Abstract
The identification of wetland vegetation is essential for environmental protection and management as well as for monitoring wetlands’ health and assessing ecosystem services. However, some limitations on vegetation classification may be related to remote sensing technology, confusion between plant species, and challenges related to inadequate data accuracy. In this paper, vegetation classification in the Yancheng Coastal Wetlands is studied and evaluated from Sentinel-2 images based on a random forest algorithm. Based on consistent time series from remote sensing observations, the characteristic patterns of the Yancheng Coastal Wetlands were better captured. Firstly, the spectral features, vegetation indices, and phenological characteristics were extracted from remote sensing images, and classification products were obtained by constructing a dense time series using a dataset based on Sentinel-2 images in Google Earth Engine (GEE). Then, remote sensing classification products based on the random forest machine learning algorithm were obtained, with an overall accuracy of 95.64% and kappa coefficient of 0.94. Four indicators (POP, SOS, NDVIre, and B12) were the main contributors to the importance of the weight analysis for all features. Comparative experiments were conducted with different classification features. The results show that the method proposed in this paper has better classification.
1. Introduction
The coastal wetlands in Jiangsu are crucial for migratory birds, providing key locations for resting, breeding, and wintering. Changes in wetland vegetation significantly affect the stability of migratory bird populations, and it is essential to closely monitor these changes and accurately assess the ecological functions and values of coastal wetlands [1,2,3,4]. Remote sensing technology is a vital tool for monitoring coastal wetlands, with high-resolution imagery offering a comprehensive understanding of vegetation’s spatial distribution [5,6]. Vegetation phenology studies the characteristics of vegetation at different growth stages throughout the year, and analyzing time-series data can reveal vegetation growth rates. High-resolution imagery is more suitable for detailed wetland vegetation studies [7]. The variation in plant life within salt marshes in coastal wetlands, with their relatively small size, suggests that higher-resolution images are more suitable for detailed investigation of wetland vegetation [8]. Meanwhile, researchers continue to conduct experiments aimed at determining the most effective features and strategies for classifying remote sensing data. Results have shown that using the red-edge band transmitted by the Sentinel-2 satellite provides a more accurate representation of the wetland vegetation properties [9,10].
However, the use of high-spatial-resolution or hyperspectral imagery for wetland vegetation monitoring is often limited by the substantial expenses associated with spatial and temporal range [11]. Consequently, an increasing interest in the investigation of coastal wetland categorization algorithms based on time-series data has emerged in recent years. One of the main advantages of conducting analyses on time series is that scholars can acquire all remote sensing image data of a region for a period of one or more years. Additionally, obtaining the significant seasonal changes characteristics in coastal wetland vegetation can help improve the classification accuracy. Wu et al. [12] examined the growth patterns of Spartina alterniflora and performed statistical analysis on the time-series data for four spectral indicators. Zhang et al. [13] developed the first 30-m detailed wetland map, known as GWL_FCS30, by using existing global wetland datasets and multisource time-series remote sensing imagery. However, the limited availability of salt marsh samples has led to an incomplete representation of some vegetation characteristics in the salt marsh area. Due to the seasonal variations in coastal wetland vegetation, relying solely on remotely sensed images taken on a single date is no longer enough for monitoring the dynamic characteristics of wetland vegetation. Instead, employing dense time-series vegetation phenology features can more effectively capture the temporal aspect of coastal wetland vegetation. “Dense time series” involve collecting all available images over a year or specific period, as opposed to a single-time image. Sun et al. [14] proposed the pixel difference time series (PDTS) technique to extract phenological features and developed a tidal filter to enhance the classification accuracy. Liu et al. [15] conducted a classification of the Yancheng Coastal Wetlands by extracting phenological features from dense time-series data. Nevertheless, their approaches solely rely on vegetation phenology for extracting coastal wetland vegetation. Given that the Sentinel-2 satellite carries a multi-spectral band providing additional remote sensing features, these features may enhance the classification process and achieve more precise classification outcomes.
Today, there are many studies conducted on coastal wetland classification by fusing multiple features, but there are few studies that really combine phenological features. We propose a random forest method that integrates spectral features, vegetation indices, and phenological features. The purpose of this experiment is to explore whether this method can obtain wetland classification products with higher accuracy. In addition, we performed a comparative analysis to evaluate how the use of multiple remote sensing features affects the precision of wetland vegetation classification. The rest of this paper is structured as follows: in Section 2, the study area, the dataset, the methodology, the random forest classification, and the accuracy assessment are presented; in Section 3, the results are obtained from the case study; Section 4 presents some discussion; finally, conclusions are given in Section 5.
2. Materials and Methods
2.1. Study Area
The study area is situated in the core zone of the Yancheng Wetland Rare Birds National Nature Reserve (33°25′0″–33°39′04″N, 120°26′40″–120°40′40″E), Dafeng District, Yancheng, Jiangsu Province, China (Figure 1). With a wide range of wetland varieties, the coastal wetland covers a total area of about 191.00 km2. Coastal wetland vegetation are a special type of wetland vegetation that has adapted to the salty–alkaline soils and salty–tidal habitats characterizing this transition zone between land and sea. Moreover, the area contains a wide range of vegetation types because it has remained untouched by human activities. Both native and introduced vegetation types coexist in this area. Native species include Phragmites australis (P. australis) and Suaeda salsa (S. salsa), while introduced species include Spartina alterniflora (S. alterniflora). The wetlands’ ecological stability is at risk due to the fast expansion of S. alterniflora.
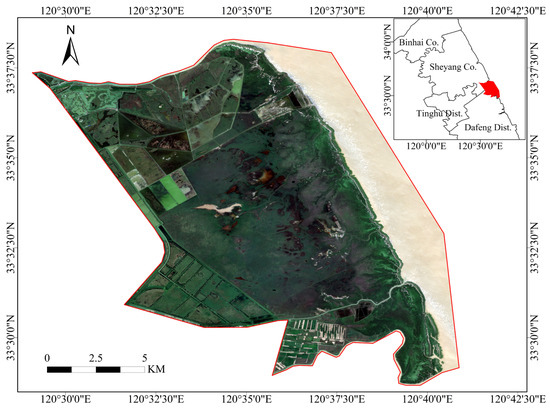
Figure 1.
Location of the study area.
2.2. Dataset
2.2.1. Sentinel-2 MSI Data
Google Earth Engine (GEE) is a cloud platform developed by Google that combines many data sources, such as Landsat, Sentinel, and other satellite data, along with meteorological, geographic, and demographic data. By utilizing the GEE cloud platform, we can expedite the acquisition of numerous observational datasets and effectively address the issue of image geometry correction. The APIs comprise numerous sophisticated algorithms and functionalities, facilitating users in efficiently processing data on a cloud platform. The remote sensing data utilized in this study were obtained from the “S2_SR_HARMONIZED” dataset available on Google Earth Engine (GEE) for the year 2022. This dataset corresponds to Sentinel-2 Level-2A (L2A) products and is sourced from the European Space Agency’s Sentinel Scientific Data Hub (https://scihub.copernicus.eu, accessed on 20 March 2024). The Sentinel-2 L2A products use spectral bands with a spatial resolution of 10 m, which include red, green, blue, and near-infrared bands. The data have been subjected to radiometric and geometric corrections, as well as atmospheric corrections, and the images are provided in the WGS84 UTM projection, including an estimated area of 110 × 110 km2.
2.2.2. Sample Data
Based on field observations and statistics, the dominant vegetation types in Yancheng include Spartina alterniflora (S. alterniflora), Suaeda salsa (S. salsa), and Phragmites australis (P. australis). Sample data were manually delimited using Google Earth and high-resolution remote sensing images, based on field observations. Using a combination of field survey sampling and image interpretation, we classified the study area in Yancheng into five categories: P. australis (PA), S. salsa (SS), S. alterniflora (SA), water, and unused land. Figure 2 illustrates the spatial arrangement of wetland habitat samples in the study area. The image shows a Sentinel-2 image taken on 10 October 2022. Sample data were generated using a uniform fishnet in ArcGIS, which was then integrated with site inspection data for annotation purposes.
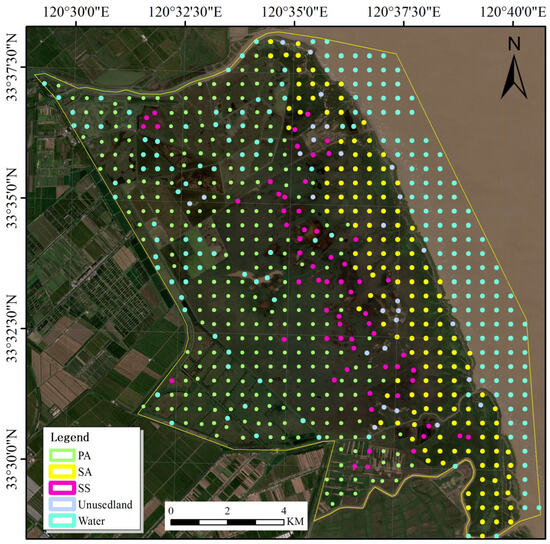
Figure 2.
Geographical distribution of the wetland sample.
2.3. Methodology
Figure 3 shows the technical procedure of wetland classification. The precise procedures encompassed the following: ① Obtaining all Sentinel-2 MSI data for the year 2022 and performing preprocessing tasks such as the removal of clouds and shadows. ② Comparing and selecting suitable spectral indices as parameters for the dense time-series dataset, combining all images within the study area into a collection with a temporal dimension to create a pixel-level time-series dataset. ③ Employing a function that aligns with the growth pattern of vegetation phenology to fit the time-series curves, resulting in smoother and more accurate curves. ④ Feature fusion involves the integration of spectral features, vegetation indices, and phenological features in order to derive wetland vegetation characteristics for classification. ⑤ By integrating field measurement data and manually annotated sample locations, the random forest algorithm was utilized to classify coastal wetland vegetation types in the research area. An accuracy assessment was then conducted to create a classification map.
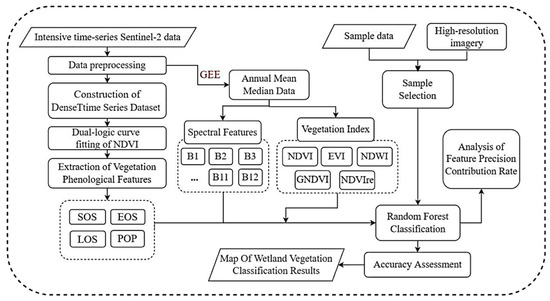
Figure 3.
General framework of wetland classification.
2.3.1. Data Preprocessing
The GEE platform utilizes the “maskCloudAndShadowsSR” method to eliminate clouds, thus guaranteeing that useable pixels are in each image. The algorithm utilizes SCL files generated by the Sentinel-2 L2A product to achieve the removal of interfering elements from the image and the retention of usable information by masking the SCL files for categories that may be identified as cloud, snow, cloud shadows, and so on. All available Sentinel-2 satellite remote sensing images were downloaded throughout the year. File names were stored using the date of image acquisition, facilitating the creation of dense time-series phenological feature curves later on. We acquired remote sensing image data from the Sentinel-2 L2A product for the entire year 2022. Specifically, 46 images were chosen, ensuring that the cloud cover was less than 80%. After cloud removal processing, 36 images were used for the reconstruction of the dense time-series dataset, as described in Table A1 in Appendix A. Simultaneously, considering the growth patterns of vegetation in coastal wetlands, the initial phase of vegetation growth was sluggish, while the latter phase more prominently exhibited the distinctive growth features of coastal wetland vegetation. The GEE time-polishing function was utilized to aggregate all high-quality pixels from June to December into a single image by considering the growth characteristics of the wetland vegetation, which was achieved by finding the median value of each pixel over all bands. The resulting image is a high-quality synthetic representation for the year 2022. Utilizing medium-digit images effectively decreases the quantity of datasets in comparison to the original images, hence producing synthetic images of superior quality that facilitate quicker and more efficient analysis to extract subsequent spectral features and commonly used vegetation indices [16].
2.3.2. Construction of Dense Time-Series Dataset
The construction of dense time-series datasets refers to the integration of all available temporal images within the study area into a collection with a temporal dimension, selecting appropriate spectral indices as parameters. In this study, NDVI is proposed as a parameter for reconstructing the dataset used for wetland vegetation classification, as its intrinsic sensitivity to coastal wetland vegetation allows for precise monitoring of plant growth patterns [17]. The formula for NDVI is as follows:
where NIR represents the near-infrared band on the S2_SR_HARMONIZED data, while RED represents the red band on the S2_SR_HARMONIZED data.
The process of dataset reconstruction involves two sequential steps: Firstly, the construction of a pixel-level dense time-series NDVI dataset, where the 36 NDVI images are processed in chronological sequence by raster manipulation to combine them into a high-dimensional raster array. Specifically, by implementing a cloud removal technique on the original S2_SR_HARMONIZED data, regions that in the original images were hidden by clouds, fog, or shadows are replaced with null values. This guarantees the accuracy and reliability of the subsequent inverse modeling of pixel-wise time-series NDVI vegetation phenology characteristics. The precision of the fitting model remains intact, eliminating any error produced by cloud and fog obstruction during the inversion process.
The second dataset corresponds temporally to the NDVI data and is an annual Julian day (DOY) time series. The Julian day is employed to record the number of days since the beginning of the year, providing a method to monitor the exact day inside a full cycle of vegetation growth. As the acquired S2_SR_ HARMONIZED dataset records image acquisition dates in the year–month–day format, the initial step involves converting these dates to Julian days, also known as DOY. Next, a time-series dataset representing the day of the year (DOY) needs to be created. This dataset should have the same size as the pixel-level dense time-series NDVI dataset. The attributes of each pixel in the images are subsequently allocated the relevant day of year (DOY) value for that specific temporal event. The above two steps are both implemented in Python.
2.3.3. Extraction of Vegetation’s Phenological Features
The curve-fitting method involves picking a function form that accurately reflects the growth patterns of the vegetation’s phenology in order to fit the time-series curve. Since the currently constructed NDVI and DOY datasets are discretely combined, Wu et al. [18] have demonstrated that obtaining continuous curves through model fitting can better capture vegetation’s phenological information. Therefore, this study aims to curve-fit the well-constructed NDVI dataset to mitigate the impact of inherent data deficiencies. Commonly used fitting models include double logistic fitting, polynomial fitting, and S-G filtering fitting. In this study, we selected the double logistic fitting function model as the preferred choice after several tests. This model combines the NDVI and DOY datasets to create a dense time-series vegetation phenology feature-fitting model based on the NDVI.
The “curve_fit” algorithm employs nonlinear least squares fitting to curve functions to find the ideal fitting model and achieve the most accurate curve. The optimization problem of the minimum multiplication can be expressed as follows:
where represents the number of data points, represents the adjusted parameter, is the function of , is the input variable, and is the actual observation value. The function must take the independent variable as the first parameter, followed by any additional required parameters.
This study adopts the logistic function model proposed by Gonsamo et al. [19] and applies it to the well-constructed NDVI and DOY datasets. This method assumes that the NDVI curve is symmetric during the rising and falling phases of the growing season, and it also presupposes that the NDVI curve is smooth, without abrupt changes or extreme values. Extreme climatic events may cause anomalies in NDVI values, and the double logistic fitting method has limited capacity to handle these outliers. The fitting model is as follows:
where represents the day of year (DOY), () is the amplitude between early summer on the plateau and the background, () is the amplitude between late summer on the plateau and the background, and are the normalized slope coefficients for spring and autumn, respectively, and and represent the DOY midpoints for the transition between the green-up and senescence periods. Based on the first, second, and third derivatives of Equation (3), phenological indices can be systematically calculated from the parameter system using the least squares method [18]. As a result, a model is created for each pixel in the research region that fits a curve to the vegetation’s phenological features using the NDVI.
Generally, the “coefficient method” [20], the “derivative method” [21], and the “threshold method” [22] are employed for extracting phenological metrics. This study used the derivative method (DES) to derive four prevalent vegetation phenology characteristics. In the derivative technique, the start of season (SOS) and the end of season (EOS) are determined by finding the DOY that corresponds to the highest and lowest values of the derivative curve-fitting model of the vegetation’s phenological features (i.e., the rate of change of the function). The peak of phenology (POP) is the specific day of the year when the maximum value is reached. Thus, these four phenological indices are determined via the derivative approach.
2.3.4. Feature Fusion of Spectral Features, Vegetation Indices, and Phenological Features
Multi-spectral bands such as red bands, green bands, blue bands, red-edge bands, near-infrared bands, and mid-infrared bands of Sentinel-2 images were extracted from GEE. Five commonly used vegetation indices for wetland classification were also selected as classification features for random forests. The selection of the five vegetation indices was based on the results of previous studies and chosen according to experience. The precise equations for computing the commonly used vegetation indices are provided in Table 1, where NDVI, GNDVI, EVI and NDVIre are all used to classify wetland vegetation [9,23,24]. GNDVI is more sensitive to chlorophyll than NDVI, which is favorable for capturing the information of wetland vegetation during the emergence period. EVI can better overcome the problem of NDVI saturating under the cover of large-scale vegetation and is able to more sensitively capture the growth and changes of vegetation. Coastal wetland vegetation is more sensitive to the red-edge information [9]; thus, NDVIre was selected. Meanwhile, NDWI was used to distinguish water bodies from other features. These indices were subsequently merged with the four vegetation phenology variables described in Section 2.3.3 to provide the complete set of input features for this experiment.

Table 1.
Common vegetation indices.
2.4. Random Forest Classification (RF)
We used the built-in simpleRandomForest() algorithm on GEE. This algorithm, conceptualized by Leo Breiman and Adele Cutler, introduced the term “random decision forests” in 1995 at Bell Laboratories [25].
The RF algorithm constructs each tree through the following steps:
- (a)
- Use N for the number of training examples (samples) and M for the number of features.
- (b)
- Choose the number of input features (m) for determining decisions at each tree node, where m is considerably less than M.
- (c)
- Employ bootstrapping by randomly sampling N times with replacement, creating a training set, and evaluating errors on the remaining unsampled examples.
- (d)
- Randomly select m features for each node and compute optimal splitting based on these features.
- (e)
- Allow each tree to grow fully without pruning.
These steps outline the construction process of each tree in the random forest algorithm. The total number of sample sites was 760, of which 280 were PA, 63 were SS, 157 were SA, 202 were water, and 28 were unused land. They were randomly divided in a 7:3 ratio for training and testing, respectively. The number of N was 70% of the total sample, and the remaining 30% was used to test the accuracy of the classification. M was set to 19, which is the sum of the spectral, vegetation index, and phenological characteristics.
After a multitude of experiments, when the number of decision trees is larger than 50, the classification model gradually tends to fit. Therefore, the number of decision trees was set to 50, and the other parameters were set to default. Since part of the data was not extracted during the sampling process, this part of the data was visualized as out-of-bag (OOB). Out-of-bag error (OOB error) generated from OOB data can not only evaluate the classification accuracy; it is also possible to calculate the importance of different feature variables (variable importance) of different feature variables for feature selection [26]. The model for assessing the importance of the characteristic variables is expressed as follows:
where VI denotes the importance of the feature variable, M is the total number of features in the sample, N is the number of decision trees generated, is the OOB error of the t-th decision tree when no noise interference is added to any feature , and is the OOB error of the t-th decision tree when noise interference is added to any feature . If the accuracy of out-of-bag data decreases significantly after adding noise randomly to a certain eigenvalue , it means that the eigenvalue has a great influence on the classification result, which also indicates high importance.
2.5. Accuracy Assessment
The assessment of wetland classification accuracy in the Yancheng National Rare Birds Nature Reserve was conducted using a combination of a confusion matrix, field inspections, and samples acquired from high-resolution images, including those obtained from Google Earth. To validate the accuracy, the overall accuracy (OA), kappa coefficient [27], producer accuracy (PA) and user accuracy (UA), which are commonly used today, were mainly selected as evaluation indices to evaluate each program. The following methods were chosen in this experiment:
where TP represents true positives, FP represents false positives, TN represents true negatives, FN represents false negatives, Po represents the observed precision, and Pe represents the expected precision of the random classification.
These evaluation metrics provide a thorough evaluation of the accuracy of wetland classification in the study area.
2.6. Statistical Significance of Classifiers’ Performance
In many remote sensing classification tasks, the comparisons of thematic map accuracy are often conducted using the same set of samples. For non-independent samples, the statistical significance of the difference between two proportions can be assessed through the McNemar test [28]. The McNemar test is based on the standardized normal test statistic. The Z-test and χ2-test are based on 2 × 2 confusion matrices. These tests were performed to test independency between two classification algorithms. The number of correctly and wrongly classified reference data pixels for two algorithms can be cross-tabulated as shown in Table 2.
where denotes the number of samples that were correctly classified by the first classification algorithm but misclassified by the second classification algorithm. Similarly, denotes the number of samples that were misclassified by the first classification algorithm and correctly classified by the second classification algorithm [29]. A difference in the classification accuracy between the confusion matrices is statistically significant (p ≤ 0.05) if the Z-value is more than 1.96 [30].

Table 2.
Cross-tabulation of the numbers of correctly and wrongly classified pixels for two algorithms.
The χ2-test is a non-parametric statistical test to determine whether the two or more classifications of the samples are independent or not. This test, if properly applied, may give us the answer by rejecting the null hypothesis or failing to reject it. If we find the value to be less than the value corresponding to our level of confidence, we can conclude that our null hypothesis is probably true. On the other hand, if our value lies over the level of confidence, we know that our χ2-test rejects the null hypothesis. Thus, we can conclude that the two classifications are dependent on one another. For the critical value defined as = 3.841, the null hypothesis is not rejected if . The McNemar test [29] is based on the standardized normal test, given as follows:
3. Results and Analysis
3.1. Analysis of Available Pixel Count
Figure 4 shows the number of available images obtained by statistically analyzing each pixel in the study area after applying the Google Earth Engine (GEE) cloud removal algorithm. In Figure 4a, the blue areas represent the regions with a lower number of available images due to variables such as clouds and rain. The minimum count of images in these areas is 15. Otherwise, the red areas indicate the regions with a higher number of available images and better quality, reaching a total of 36 images. Figure 4b illustrates the data availability for each month based on the selected remote sensing images. By creating a dense time-series dataset, the inclusion of images for every month of the year is ensured. This method solves the problem of limited availability of images during the summer season due to weather conditions. It ensures the accuracy of the fitting curves for vegetation phenology features by using vegetation indices. The construction of a densely populated time-series dataset for the whole year provides a more complete and unbiased depiction of the real land cover categories in a specific area, hence bolstering the reliability of classification outcomes.
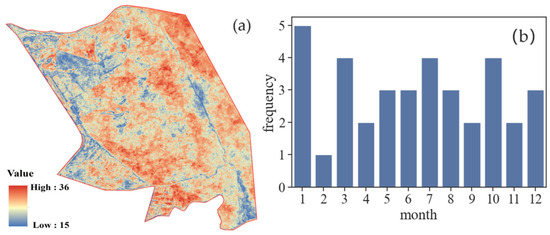
Figure 4.
Analysis of available pixels: (a) pixel-wise count of available images; (b) monthly count of available images.
3.2. Analysis of Vegetation Phenology Models’ Fitted Curves Based on NDVI
Figure 5 shows the fitted curves of the NDVI vegetation phenology model for typical coastal vegetation. The DOY is represented on the horizontal axis, while the corresponding fitted values of NDVI for the vegetation on that specific day of the year are represented on the vertical axis. The picture displays three separate curves, representing each of the fitted models for three common vegetation types found in the coastal wetlands of Jiangsu: PA, SS, and SA. The three colored empty dots in the figure depict the average NDVI values computed from all sample points for each wetland vegetation type on the specific DOY that corresponds to the remote sensing images obtained by the Sentinel-2 satellite. Given the temporal discontinuity, this paper uses a dual logistic fitting approach to calculate the associated NDVI curves. This methodology captures the fluctuations in the growth of a particular type of vegetation during a year, encompassing one complete growth cycle. This procedure eliminates the impact of specific data and random factors, and it reveals objective patterns of vegetation throughout its entire life cycle. In order to make it easier for readers to understand the vegetation’s phenological information, three kinds of vegetation phenology parameters (SOS, EOS, and POP) are represented in Figure 5, and the phenological parameters here should actually be the DOY values corresponding to their horizontal coordinates.
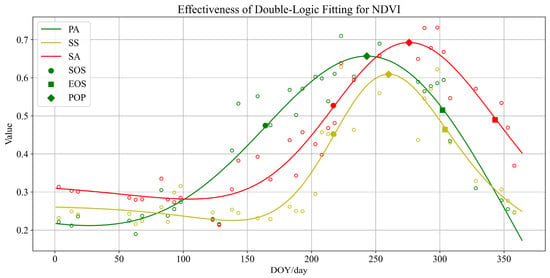
Figure 5.
Analysis of fitted curves for NDVI phenological models of vegetation.
PA demonstrated the earliest growth period, beginning rapid development in early April, achieving its maximum size in July and August, and quickly declining from mid-September, with almost full deterioration by the end of November. Conversely, SS and SA both started growing quickly in June, reached their highest growth rate in September, and began to deteriorate in October. By the end of November, SS was mostly withered, although SA had not yet fully deteriorated.
Figure 6 shows the boxplots of four widely used vegetation phenology indices (SOS, EOS, LOS, and POP) obtained by the derivative approach, based on the fitting curves of the NDVI vegetation phenology model as described in Section 2.3.3. The x-axis indicates three different vegetation types, while the y-axis corresponds to the DOY for the corresponding phenological traits. Clearly, PA’s SOS occurs significantly earlier than the SOS of the other two vegetation types, making it easily recognizable. The efficacy of EOS and LOS is not as evident, due to the difficulty in distinguishing data distribution across the three vegetation types. The data obtained from the POP analysis clearly demonstrate that PA exhibits the lowest POP, followed by SS, while SA has the highest POP. Examining the widths of the boxplots for the three vegetation types, it is evident that the box representing SS has the least variation in its upper and lower boundaries. This suggests that SS has the least variability and a more tightly clustered distribution of data.
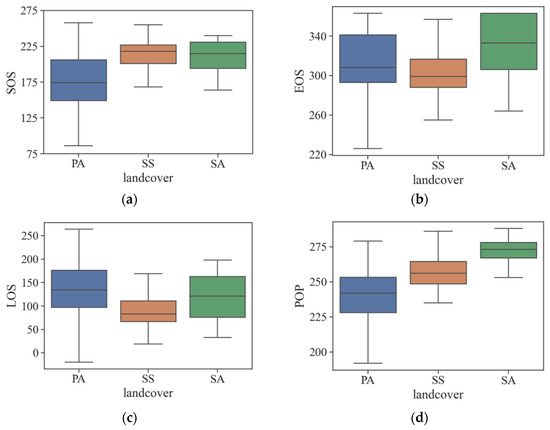
Figure 6.
Analysis of phenological characteristics of typical vegetation in coastal wetlands: (a) SOS; (b) EOS; (c) LOS; (d) POP.
3.3. Classification Results and Accuracy Evaluation
Figure 7 shows the classification map created by combining multisource features, such as spectral features, vegetation indices, and phenological features. This fusion was achieved using the random forest classification technique. The classification had an overall accuracy of 95.64%, along with a kappa coefficient of 0.94. Figure 7 illustrates the spatial arrangement of coastal wetland vegetation, showcasing its growing spread from inland regions to the beach. The vegetation succession from inside to the external regions occurs in the order of PA, SS, and SA.
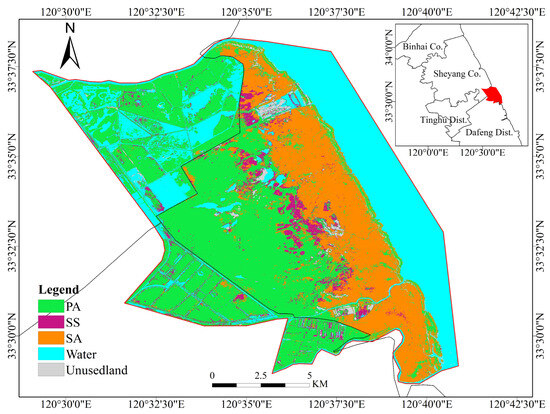
Figure 7.
Classification of typical vegetation in coastal wetlands.
The distribution of SS is divided due to the erosion caused by SA. Field investigations revealed that the central area experiences minimal human impact, resulting in dense growth of SS on specific roadways in the region. This phenomenon can be attributed to the fact that some elongated roads are frequently categorized as SS on the classification map. Due to the varying salt tolerance levels of different types of land cover and the fluctuations in soil salinity, PA, which has a lower salt tolerance, tends to grow further inland. Conversely, SA and SS, which have more salt tolerance, occur together in specific regions and are found near the ocean. Near the coastline, there were occasional cases where SA was misclassified as PA. The presence of SA in both submerged and non-submerged forms for a long time may be due to the tidal influence. This leads to inaccuracies in extracting phenological information. White waves captured by satellite imagery can also affect the identification of SA near the coastline.
The accuracy assessment of the classification results, as shown in Figure 8, was performed by calculating a confusion matrix. Here, the remaining 30% of samples that were not used for training were used to assess the accuracy. The figure shows the producer accuracy of different land cover classes. All other categories had classification accuracies beyond 90%, while the SS category had an accuracy of 75.26%. This indicates that, in general, all land cover types can be classified relatively well, with relatively few cases of misclassification. A portion of the SS was erroneously partitioned into PA, resulting in a blending of image features that are challenging to discern. Additionally, a portion of the SA was mistakenly divided into PA and the water column, potentially influenced by the tidal movements along the coastline.
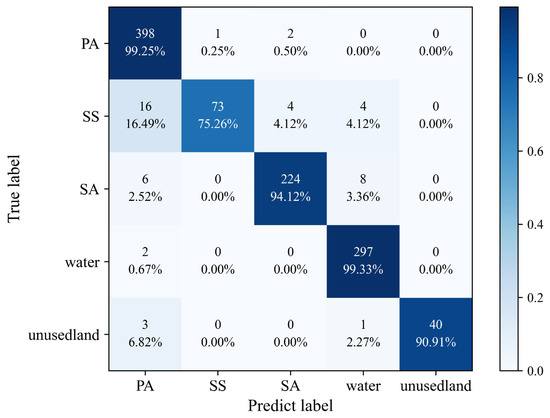
Figure 8.
Confusion matrix for classification.
3.4. Feature Contribution Analysis
Figure 9 shows the plot of the feature contribution analysis derived from the random forest training. Above the bar chart, the displayed values represent the contribution weights of each respective feature. The y-axis of the graph reflects the levels of contribution, while the x-axis represents 19 input features, arranged in descending order based on their contribution. A higher contribution indicates a more significant influence on the classification by that particular attribute. The five most important characteristics, sorted from highest to lowest, were POP, SOS, NDVIre, Band12, and Band11. The significant weight assigned to the POP illustrates its vital significance in the classification task. This is further supported by the box-and-line plot of POP in Figure 6, which demonstrates its ability to effectively differentiate coastal wetland vegetation. SOS is listed as the second most desirable band, and NDVIre is placed third. The B12 and B11 bands are the mid-infrared bands on the Sentinel-2 satellite. This indicates that the mid-infrared band may have an advantage in capturing information about the features related to coastal wetlands in Yancheng Rare Bird Wetland National Nature Reserve, Jiangsu Province. The importance of Band6, the red-edge band, is not ranked high, while Band5, also a red-edge band, and NDVIre, which contains a red-edge band, are ranked high in the importance weighting values (third and sixth, respectively). Therefore, specific red-edge bands may be more effective in capturing coastal wetland feature information. The Band8 and EOS contribution values ranked last. This may be due to the fact that the wetland vegetation all showed a similar green color during its peak growth period, and Band8 had difficulties capturing the difference between them. On the other hand, EOS may not be applicable as a phenological feature for the classification of coastal wetland vegetation.
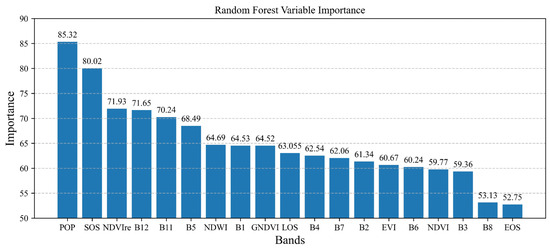
Figure 9.
Random forest variable importance.
We then selected the top seven features based on feature importance contribution (phenological features: POP, SOS; spectral features: B12, B11, B5; vegetation index features: NDVIre, NDWI) and performed random forest classification under identical conditions. The final classification accuracy reached 95.37%, with a kappa coefficient of 0.93. This resulted in high classification accuracy and also provided a method for simplifying input features for classification.
3.5. Comparison of Multiple Feature Fusion Methods
To validate the usability of this method, we conducted a classification comparison using different feature combinations as input features. We devised a total of seven schemes with various combinations of features by combining spectral features, vegetation indices, and phenological features in different ways. These methods include Scheme 1: using only spectral features (SP), Scheme 2: using only vegetation indices (VI), Scheme 3: using only phenological features (PH), Scheme 4: using spectral features and vegetation indices together (SP+VI), Scheme 5: using spectral features and phenological features together (SP+PH), Scheme 6: using vegetation indices and phenological features together (VI+PH), and Scheme 7: using spectral features, vegetation indices, and phenological features together (SP+VI+PH). The classification outcomes are presented in Table 3 and Figure 10. The classification results indicate that the overall accuracy of employing only phenological features for classification is 82.67%, which corresponds to the lowest kappa coefficient. The multisource feature fusion technique employed, which represents Scheme 7 in this work, attained the highest classification accuracy, exhibiting an overall accuracy of 95.64% and a kappa coefficient of 0.94. Table 3 shows the PA1 (producer accuracy) and UA (user accuracy) for all categories for the seven different scenarios. Among all five land cover categories, the PA1 and UA values for Scheme 7 were generally at a fairly high level of classification accuracy in each category, and this Scheme obtained the highest PA1 and UA for PA among all of the scenarios. Scheme 5 had the second highest classification accuracy and was very similar to Scheme 7 (OA of 95.46, kappa coefficient of 0.94). Scheme 3 performed poorly in the classification of SS and UN (unutilized land). Although classification based only on phenological data had the lowest accuracy, a comparison of the two methods of Scheme 4 (SP+VI) and Scheme 7 (SP+VI+PH) showed that the classification accuracy generally improved after integrating phenological features. Furthermore, by examining the classification result maps of several approaches in Figure 10, it is evident that Scheme 7 (SP+VI+PH) demonstrates the highest classification accuracy, closely resembling the real land cover distribution. While the classification accuracy using only spectral features (SP) was high, Figure 10a demonstrates that specific regions of SA were misclassified as PA, contradicting the real situation. Hence, relying only on classification accuracy to assess the real quality of classification is inadequate.

Table 3.
Comparative analysis of various feature fusion methods.
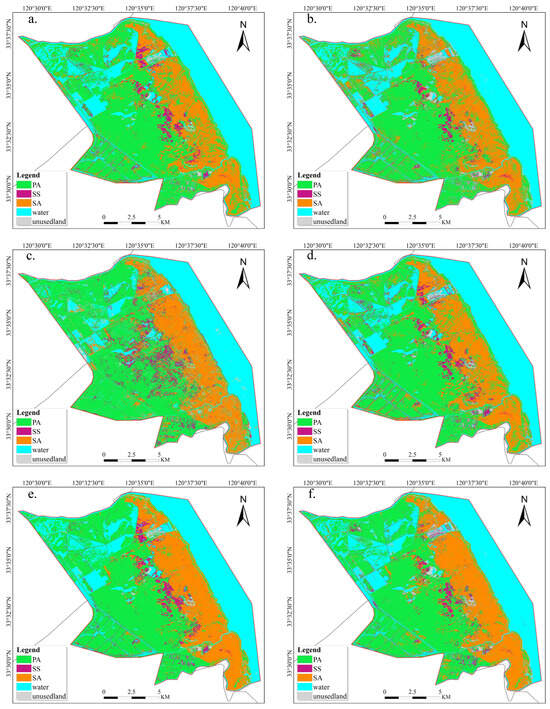
Figure 10.
Comparison of different feature classification maps: (a) RF classification based on spectral features; (b) RF classification based on vegetation indices; (c) RF classification based on phenological characteristics; (d) RF classification based on spectral features and vegetation indices; (e) RF classification based on spectral and phenological features; (f) RF classification based on vegetation indices and phenological characteristics; (g) RF classification based on spectral indices, vegetation indices, and phenological characteristics; (h) remote sensing imagery captured on 24 November 2022.
Figure 10h presents a true-color composite remote sensing image observed by Sentinel-2 on 24 November 2022. In this image, the magenta-colored areas correspond to the SS, with the PA in a predominantly withered state during this period. The green areas near the coast represent vegetation classified as SS. This image serves for comparing the accuracy of classification results obtained by various schemes. By conducting on-site investigations and analyzing high-resolution satellite images, we compared the categorization map results of these seven combinations. Figure 10a,b,d reveal a high occurrence of misclassifications of PA as SA near the shoreline of the study area. Figure 10c shows that, in the central area, there were many instances when areas of PA were misidentified as SS. This led to the lowest overall classification accuracy compared to all other combinations. The identification of the experimental area containing SS in the northeastern half of the study area was inadequate, as shown in Figure 10f. Figure 10e,g exhibit comparable classifications, achieving the best accuracy in categorization.
Since all methods use sample data from the same study area, assessing accuracy through confusion matrices and kappa coefficients is inadequate. It was critical to determine whether there were substantial statistical differences between the methods. Table 4 lists the Z-test values and χ2-test values used for pairwise comparisons of classification algorithms. Statistically, all combinations, except Scheme 7 (RF with SP+VI+PH) vs. Scheme 5 (RF with SP+PH), had Z-values greater than 1.96 and χ2-values above the basic threshold of 3.841. This indicates that there is a significant difference between these possible combinations. The method proposed in this study (Scheme 7) showed higher classification accuracy at a 95% confidence level. However, the Z-test and χ2-test statistics indicated no significant difference between Scheme 7 vs. Scheme 5 in classification. The vegetation index selected in this paper may not play a key role in the classification of coastal wetlands, and the difference can be reflected by adding more and more effective vegetation indices (VIs).

Table 4.
Statistical significance of differences in classification accuracy between two different algorithms.
3.6. Misclassification Analysis Based on Land-Use Change Mapping
While the overall accuracy obtained from the classification results of the seven schemes suggests that the proposed method in this study can further improve accuracy, it is challenging to visibly observe the differences in the classification results from the classification result maps. In order to visually highlight the differences in classification results among other schemes, this study assumes the classification results obtained by the proposed Scheme 7 (RF with SP+VI+PH) as the ground truth. Land-use change maps and error classification maps were generated by comparing these results with those obtained by other schemes, as shown in Figure 11. From Figure 11a,b,d, it can be observed that in Scheme 7, the area classified as SA is misclassified as PA in Schemes 1, 2, and 4. Combining this observation with Figure 10h, we can conclude that PA does not grow in areas close to the coast. Therefore, the classification performance of these feature combination methods is not as good as that of Scheme 7 (RF with SP+VI+PH). Figure 11c reveals a significant difference between the classification results of Scheme 7 and Scheme 2, indicating that using only phenological features for classification may not necessarily meet the requirements of land cover classification tasks near the coastal wetlands. In Figure 11e, it can be observed that Scheme 5 and Scheme 7 yield the most similar classification results, with classification accuracies of 95.46% and 95.64%, respectively. This further illustrates that there is no significant difference between these two experimental protocols. Figure 11f suggests that Scheme 6 may misclassify some SS as PA, indicating that this combination method may not be very effective in distinguishing between PA and SS in mixed habitats.
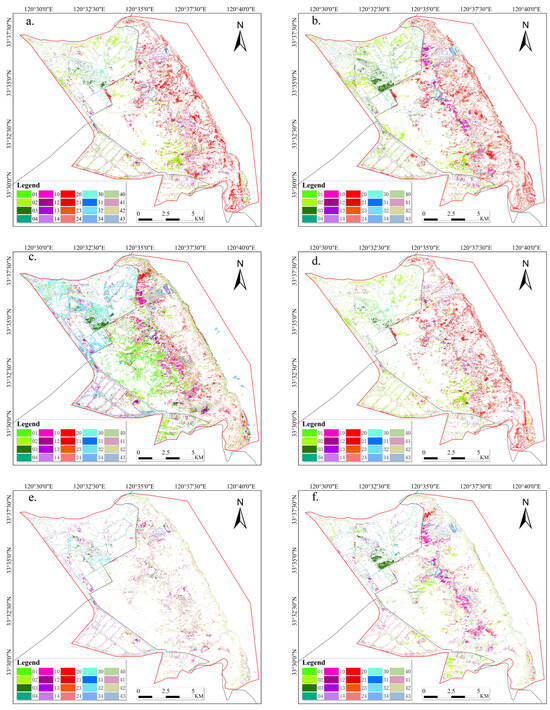
Figure 11.
Misclassification analysis based on land-use change mapping ((a) land-use change from Scheme 7 to Scheme 1; (b) land-use change from Scheme 7 to Scheme 2; (c) land-use change from Scheme 7 to Scheme 3; (d) land-use change from Scheme 7 to Scheme 4; (e) land-use change from Scheme 7 to Scheme 5; (f) land-use change from Scheme 7 to Scheme 6), where 0 represents PA, 1 represents SS, 2 represents SA, 3 represents water area, and 4 represents unused land; “01” represents the land use from PA to SS; the others are in a similar fashion.
3.7. Analysis of Misclassification Results
To solve the problem of misclassification of SA in the classification map near the coastline, we chose a subset of SA samples located near the coastline and samples located far from the coastline (as depicted in Figure 12a). The satellite image depicted in Figure 12a is a standard false-color composite image that was derived from the scene acquired on 18 May. The presence of SA is indicated by the dark red spots near the coastline in this map, illustrating its growth in those specific regions. Simultaneously, this investigation acquired the average NDVI values over time at the specific locations of the chosen samples. A dense time-series NDVI fitting model was utilized to produce NDVI fitting curves for SA at locations both close to and distant from the coastline.
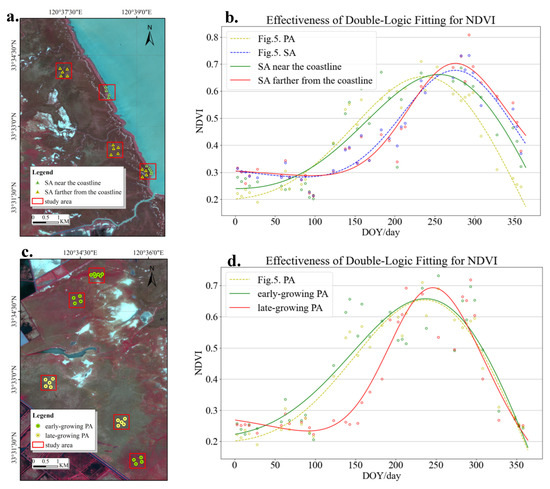
Figure 12.
Analysis of wetland vegetation misclassification.
In Figure 11b, the analysis shows the comparison of the curves for PA and SA from Figure 5 with the NDVI fitting curves for sample points in Figure 12a. Figure 12b illustrates that the regression curve of SA is shifted towards the left along the shoreline, in contrast to SA situated farther from the coast. Moreover, the highest possible value of NDVI is decreased, leading to a more accurate alignment with the NDVI fitting curve of PA. The cyclic inundation of vegetation in the intertidal zone, driven by tidal fluctuations, leads to changes in phenological traits. Because of the limited and scarce plant life in this area, the young shoots are highly susceptible to tidal influences and can be misidentified as similar species at different growth stages [14]. The misidentification of SA along the shoreline as PA is caused by the decreased extent of its phenological traits.
Similarly, we noticed discrepancies in the developmental trends of PA in various areas within the coastal wetland. Figure 12c illustrates the comparison of the NDVI fitting curves between early-growing PA and late-growing PA, as shown in Figure 12d. PA that grows later in the season demonstrates greater specificity, typically commencing growth one to two months after the PA that grows earlier in the season. Furthermore, its rate of growth is significantly higher, with less variation in the period of decline as compared to the early-growing PA. The observed variation can be attributed to environmental factors such as vegetation attributes, climatic conditions, soil salinity, and vegetation complexity in the growing region.
4. Discussion
4.1. Comparison with Previous Works
A single temporal remote sensing image for wetland classification has achieved good results [31], while the diverse growth and change characteristics of wetland vegetation cannot be fully captured by a single temporal phase. Therefore, this study considers a method introducing phenological features to more comprehensively capture information about wetland vegetation. Chao [14] and Liu [15] used phenological features such as SOS (start of season), EOS (end of season), MOS (middle of season, equivalent to POP in this study), MV (maximum value), and SV (start value) for wetland classification. Although the phenological parameters extracted in this study may differ slightly, we emphasize the integration of phenological information with various common spectral indices and vegetation indices to achieve better classification results. From the comparison between Scheme 3 and Scheme 7 in Section 3.5, it can be concluded that relying solely on phenological features for coastal wetland classification tasks may not achieve higher accuracy, due to factors such as regional characteristics, image quality, and human activities. The random forest classification method, combining multiple features, captures more details in wetlands, resulting in better classification results and demonstrating the feasibility of the proposed method in this study. The specific selection of phenological features for better classification results often shows specificity in different study areas and requires analysis based on actual conditions. This study did not delve deeper into this aspect, and the experiment only explored whether adding phenological features to time-series remote sensing data could improve the overall classification accuracy. Zhang [9] combined multiple features, including spectral features, vegetation features, and texture features, for integrated classification. The results indicated that texture features contributed little to the overall classification and could be excluded from wetland classification. Therefore, Zhang’s method can be considered similar to Scheme 4 in Section 3.5. Comparisons between Scheme 4 and other schemes, as well as the contribution weights of the phenological features shown in Figure 9, reveal that the random forest classification method proposed in this study, “SP+VI+PH”, achieves better classification accuracy. Depending on the scale of wetland classification and the requirements for land-use types, utilizing conventional remote sensing data acquired from June to September may not be suitable for vegetation classification in the coastal wetlands of Jiangsu. For the two common vegetation types in these wetlands, SS and SA, the optimal observation month is November.
4.2. Shortcomings and Future Plans
In the initial image selection, this study used 22 scenes of Sentinel-2 images with low cloud cover throughout the year 2022 for extracting phenological features. Later, by applying a cloud masking algorithm to remove interference such as clouds and shadows, the number of images was increased to 36 scenes. The overall classification accuracy significantly improved, indicating that the inclusion of additional remote sensing images allows for a more comprehensive capture of wetland vegetation’s growth characteristics. However, the removal of clouds and shadows inevitably results in the loss of information within the excluded areas. For example, November is the optimal period for observing SS, while, because of the reasons discussed above, the lack of complete information on SS in some regions might result in discrepancies and mistakes when extracting phenological characteristics for the same type of land cover. During the field investigation, we observed that interventions by national policies could potentially influence the classification results of local wetlands. For instance, in the process of national interventions to control the growth of SA on coastal mudflats, there were instances where an area previously covered by SA suddenly transformed into bare land. This occurred due to local policies involving the use of excavators to remove SA from the region. Such interventions can lead to inaccuracies in the extraction of phenological features, making it challenging to determine the accurate classification of land cover types.
This paper introduces a method that extracts phenological features by observing the entire annual growth cycle of wetland vegetation, without considering the influence of environmental factors on the classification results. Figure 10c,d elucidates that the impact of environmental factors may lead to growth variations among vegetation of the same species. In the next step, we plan to further refine the method, such as addressing different geographical locations and soil salinity for the same vegetation type by augmenting the classification categories. Additionally, we will consider factors like variations in annual precipitation and temperature changes across different years. These refinements are intended to optimize the model further. The extraction process of vegetation’s phenological features during the experiment was time-consuming because it involved extracting features pixel by pixel. This could impose limitations on the classification of large-scale wetlands.
Although common vegetation indices contribute to wetland classification, there are no significant differences between the classification methods of Scheme 7 and Scheme 5. Both schemes achieve a high level of classification accuracy, although Scheme 7 is the best as far as classification accuracy is concerned, according to several experiments. Subsequently, we will hopefully be able to further demonstrate the differences between Scheme 7 and Scheme 5 by introducing some new vegetation indices.
It is worthwhile to study whether the proposed method could be migrated to other coastal wetland areas. Hao et al. [32] demonstrated that when the time series of observed images is long enough, the classification results of even migrated samples are similar to the classification accuracy obtained from the training of locally measured samples. In this paper, the method captures the annual time-series image information to extract vegetation’s phenological features, which can comprehensively capture the differences in the growth of vegetation in different wetlands. However, the climate change caused by latitude differences alters the vegetation’s growth cycle. This will affect the accuracy of classification during migration. This method is mainly applicable to other sample sites that have similar vegetation to the types found in the coastal wetlands in this paper, and for other new vegetation types it is necessary to re-observe the growth change rules and obtain certain a priori knowledge before classification. This is because new vegetation types may need to change the length of the time series of extracted features. At the same time, new vegetation types may cause similar NDVI time series with other vegetation at a certain stage, so in the process of generalizing to other coastal wetlands, we can consider trying to extract multiple vegetation indices for time series and increasing the selection of phenological features. These efforts will require more in-depth methodological design and research. Subsequent research will also explore the use of this approach to derive multi-year categorization outcomes for coastal wetlands, assess alterations and patterns in habitats, and investigate the underlying factors contributing to these modifications.
5. Conclusions
This study provides an effective technical approach for the protection and management of coastal wetland ecosystems by constructing a dense time series of remote sensing data. This method not only reveals the complex characteristics of coastal wetland habitats but also offers precise classification and mapping results for the restoration of ecological wetlands and bird conservation. Through comprehensive analysis of the spectral features, vegetation index features, and phenological features of coastal wetland plants, this research enhances our understanding of these critical ecological areas, providing valuable information for biodiversity conservation and ecosystem service assessment. The application of the random forest method demonstrates the immense potential of machine learning in natural resource management and environmental protection, laying a solid foundation for future research and conservation efforts in coastal wetlands. The primary discoveries are summarized as follows:
- A classified map of the core zone of the Yancheng Wetland Rare Birds National Nature Reserve was obtained for the year 2022, with an overall classification accuracy of 95.64% and a kappa coefficient of 0.94.
- The combination of spectral features, vegetation indices, and phenological characteristics produced the highest level of accuracy in classification. POP, SOS, NDVIre, and mid-infrared bands (Band12 and Band11) were useful for the classification of coastal wetlands.
- The influence of tidal fluctuations on SA along the shoreline was not considered in this experiment. The misclassification of SA near the coastline in the categorization map was caused by its long-term submersion, partial submersion, and non-submersion. By comparing SA plants located near the coastline with those located farther away, we showed that the phenological magnitude of SA near the coastline was relatively smaller. This finding helps to explain why these plants are more likely to be misidentified as PA.
- Different regions within the core zone of the Yancheng Wetland Rare Birds National Nature Reserve exhibit different development patterns of PA. There is a potential 1–2-month disparity in growth between early- and late-growing PA. The variation in vegetation cover can be explained by different factors, such as vegetation characteristics, soil salinity, climate changes, and the intricate nature of the vegetation.
Author Contributions
Conceptualization, S.J. and Y.W.; methodology, Y.W. and S.J.; software, Y.W.; validation, S.J. and Y.W.; formal analysis, Y.W.; investigation, Y.W.; resources, Y.W. and S.J.; data curation, Y.W.; writing—original draft preparation, Y.W.; writing—review and editing, S.J. and G.D.; visualization, Y.W.; supervision, S.J.; project administration, S.J.; funding acquisition, S.J. and G.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Jiangsu Marine Science and Technology Innovation Project, grant number JSZRHYKJ202202.
Data Availability Statement
Sentinel-2 images were provided by the ESA Copernicus Data Center (https://scihub.copernicus.eu/dhus/#/home, accessed on 3 March 2023).
Acknowledgments
We would like to express our gratitude to the European Space Agency (ESA) for granting us access to the time-series Sentinel-2 Level-2A MSI product data. We would like to express our gratitude to the Jiangsu Provincial Forestry Bureau, the Yancheng National Nature Reserve for Rare Birds, and all of the individuals involved in the reserve for their valuable support and cooperation.
Conflicts of Interest
The authors declare no conflicts of interest.
Appendix A

Table A1.
Sentinel-2 MSI data.
Table A1.
Sentinel-2 MSI data.
| Name | Date | Cloud Cover (%) |
|---|---|---|
| S2_SR_HARMONIZED_20220103T024121 | 3 January 2022 | 16.93 |
| S2_SR_HARMONIZED_20220108T024059 | 8 January 2022 | 29.19 |
| S2_SR_HARMONIZED_20220113T024051 | 13 January 2022 | 16.13 |
| S2_SR_HARMONIZED_20220118T024029 | 18 January 2022 | 20.87 |
| S2_SR_HARMONIZED_20220128T023949 | 28 January 2022 | 47.73 |
| S2_SR_HARMONIZED_20220227T023639 | 2 February 2022 | 4.41 |
| S2_SR_HARMONIZED_20220304T023611 | 4 March 2022 | 16.66 |
| S2_SR_HARMONIZED_20220309T023549 | 9 March 2022 | 18.62 |
| S2_SR_HARMONIZED_20220324T023551 | 24 March 2022 | 0 |
| S2_SR_HARMONIZED_20220329T023549 | 29 March 2022 | 3.76 |
| S2_SR_HARMONIZED_20220403T023551 | 3 April 2022 | 3.03 |
| S2_SR_HARMONIZED_20220408T023549 | 8 April 2022 | 1.59 |
| S2_SR_HARMONIZED_20220503T023551 | 3 May 2022 | 0.09 |
| S2_SR_HARMONIZED_20220518T023549 | 18 May 2022 | 13.98 |
| S2_SR_HARMONIZED_20220523T023601 | 23 May 2022 | 0 |
| S2_SR_HARMONIZED_20220602T023601 | 2 June 2022 | 19.08 |
| S2_SR_HARMONIZED_20220607T023549 | 7 June 2022 | 0.41 |
| S2_SR_HARMONIZED_20220617T023529 | 17 June 2022 | 35.13 |
| S2_SR_HARMONIZED_20220702T023541 | 2 July 2022 | 41.39 |
| S2_SR_HARMONIZED_20220712T023541 | 12 July 2022 | 66.25 |
| S2_SR_HARMONIZED_20220722T023541 | 22 July 2022 | 22.27 |
| S2_SR_HARMONIZED_20220727T023529 | 27 July 2022 | 30.29 |
| S2_SR_HARMONIZED_20220801T023541 | 1 August 2022 | 32.59 |
| S2_SR_HARMONIZED_20220821T023541 | 21 August 2022 | 0.07 |
| S2_SR_HARMONIZED_20220826T023529 | 26 August 2022 | 61.19 |
| S2_SR_HARMONIZED_20220910T023541 | 10 September 2022 | 73.93 |
| S2_SR_HARMONIZED_20220930T023541 | 30 September 2022 | 55.15 |
| S2_SR_HARMONIZED_20221010T023621 | 10 October 2022 | 1.52 |
| S2_SR_HARMONIZED_20221015T023649 | 15 October 2022 | 41.48 |
| S2_SR_HARMONIZED_20221020T023731 | 20 October 2022 | 34.13 |
| S2_SR_HARMONIZED_20221025T023759 | 25 October 2022 | 4.44 |
| S2_SR_HARMONIZED_20221104T023849 | 4 November 2022 | 19.96 |
| S2_SR_HARMONIZED_20221124T024029 | 24 November 2022 | 21.01 |
| S2_SR_HARMONIZED_20221214T024119 | 14 December 2022 | 11.05 |
| S2_SR_HARMONIZED_20221219T024121 | 19 December 2022 | 1.84 |
| S2_SR_HARMONIZED_20221224T024119 | 24 December 2022 | 39.70 |
References
- Duan, H.L.; Yu, X.B. Research on Dynamic Changes of Endangered Waterbird Habitats in the Yellow and Bohai Seas. Acta Ecol. Sin. 2023, 43, 6354–6363. [Google Scholar]
- Mohseni, F.; Amani, M.; Mohammadpour, P.; Kakooei, M.; Jin, S.; Moghimi, A. Wetland mapping in Great Lakes using Sentinel-1/2 time-series imagery and DEM data in Google Earth Engin. Remote Sens. 2023, 15, 3495. [Google Scholar] [CrossRef]
- Duarte, C.M.; Losada, I.J.; Hendriks, I.E.; Mazarrasa, I.; Marbà, N. The role of coastal plant communities for climate change mitigation and adaptation. Nat. Clim. Change 2013, 3, 961–968. [Google Scholar] [CrossRef]
- Hou, X.J.; Feng, L.; Cchen, X.L.; Zhang, Y. Dynamics of the wetland vegetation in large lakes of the Yangtze Plain in response to both fertilizer consumption and climatic changes. ISPRS J. Photogramm. Remote Sens. 2018, 141, 148–160. [Google Scholar] [CrossRef]
- Ning, X.G.; Chang, W.T.; Wang, H.; Zhang, H.C.; Zhu, Q.D. Wetland Information Extraction in the Heilongjiang River Basin Using Google Earth Engine and Multi-source Remote Sensing Data. J. Remote Sens. 2022, 26, 386–396. [Google Scholar]
- Chen, B.; Chen, L.; Huang, B.; Michishita, R.; Xu, B. Dynamic monitoring of the Poyang Lake wetland by integrating Landsat and MODIS observations. ISPRS J. Photogramm. Remote Sens. 2018, 139, 75–87. [Google Scholar] [CrossRef]
- Fan, D.Q.; Zhao, X.S.; Zhu, W.Q.; Zheng, Z. Review on Factors Affecting the Accuracy of Plant Phenology Remote Sensing Monitoring. Prog. Geogr. 2016, 35, 304–319. [Google Scholar]
- Liu, R.Q. Coastal Wetland Classification Based on Time Series Remote Sensing Images and Vegetation Phenological Characteristics. Master’s Thesis, Ningbo University, Ningbo, China, 2022. [Google Scholar]
- Zhang, L.; Gong, Z.N.; Wang, Q.W.; Jin, D.D.; Wang, X. Wetland mapping of Yellow River Delta wetlands based on multi-feature optimization of Sentinel-2 images. J. Remote Sens. 2019, 23, 313–326. [Google Scholar] [CrossRef]
- Zheng, H.; Chen, X.T.; Song, L.J.; Fan, J.H.; Yang, X.W.; Song, J.R.; Song, T.L.; Liu, M.Y. Research on the Extraction Method of Spartina alterniflora Information in Coastal Wetlands Based on Google Earth Engine (GEE). J. Chifeng Univ. Nat. Sci. Ed. 2022, 38, 26–31. [Google Scholar]
- Sun, W.W.; Liu, W.W.; Wang, Y.M.; Zhao, R.; Huang, M.; Wang, Y.; Yang, G.; Meng, X. Progress and Prospects of Global Wetland Hyperspectral Remote Sensing Research from 2010 to 2022. J. Remote Sens. 2023, 27, 1281–1299. [Google Scholar]
- Wu, Y.; Xiao, X.; Chen, B.; Ma, J.; Wang, X.; Zhang, Y.; Zhao, B.; Li, B. Tracking the phenology and expansion of Spartina alterniflora coastal wetland by time series MODIS and Landsat images. Multimed. Tools Appl. 2020, 79, 5175–5195. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, L.; Zhao, T.; Chen, X.; Lin, S.; Wang, J.; Mi, J.; Liu, W. GWL_FCS30: Global 30 m wetland map with fine classification system using multi-sourced and time-series remote sensing imagery in 2020. Earth Syst. Sci. Data Discuss. 2022, 15, 265–293. [Google Scholar] [CrossRef]
- Chao, S.; Li, J.; Liu, Y.; Liu, Y.; Liu, Y.; Liu, R. Plant species classification in salt marshes using phenological parameters derived from Sentinel-2 pixel-differential time-series. Remote Sens. Environ. 2021, 256, 112320. [Google Scholar]
- Liu, R.Q.; Li, J.L.; Sun, C.; Sun, W.W.; Cao, L.D.; Tian, P. Vegetation Classification of Yancheng Coastal Wetlands Based on Sentinel-2 Remote Sensing Time Series Phenological Features. Acta Geogr. Sin. 2021, 76, 1680–1692. [Google Scholar]
- Tassi, A.; Vizzari, M. Object-oriented LULC classification in Google earth engine combining SNIC, GLCM, and machine learning algorithms. Remote Sens. 2020, 12, 3776. [Google Scholar] [CrossRef]
- Taddeo, S.; Dronova, I.; Depsky, N. Spectral vegetation indices of wetland greenness: Responses to vegetation structure, composition, and spatial distribution. Remote Sens. Environ. 2019, 234, 111467. [Google Scholar] [CrossRef]
- Wu, X.F.; Hua, S.H.; Zhang, S.; Gu, L.; Ma, C.; Li, C. Extraction of Winter Wheat Distribution Information Based on Multi-Phenological Feature Indices from Sentinel-2 Data. Trans. Chin. Soc. Agric. Mach. 2023, 54, 207–216. [Google Scholar]
- Gonsamo, A.; Chen, J.M.; D’Odorico, P. Deriving land surface phenology indicators from CO2 eddy covariance measurements. Ecol. Indic. 2013, 29, 203–207. [Google Scholar] [CrossRef]
- Rogers, C.; Chen, J.M.; Croft, H.; Gonsamo, A.; Luo, X.; Bartlett, P.; Staebler, R.M. Daily leaf area index from photosynthetically active radiation for long term records of canopy structure and leaf phenology. Agric. For. Meteorol. 2021, 304, 108407. [Google Scholar] [CrossRef]
- Zhang, Q.; Kong, D.; Shi, P.; Singh, V.P.; Sun, P. Vegetation phenology on the Qinghai-Tibetan Plateau and its response to climate change (1982–2013). Agric. For. Meteorol. 2018, 248, 408–417. [Google Scholar] [CrossRef]
- Wu, C.; Peng, D.; Soudani, K.; Siebicke, L.; Gough, C.M.; Arain, M.A.; Bohrer, G.; Lafleur, P.M.; Peichl, M.; Gonsamo, A.; et al. Land surface phenology derived from normalized difference vegetation index (NDVI) at global FLUXNET sites. Agric. For. Meteorol. 2017, 233, 171–182. [Google Scholar] [CrossRef]
- Prasad, P.; Loveson, V.J.; Kotha, M. Probabilistic coastal wetland mapping with integration of optical, SAR and hydro-geomorphic data through stacking ensemble machine learning model. Ecol. Inform. 2023, 77, 102273. [Google Scholar] [CrossRef]
- Wen, L.; Mason, T.J.; Ryan, S.; Ling, J.E.; Saintilan, N.; Rodriguez, J. Monitoring long-term vegetation condition dynamics in persistent semi-arid wetland communities using time series of Landsat data. Sci. Total Environ. 2023, 905, 167212. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Genuer, R.; Poggi, J.M.; Tuleau-Malot, C. Variable selection using random forests. Pattern Recognit. Lett. 2010, 31, 2225–2236. [Google Scholar] [CrossRef]
- Congalton, R.G. Remote sensing and geographic information system data integration: Error sources and research issues. Photogramm. Eng. Remote Sens. 1991, 57, 677–687. [Google Scholar]
- Foody, G.M. Thematic map comparison. Photogramm. Eng. Remote Sens. 2004, 70, 627–633. [Google Scholar] [CrossRef]
- Agresti, A. An Introduction to Categorical Data Analysis; Wiley: New York, NY, USA, 1996. [Google Scholar]
- Congalton, R.G.; Oderwald, R.G.; Mead, R.A. Assessing Landsat classification accuracy using discrete multivariate statistical techniques. Photogramm. Eng. Remote Sens. 1983, 49, 1671–1678. [Google Scholar]
- Xing, H.; Niu, J.; Feng, Y.; Hou, D.; Wang, Y.; Wang, Z. A coastal wetlands mapping approach of Yellow River Delta with a hierarchical classification and optimal feature selection framework. Catena 2023, 223, 106897. [Google Scholar] [CrossRef]
- Hao, P.; Di, L.; Zhang, C.; Guo, L. Transfer Learning for Crop classification with Cropland Data Layer data (CDL) as training samples. Sci. Total Environ. 2020, 733, 138869. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).