell 2017, 169, 381–405. [Google Scholar] [CrossRef]
- Zhang, X.; Li, L.; Butcher, J.; Stintzi, A.; Figeys, D. Advancing functional and translational microbiome research using meta-omics approaches. Microbiome 2019, 7, 154. [Google Scholar] [CrossRef]
- Rosshart, S.P.; Herz, J.; Vassallo, B.G.; Hunter, A.; Wall, M.K.; Badger, J.H.; McCulloch, J.A.; Anastasakis, D.G.; Sarshad, A.A.; Leonardi, I.; et al. Laboratory mice born to wild mice have natural microbiota and model human immune responses. Science 2019, 365, eaaw4361. [Google Scholar] [CrossRef]
- Chandrasekaran, P.; Weiskirchen, S.; Weiskirchen, R. Effects of Probiotics on Gut Microbiota: An Overview. Int. J. Mol. Sci. 2024, 25, 6022. [Google Scholar] [CrossRef] [PubMed]
- Derrien, M.; Alvarez, A.S.; de Vos, W.M. The Gut Microbiota in the First Decade of Life. Trends Microbiol. 2019, 27, 997–1010. [Google Scholar] [CrossRef] [PubMed]
- Reynoso-Garcia, J.; Miranda-Santiago, A.E.; Melendez-Vazquez, N.M.; Acosta-Pagan, K.; Sanchez-Rosado, M.; Diaz-Rivera, J.; Rosado-Quinones, A.M.; Acevedo-Marquez, L.; Cruz-Roldan, L.; Tosado-Rodriguez, E.L.; et al. A complete guide to human microbiomes: Body niches, transmission, development, dysbiosis, and restoration. Front. Syst. Biol. 2022, 2, 951403. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Wong, P.Y.; Wang, Q.; Wong, H.Y.; Huang, T.; Cui, C.; Zhang, N.; Cheung, W.H.; Wong, R.M.Y. Short-chain fatty acids enhance muscle mass and function through the activation of mTOR signalling pathways in sarcopenic mice. J. Cachexia Sarcopenia Muscle 2024, 15, 2387–2401. [Google Scholar] [CrossRef]
- Liu, X.; Xu, M.; Wang, H.; Zhu, L. Role and Mechanism of Short-Chain Fatty Acids in Skeletal Muscle Homeostasis and Exercise Performance. Nutrients 2025, 17, 1463. [Google Scholar] [CrossRef]
- Zhu, J.; Peng, F.; Yang, H.; Luo, J.; Zhang, L.; Chen, X.; Liao, H.; Lei, H.; Liu, S.; Yang, T.; et al. Probiotics and muscle health: The impact of Lactobacillus on sarcopenia through the gut-muscle axis. Front. Microbiol. 2025, 16, 1559119. [Google Scholar] [CrossRef]
- Zhao, J.; Huang, Y.; Yu, X. A Narrative Review of Gut-Muscle Axis and Sarcopenia: The Potential Role of Gut Microbiota. Int. J. Gen. Med. 2021, 14, 1263–1273. [Google Scholar] [CrossRef]
- Frampton, J.; Murphy, K.G.; Frost, G.; Chambers, E.S. Higher dietary fibre intake is associated with increased skeletal muscle mass and strength in adults aged 40 years and older. J. Cachexia Sarcopenia Muscle 2021, 12, 2134–2144. [Google Scholar] [CrossRef]
- Takahashi, F.; Hashimoto, Y.; Kaji, A.; Sakai, R.; Kawate, Y.; Okamura, T.; Okada, H.; Kitagawa, N.; Nakanishi, N.; Majima, S.; et al. Dietary Fiber Intake Is Related to Skeletal Muscle Mass, Body Fat Mass, and Muscle-to-Fat Ratio Among People With Type 2 Diabetes: A Cross-Sectional Study. Front. Nutr. 2022, 9, 881877. [Google Scholar] [CrossRef]
- Kang, H.; You, H.J.; Lee, G.; Lee, S.H.; Yoo, T.; Choi, M.; Joo, S.K.; Park, J.H.; Chang, M.S.; Lee, D.H.; et al. Interaction effect between NAFLD severity and high carbohydrate diet on gut microbiome alteration and hepatic de novo lipogenesis. Gut. Microbes 2022, 14, 2078612. [Google Scholar] [CrossRef]
- Lee, G.; You, H.J.; Bajaj, J.S.; Joo, S.K.; Yu, J.; Park, S.; Kang, H.; Park, J.H.; Kim, J.H.; Lee, D.H.; et al. Distinct signatures of gut microbiome and metabolites associated with significant fibrosis in non-obese NAFLD. Nat. Commun. 2020, 11, 4982. [Google Scholar] [CrossRef]
- You, H.J.; Si, J.; Kim, J.; Yoon, S.; Cha, K.H.; Yoon, H.S.; Lee, G.; Yu, J.; Choi, J.S.; Jung, M.; et al. Bacteroides vulgatus SNUG 40005 Restores Akkermansia Depletion by Metabolite Modulation. Gastroenterology 2023, 164, 103–116. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karekezi, J.; Kim, H.; Dusabimana, T.; Nugroho, T.A.; Ndahigwa, E.N.; So, Y.J.; Kim, J.; Kim, T.-R.; Sohn, M.; Miao, J.; et al. Lactiplantibacillus plantarum LM1001 Supplementation Attenuates Muscle Atrophy and Function Decline in Aged Mice. Nutrients 2025, 17, 3156. https://doi.org/10.3390/nu17193156
Karekezi J, Kim H, Dusabimana T, Nugroho TA, Ndahigwa EN, So YJ, Kim J, Kim T-R, Sohn M, Miao J, et al. Lactiplantibacillus plantarum LM1001 Supplementation Attenuates Muscle Atrophy and Function Decline in Aged Mice. Nutrients. 2025; 17(19):3156. https://doi.org/10.3390/nu17193156
Chicago/Turabian StyleKarekezi, Jacques, Hwajin Kim, Theodomir Dusabimana, Tatang Aldi Nugroho, Edvard Ntambara Ndahigwa, Yoon Ju So, Juil Kim, Tae-Rahk Kim, Minn Sohn, Ji Miao, and et al. 2025. "Lactiplantibacillus plantarum LM1001 Supplementation Attenuates Muscle Atrophy and Function Decline in Aged Mice" Nutrients 17, no. 19: 3156. https://doi.org/10.3390/nu17193156
APA StyleKarekezi, J., Kim, H., Dusabimana, T., Nugroho, T. A., Ndahigwa, E. N., So, Y. J., Kim, J., Kim, T.-R., Sohn, M., Miao, J., Moon, Y., & Park, S. W. (2025). Lactiplantibacillus plantarum LM1001 Supplementation Attenuates Muscle Atrophy and Function Decline in Aged Mice. Nutrients, 17(19), 3156. https://doi.org/10.3390/nu17193156








