Abstract
Background: Nonsyndromic cleft lip with or without palate (NSCL/P) is a multifactorial and common birth malformation caused by genetic and environmental factors, as well as by teratogens. Genome-wide association studies found genetic variations with modulatory effects of NSCL/P formation in Chinese and Iranian populations. We aimed to identify the susceptibility of single-nucleotide polymorphisms (SNPs) to nonsyndromic cleft lip with or without palate in the Indian population. Material and Methods: The present study was conducted on NSCL/P cases and controls. Genomic DNA was extracted from peripheral blood and Axiom- Precision Medicine Research Array (PMRA) was performed. The Axiom-PMRA covers 902,527 markers and several thousand novel risk variants. Quality control-passed samples were included for candidate genetic variation identification, gene functional enrichment, and pathway and network analysis. Results: The genome-wide association study identified fourteen novel candidate gene SNPs that showed the most significant association with the risk of NSCL/P, and eight were predicted to have regulatory sequences. Conclusion: The GWAS study showed novel candidate genetic variations in NSCL/P formations. These findings contribute to the understanding of genetic predisposition to nonsyndromic cleft lip with or without palate.
1. Introduction
Orofacial clefts (OFCs) are the second most common congenital birth defect in humans [1]. Despite the progress in surgical treatment, the disease has lifelong consequences for the health and social integration of affected people. Worldwide incidence of cleft lip and palate is 1 per 700 live births. However, race, ethnicity, geographical regions, environmental factors and socioeconomic status affect the occurrence of CL/P [2]. The rate of incidences is higher in Asians (European ancestry), and lower in African populations [3]. Proper management of OFCs requires multidisciplinary care and places a significant burden on not only the affected, but also their family, social, and healthcare systems [4]. Therefore, there is great interest in identifying noncleft phenotypic markers for predicting risk in general populations and developing prevention strategies.
OFCs subtypes are defined according to the affected anatomical structures, and can occur as a part of the Mendelian syndrome and isolated nonsyndromic form. Nonsyndromic Orofacial Cleft (NSOFC) is the most common form of Orofacial Clefts, with approximately 70% of cases [1].
Nonsyndromic cleft lip and palate is further classified based on structural morphology—nonsyndromic cleft lip and palate (NSCLP), nonsyndromic cleft lip only (NSCLO) and cleft palate (NSCP) [5]. The causes of NSCLP are multifactorial, the involvement of genetic risk factors, exposure to teratogens and possible genetic–environmental interactions are linked with disease susceptibility. The estimated contribution of all integrated genetic factors to NSCL/P is 90% [6]. M. T. Colobourne et al. stated that CLP and isolated CP cases are governed by environmental factors, and hence it has beocme a necessity explore the interconnection of candidate genes and environmental factors associated with the disease progression. M.T Colobourne et al., in a review article, also highlighted the association of inheritance pattern and genetic loci with this multifactorial disorder [7]. A recent study conducted by Paradowska-Stolarz et al., in 2015, also highlighted that the genetic alternations in the sequences of muscle segment homeobox gene (MSX1) is influenced by environmental factors, thereby leading to the development of deformities including cleft lip palate [8]. Previous studies have also highlighted that interaction of paired box gene 9 (PAX9) with numerous genes playing important roles in different pathways gives rise to such deformities, especially during palate elevation and fusion events [9].
In the past, linkage and candidate gene association studies were performed with traditional genomic techniques. These studies have only identified two common genetic variants that are risk factors for NSCLP. Risk loci identified in the meta-analysis of linkage studies are interferon regulatory factor 6 IRF6 (1q32.2) and forkhead box E1 FOXE1 (9q22.33) [10,11]. Several significant genomic findings in NSCLP have arisen from high-throughput tools such as genome-wide association (GWAS) in the recent genomic era. Studies so far have identified a total of 40 genome-wide significant risk loci for NSLC/P in various populations. Of these, 37 loci were detected by GWAS, metadata analysis of GWAS, or subsequent studies [12,13,14,15,16,17,18,19,20,21,22,23,24]. In the genomic era, new sequencing techniques including whole-exome sequencing have identified cadherin-1 (CHD1) and some other new susceptible genes in NSCLP [25,26,27].
The racial differences must be considered for the fundamental identification of genomic susceptibility to complex diseases in a multiethnic population [28]. There is no GWAS study that has been performed and reported on the Indian population. An advancement in genomic array techniques, the Axiom-Precision Medicine Research Array (PMRA) is comprehensive genotyping array based on South Asian and East Asian populations, containing more than 750,000 markers, including common and rare disease-associated variants catalogued in public databases such as the GWAS catalog and ClinVar (Thermo Fisher Scientific, Walthman, MA, USA). In the present study, we tried to identify the genetic variants predicting nonsyndromic cleft lip and palate (NSCL/P) in the Indian population. The best of our knowledge, this is the first GWAS report that aims to provide a better understanding of NSCLP formation with Asia-specific PMRA.
2. Material & Methods
2.1. Sample Collection
In this study, clinically confirmed nonsyndromic cleft lip and palate (N = 86; age of patients ranging from 0 to 2 years) patients and age–sex-matched patients ranging from 0 to 2 years, including male and female healthy controls (N = 10) were recruited for the study, after signing an informed consent form. Patients were recruited from Medical Genetics OPD, Sanjay Gandhi Postgraduate Institute of Medical Sciences, Lucknow (India) and Plastic, craniofacial & microsurgery OPD, Vivekananda polyclinic & Institute of Medical Sciences, Lucknow (UP). The exclusion criteria for NSCLP cases were patients with any other history of developmental disorder, syndromic forms of NSCLP (e.g., eye, brain, limb anomalies and cardiac defects). The healthy controls were Indian-origin children without family history of orofacial cleft.
The study was approved by the Institutional Medical Research Ethics Committee of the Sanjay Gandhi Postgraduate Institute of Medical Sciences, India, (2018-107-EMP-EXP), and a written informed consent was taken from the guardians.
Genotyping was conducted for 86 cases and 10 control samples using Affymetrix PMRA array.
2.2. DNA Isolation
Genomic DNA was extracted from 3 mL peripheral venous blood samples of patient and controls, with DNA extracted by using standard phenol–chloroform method (Nasiri, 2005). The quality of DNA was assessed on 1% agarose gel electrophoresis, and quantity of DNA was measured by NanoDropTM.
2.3. AxiomTM PMRA Assay
The NSCLP patient samples were genotyped using the AxiomTM Asia-precision medicine Research Array (PMRA) kit (Thermo Fisher Scientific, Waltham, MA, USA) for genotyping, using the Affymatrix GeneTitan platform. It contains over 750,000 SNPs including 50,000 novel markers covering South and East Asian populations based on the human genome version 19 (build 37). Target probes were prepared using high-quality DNA (20 ng) in each well according to the standard protocol of the Affymatrix Axiom 2.0 assay. In addition, DNA samples were amplified, fragmented and hybridised on a chip, followed by single-base expansion via DNA ligation and signal modification. Samples were stained and scanned using Affymatrix GeneTitan multichannel instrument.
2.4. Quality Control
Axiom Analysis suite version 1.0 (http://media.affymetrix.com/support/downloads/manuals/Axiom_analysis_suite_user_guide.pdf, Last accessed 13 September 2020) was used for detection of allele-specific SNPs, and a best-practice workflow was used for quality-control analysis of the dataset. A total of 845,858 out of 888,799 variants, including small insertions/deletions (indels), passed the quality-control test with a genotype call rate of ≥ 95%. Samples with DQC < 0.82 and QC call rate < 97% were not considered for further analysis. SNP QC was performed under default parameters for humans, that is, cr-cutoff ≥ 95; fld-cutoff ≥ 3.6; het-so-cutoff ≥ 0.1; het-so-otv-cutoff ≥ 0.3; hom-ro-1-cutoff ≥ 0.6; hom-ro-2-cutoff ≥ 0.3.
2.5. Allele Calling and Data Analysis
The genotyping workflow was used to perform genotyping of the imported .CELL files. Finally, the summary-only workflow was used to summarise the intensity details of the probe set for use in further analysis tools. Plink software was used to perform the GWAS analysis and Haploview was used for generating Manhattan plot.
3. Results
A total of eight hundred ten thousand nine hundred six single-nucleotide variants were genotyped, with a genotyping rate of 0.99. The data were filtered for missing genotypes, and one lakh two thousand eight hundred eighty-seven SNPs were removed. Similarly, three lakhs seventy-one thousand eight hundred and ten SNPs with a minor allele frequency of less than 0.01 and 278 SNPs deviating Hardy–Weinberg equilibrium, with a p-value of 0.001, were removed from further analysis. Three lakhs thirty-five thousand nine hundred seventy-one SNPs passed the filtering for analysis.
GWAS results: Of three lakhs thirty-five thousand nine hundred seventy-one SNPs analysed for their genetic association with the OFCs, rs36019844 showed a significant association (FDR Benjamini and Hochberg p-value of 0.001688, Figure 1). Additionally, 13 other SNPs were also found to be associated with the OFC (Table 1). Among these 13 SNPs, 6 of them were in the intronic region of the genes ZNF503 (rs74146603), SLC35F3 (rs146076295), ENTPD1 (rs117864318), PIP4K2A (rs80087712), and EHBP1 (rs115646634).
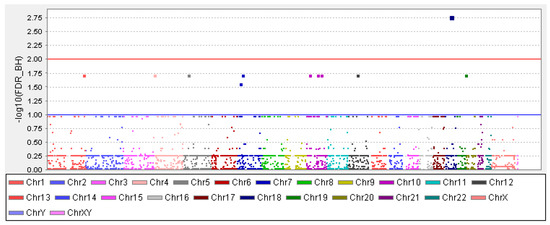
Figure 1.
Manhattan plot with a blue line and red line indicating the threshold for genome-wide significance. The x-axis has the chromosome numbers and -log 10 FDR–BH (false discovery rate–Benjamini and Hochberg) adjusted p-value in the y-axis.

Table 1.
List of SNPs that showed a significant association with an orofacial cleft in GWAS.
Since significant SNPs are found in the intronic and intergenic region, we predicted a gene regulatory element’s presence using RegulomeDB. Out of 14 SNPs, 8 were predicted to have regulatory sequences in them, as given in Table 2. The list of deciphered morbid genes that are located in the 1 mb region before and after the significant SNPs are shown in Table 3.

Table 2.
RegulomeDB score for the SNPs that showed significance in the GWAS analysis.

Table 3.
List of deciphered morbid genes that are located in the 1 mb region before and after the significant SNPs.
4. Discussion
Orofacial clefts are a developmental anomaly that occur during pregnancy. The body tissue that makes up the lips and palate fails to join completely before birth. Such failure results in a small-to-large opening originating in the lip and going through the nose. The clefting can be unilateral or bilateral. The clefting in newborns disturbs their feeding and speaking ability.
Though numerous studies are available to explain the aetiology of the CLP, we do not have a clear understanding of it. This poor understanding is because of the multifactorial nature of the condition. To a large extent, a genetic cause for the syndromic form of CLP is identifiable. However, identifying the causative factor in isolated cases is challenging even though numerous genetic variants have been attributed to CL/P [29]. Therefore, this study was undertaken to find the genetic variants underlying the isolated cases of CL/P using Affymetrix PMRA array.
In this study, we performed a genome-wide analysis of SNPs using the Affymetrix PMRA array in a cohort of 86 cases and 10 control subjects. We identified 14 SNPs with statistical significance that are distributed across chromosomes 1, 2, 4, 5 (2 SNPs), 7 (2 SNPs), 10 (3 SNPs), 12, 17, 18 and 19. Except for rs80357922, all other variants were located in the intronic or intergenic region.
The most statistically significant signal was obtained for rs36019844, a variant located in MIR924HG, or Long Intergenic Non-Protein-Coding RNA gene. Unfortunately, until now, no regulatory function of MIR924HG has been characterized. We tried to identify any gene that was located 1 MB upstream and downstream of the identified SNP. The MOCOS gene was found, and it encodes for Molybdenum Cofactor Sulphur-transferase enzyme, involved in the activation of the xanthine dehydrogenase (XDH) and aldehyde oxidase (AO) enzymes. Xanthinuria Type II and Xanthinuria are diseases associated with the MOCOS gene.
Interestingly, the BRCA1 gene showed a significant association, emphasizing that subjects born with CLP are susceptible to cancer and have shorter lifespan [30,31,32]. The genes that regulate development are frequently associated with cancers and variations in genes critical functions, such as DNA damage and repair, including BRCA1, increase susceptibility to NSCLP [33,34]. In a Denmark-based study, it was reported that breast and brain cancers are frequent among female and primary lung cancer among male CLP subjects [35]. A recent study by Li A. et al., in 2019, attempted to investigate the association between transcription factors, miRNA’s and cleft lip genes by constructing miRNA–TF coregulatory networks, and identified 10 crucial genetic markers in the signalling pathways regulating pluripotency of stems cells. The study further concluded by highlighting the critical role of miRNAs, namely hsa-mir-27b and hsa-mir-497, in triggering the expression of the Wnt pathway, which is involved in the occurrence of cleft lip [36].
Most identified SNPs are in the intronic and intergenic region, and we tried to find out whether such regions have regulatory elements that may act as cis-regulatory elements for nearby genes. RegulomeDB score was estimated for each of the SNPs. Four SNPs (rs113361480, rs10254958, rs74146603 and rs80087712) had a rank of 3a, which signifies that they are probably a TF-binding site, DNase peak and a motif element, while rs146076295, rs116146139, rs80357922 and rs8102243 had a rank of 4, which indicates that these SNPs are associated with transcription factor binding and DNase peaks.
5. Conclusions
Our study demonstrated that rs36019844 at loci 18q12.2 showed the highest statistical significance for its association with the orofacial cleft. Following which, 13 other SNPs (Table 1) showed significance. However, our study also highlighted the association of BRACA1 gene with CLP subjects, thus further providing the emphasis on the risk of cancer development in such cases.
Limitation of Study
The current study is region-centred, however, to establish strong evidence for the association, further detailed analysis of a larger cohort in different ethnic groups is essential.
Author Contributions
Conceptualization, K.K.A. and S.A.; methodology, K.K.A.; formal analysis, K.K.A. and S.M.; investigation, K.K.A. and S.M.; resources, K.K.A., S.A. and A.A. (Amit Agarwal); writing-original draft preparation, K.K.A., S.M.; writing-review and editing, K.K.A., S.M., A.A. (Ambreen Asim) and S.A.; supervision, S.A. and A.A. (Amit Agarwal); funding acquisition, K.K.A. and S.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Department of Science and Technology-Inspire Fellowship programme (Kapil Kumar Avasthi/IF160538) by the Government of India.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Ethics Committee of Sanjay Gandhi Postgraduate Institute of Medical Sciences, Lucknow (India) (Approval No. 2018-107-EMP-EXP).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Data are available upon reasonable request.
Acknowledgments
The author would like to thank DST-INSPIRE, Ministry of Science and Technology, India Sanjay Gandhi Post Graduate Institute of Medical Sciences SGPGIMS, Lucknow, Uttar Pradesh and the smile train foundation.
Conflicts of Interest
The authors declare that they have no competing interests.
References
- Mangold, E.; Ludwig, K.U.; Nöthen, M.M. Breakthroughs in the genetics of orofacial clefting. Trends Mol. Med. 2011, 17, 725–733. [Google Scholar] [CrossRef] [PubMed]
- Gorlin, R.J.; Cohen, M.M., Jr.; Hennekam, R.C.M. Syndromes of the Head and Neck; Oxford University Press: New York, NY, USA, 2001. [Google Scholar]
- Carroll, K.; Mossey, P.A. Anatomical Variations in Clefts of the Lip with or without Cleft Palate. Plast. Surg. Int. 2012, 2012, 542078. [Google Scholar] [CrossRef][Green Version]
- Mossey, P.; Little, J. Addressing the challenges of cleft lip and palate research in India. Indian J. Plast. Surg. 2009, 42, S9–S18. [Google Scholar] [CrossRef] [PubMed]
- Harville, E.W.; Wilcox, A.J.; Lie, R.T.; Vindenes, H.; Abyholm, F. Cleft lip and palate versus cleft lip only: Are they distinct defects? Am. J. Epidemiol. 2005, 162, 448–453. [Google Scholar] [CrossRef] [PubMed]
- Grosen, D.; Bille, C.; Petersen, I.; Skytthe, A.; von Bornemann Hjelmborg, J.; Pedersen, J.K.; Murray, J.C.; Christensen, K. Risk of oral clefts in twins. Epidemiology 2011, 22, 313–319. [Google Scholar] [CrossRef]
- Cobourne, M.T. The complex genetics of cleft lip and palate. Eur. J. Orthod. 2004, 26, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Paradowska-Stolarz, A. MSX1 gene in the etiology orofacial deformities. Postepy Hig. Med. Dosw. (Online) 2015, 69, 1499–1504. [Google Scholar] [PubMed]
- Li, R.; Chen, Z.; Yu, Q.; Weng, M.; Chen, Z. The Function and Regulatory Network of Pax9 Gene in Palate Development. J. Dent. Res. 2019, 98, 277–287. [Google Scholar] [CrossRef] [PubMed]
- Rahimov, F.; Program, N.C.S.; Marazita, M.L.; Visel, A.; Cooper, M.E.; Hitchler, M.J.; Rubini, M.; Domann, F.; Govil, M.; Christensen, K.; et al. Disruption of an AP-2alpha binding site in an IRF6 enhancer is associated with cleft lip. Nat. Genet. 2008, 40, 1341–1347. [Google Scholar] [CrossRef] [PubMed]
- Moreno, L.M.; Mansilla, M.A.; Bullard, S.A.; Cooper, M.E.; Busch, T.D.; Machida, J.; Johnson, M.K.; Brauer, D.; Krahn, K.; Daack-Hirsch, S.; et al. FOXE1 association with both isolated cleft lip with or without cleft palate, and isolated cleft palate. Hum. Mol. Genet. 2009, 18, 4879–4896. [Google Scholar] [CrossRef] [PubMed]
- Birnbaum, S.; Ludwig, K.U.; Reutter, H.; Herms, S.; Steffens, M.; Rubini, M.; Baluardo, C.; Ferrian, M.; De Assis, N.A.; Alblas, M.A.; et al. Key susceptibility locus for nonsyndromic cleft lip with or without cleft palate on chromosome 8q24. Nat. Genet. 2009, 41, 473–477. [Google Scholar] [CrossRef] [PubMed]
- Grant, S.F.; Wang, K.; Zhang, H.; Glaberson, W.; Annaiah, K.; Kim, C.E.; Bradfield, J.P.; Glessner, J.; Thomas, K.A.; Garris, M.; et al. A genome-wide association study identifies a locus for nonsyndromic cleft lip with or without cleft palate on 8q24. J. Pediatr. 2009, 155, 909–913. [Google Scholar] [CrossRef] [PubMed]
- Mangold, E.; Ludwig, K.U.; Birnbaum, S.; Baluardo, C.; Ferrian, M.; Herms, S.; Reutter, H.; de Assis, N.A.; Chawa, T.A.; Mattheisen, M.; et al. Genome-wide association study identifies two susceptibility loci for nonsyndromic cleft lip with or without cleft palate. Nat. Genet. 2010, 42, 24–26. [Google Scholar] [CrossRef]
- Beaty, T.H.; Murray, J.C.; Marazita, M.L.; Munger, R.G.; Ruczinski, I.; Hetmanski, J.B.; Liang, K.Y.; Wu, T.; Murray, T.; Fallin, M.D. A genome-wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4. Nat. Genet. 2010, 42, 525–529. [Google Scholar] [CrossRef]
- Ludwig, K.U.; Mangold, E.; Herms, S.; Nowak, S.; Reutter, H.; Paul, A.; Becker, J.; Herberz, R.; Chawa, T.A.; Nasser, E.; et al. Genome-wide meta-analyses of nonsyndromic cleft lip with or without cleft palate identify six new risk loci. Nat. Genet. 2012, 44, 968–971. [Google Scholar] [CrossRef]
- Beaty, T.H.; Taub, M.A.; Scott, A.F.; Murray, J.C.; Marazita, M.L.; Schwender, H.; Parker, M.; Hetmanski, J.B.; Balakrishnan, P.; Mansilla, M.A.; et al. Confirming genes influencing risk to cleft lip with/without cleft palate in a case-parent trio study. Hum Genet. 2013, 132, 771–781. [Google Scholar] [CrossRef]
- Sun, Y.; Huang, Y.; Yin, A.; Pan, Y.; Wang, Y.; Wang, C.; Du, Y.; Wang, M.; Lan, F.; Hu, Z.; et al. Genome-wide association study identifies a new susceptibility locus for cleft lip with or without a cleft palate. Nat. Commun. 2015, 6, 6414. [Google Scholar] [CrossRef]
- Ludwig, K.U.; Ahmed, S.T.; Böhmer, A.C.; Sangani, N.B.; Varghese, S.; Klamt, J.; Schuenke, H.; Gültepe, P.; Hofmann, A.; Rubini, M.; et al. Meta-analysis Reveals Genome-Wide Significance at 15q13 for Nonsyndromic Clefting of Both the Lip and the Palate, and Functional Analyses Implicate GREM1 As a Plausible Causative Gene. PLoS Genet. 2016, 12, e1005914. [Google Scholar] [CrossRef] [PubMed]
- Leslie, E.J.; Carlson, J.; Shaffer, J.R.; Feingold, E.; Wehby, G.; Laurie, C.A.; Jain, D.; Laurie, C.C.; Doheny, K.F.; McHenry, T.; et al. A multi-ethnic genome-wide association study identifies novel loci for non-syndromic cleft lip with or without cleft palate on 2p24.2, 17q23 and 19q13. Hum. Mol. Genet. 2016, 25, 2862–2872. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Zuo, X.; He, M.; Gao, J.; Fu, Y.; Qin, C.; Meng, L.; Wang, W.; Song, Y.; Cheng, Y.; et al. Genome-wide analyses of non-syndromic cleft lip with palate identify 14 novel loci and genetic heterogeneity. Nat. Commun. 2017, 8, 14364. [Google Scholar] [CrossRef] [PubMed]
- Mostowska, A.; Gaczkowska, A.; Żukowski, K.; Ludwig, K.U.; Hozyasz, K.; Wojcicki, P.; Mangold, E.; Böhmer, A.C.; Heilmann-Heimbach, S.; Knapp, M.; et al. Common variants in DLG1 locus are associated with non-syndromic cleft lip with or without cleft palate. Clin. Genet. 2018, 93, 784–793. [Google Scholar] [CrossRef]
- Ludwig, K.U.; Böhmer, A.C.; Bowes, J.; Nikolić, M.; Ishorst, N.; Wyatt, N.; Hammond, N.L.; Gölz, L.; Thieme, F.; Barth, S.; et al. Imputation of orofacial clefting data identifies novel risk loci and sheds light on the genetic background of cleft lip ± cleft palate and cleft palate only. Hum. Mol. Genet. 2017, 26, 829–842. [Google Scholar] [CrossRef] [PubMed]
- Leslie, E.J.; Carlson, J.C.; Shaffer, J.R.; Butali, A.; Buxó, C.J.; Castilla, E.E.; Christensen, K.; Deleyiannis, F.W.B.; Field, L.L.; Hecht, J.T.; et al. Genome-wide meta-analyses of nonsyndromic orofacial clefts identify novel associations between FOXE1 and all orofacial clefts, and TP63 and cleft lip with or without cleft palate. Hum. Genet. 2017, 136, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Bureau, A.; Parker, M.; Ruczinski, I.; Taub, M.A.; Marazita, M.L.; Murray, J.C.; Mangold, E.; Noethen, M.M.; Ludwig, K.U.; Hetmanski, J.B.; et al. Whole exome sequencing of distant relatives in multiplex families implicates rare variants in candidate genes for oral clefts. Genetics 2014, 197, 1039–1044. [Google Scholar] [CrossRef]
- Cox, L.L.; Cox, T.C.; Uribe, L.M.M.; Zhu, Y.; Richter, C.T.; Nidey, N.; Standley, J.M.; Deng, M.; Blue, E.; Chong, J.X.; et al. Mutations in the Epithelial Cadherin-p120-Catenin Complex Cause Mendelian Non-Syndromic Cleft Lip with or without Cleft Palate. Am. J. Hum. Genet. 2018, 102, 1143–1157. [Google Scholar] [CrossRef] [PubMed]
- Cox, T.C.; Lidral, A.C.; McCoy, J.C.; Liu, H.; Cox, L.L.; Zhu, Y.; Anderson, R.D.; Uribe, L.M.M.; Anand, D.; Deng, M.; et al. Mutations in GDF11 and the extracellular antagonist, Follistatin, as a likely cause of Mendelian forms of orofacial clefting in humans. Hum. Mutat. 2019, 40, 1813–1825. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Shu, Y.; Cai, Y.D. Genetic differences among ethnic groups. BMC Genom. 2015, 16, 1093. [Google Scholar] [CrossRef] [PubMed]
- Dixon, M.J.; Marazita, M.L.; Beaty, T.H.; Murray, J.C. Cleft lip and palate: Understanding genetic and environmental influences. Nat. Rev. Genet. 2011, 12, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Christensen, K.; Juel, K.; Herskind, A.M.; Murray, J.C. Long term follow up study of survival associated with cleft lip and palate at birth. BMJ 2004, 328, 1405. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]
- Vieira, A.R. Unraveling human cleft lip and palate research. J. Dent. Res. 2008, 87, 119–125. [Google Scholar] [CrossRef]
- Kobayashi, G.S.; Alvizi, L.; Sunaga, D.Y.; Francis-West, P.; Kuta, A.; Almada, B.V.P.; Ferreira, S.G.; De Andrade-Lima, L.C.; Bueno, D.F.; Raposo-Amaral, C.E.; et al. Susceptibility to DNA damage as a molecular mechanism for non-syndromic cleft lip and palate. PLoS ONE. 2013, 8, e65677. [Google Scholar]
- Mostowska, A.; Hozyasz, K.; Wójcicki, P.; Galas-Filipowicz, D.; Lasota, A.; Dunin-Wilczyńska, I.; Lianeri, M.; Jagodzinski, P. Genetic variants in BRIP1 (BACH1) contribute to risk of nonsyndromic cleft lip with or without cleft palate. Birth. Defects Res. A Clin. Mol. Teratol. 2014, 100, 670–678. [Google Scholar] [CrossRef]
- Bille, C.; Winther, J.F.; Bautz, A.; Murray, J.C.; Olsen, J.; Christensen, K. Cancer risk in persons with oral cleft—A population-based study of 8,093 cases. Am. J. Epidemiol. 2005, 161, 1047–1055. [Google Scholar] [CrossRef]
- Li, A.; Qin, G.; Suzuki, A. Network-based identification of critical regulators as putative drivers of human cleft lip. BMC Med. Genomics. 2019, 12, 16. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).