Abstract
Understanding the function of a locus is an issue in molecular biology. Although numerous molecular data have been generated in the last decades, it remains difficult to grasp how these data are related at a locus. In this study, we describe an analytical workflow that can solve this problem using the knowledge available at the single-nucleotide polymorphism (SNP) level. The underlying algorithm uses SNPs as connectors to link biological entities and identify correlations between them through a joint bioinformatics/statistics approach. We demonstrate its application in finding the mechanism whereby a mutation causes a phenotype and in revealing the path whereby a gene is regulated and impacts a phenotype. We translate our workflow into publicly available shell scripts. Our approach provides a basic framework to solve the information overload problem in biology surrounding the annotation of a locus and is a step toward repurposing GWAS data for new applications.
1. Introduction
Over the past several years, high-throughput studies have generated omics data for various biological entities, including functional features and phenotypes. These data are usually generated, analyzed and published separately due to logistical/technical restraints. Genome browsers then add these data to their repertoire and make them available as annotation tracks. This, however, has created an information overload problem. For example, a biologist that wishes to know the function of a locus starts the task by looking at a stack of annotation tracks provided by a genome browser and investigates their connections visually. However, this is a tedious task and often not fulfilling. In this study, we describe a different approach that statistically evaluates the relationship between biological entities that converge at a locus and provides the user with a report for decision making.
Our approach relies on summary association statistics from GWAS studies, which are becoming increasingly available for various functional features and phenotypes. GWAS studies uniformly report their association findings with regard to SNPs. Therefore, SNPs can be used as common identifiers to investigate correlation between biological entities. This is valuable because, by scanning various entities converging at a locus and identifying those that interact, we can understand the function of a locus, which could represent a rare variant (mutation), a gene or an unannotated genomic region. In the next sections, we describe our approach and demonstrate its application through known examples for a rare variant and a gene. We provide freely available shell scripts that carry out the tasks and generate the report.
2. Methods
Advances in sequencing technology allow us to capture rare variants efficiently; however, often, it remains elusive among the identified variants which variant causes the phenotype. Furthermore, it is important to know the mechanism whereby a rare variant triggers a phenotypic change for therapeutic purposes.
From the molecular perspective, a rare variant is expected to cause a phenotype by disrupting a functional feature, i.e.,
Rare variant → Functional feature → Phenotype
Therefore, if we detect that the above condition is true for a variant;
- (i)
- We have identified the variant that causes the phenotype
- (ii)
- We have identified the underlying mechanism
However, how can we relate a rare variant to a functional feature and the functional feature to the phenotype? The solution is SNPs. Traditionally, SNPs have been used to study both the genetics of phenotypes and the genetics of functional features. Therefore, if we can find a set of SNPs in a region that meet the following criteria, we have fulfilled our aims ((i) and (ii)):
- (a)
- The location of the rare variant overlaps with the coordinate of the SNP set
- (b)
- The SNP set is associated with the functional feature and the phenotype
- (c)
- The functional feature causes the phenotype
Based on these requirements, we devise the following analytical pipeline (Figure 1). We review each step and the reasoning behind it below.
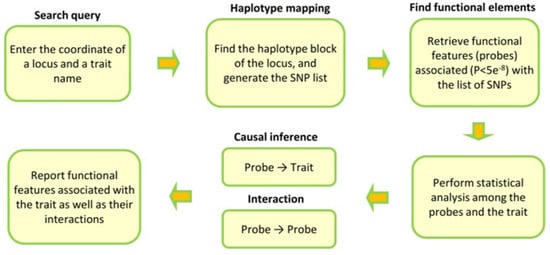
Figure 1.
The design of our workflow to understand the function of a locus and its impact on a phenotype. The search starts by finding the haplotype block that the queried locus resides within. Our algorithm then uses SNPs in the haplotype block to find functional features (probes) associated with them (p < 5 × 10−8). The next step is to test the association between the probes and the trait using Mendelian randomization (MR). Our algorithm also uses MR to test whether the identified probes act together (interaction) or independently. The final step is to generate a report that summarizes the nature and the magnitude of association between each probe and the trait as well as between the probes. In Figures 2 and 3, we provide examples and interpret the meaning of the statistics. A detailed description of each step is provided in the Section 2.
Haplotype mapping: The search starts by finding the haplotype block that the queried mutation resides within. After finding the haplotype block, our algorithm retrieves the list of SNPs within the block to identify corresponding functional features. A haplotype block is a genomic region whereby SNPs within it are in linkage disequilibrium (LD). Haplotype mapping has been traditionally used to identify genes for Mendelian diseases. A mutation that arises in a block is expected to cause the disease by impacting the function of the block. Therefore, haplotype mapping is a viable step towards narrowing the search space for functional features.
We estimated the boundaries of human haplotypes using PLINK (v 1.9) [1], based on the definition of blocks suggested by Gabriel et al. [2] and using the European sample (n = 503) of the 1000 Genomes Project. The majority of the existing GWAS data come from studies conducted in European populations and choosing a different population could cause downstream issues such as population stratification; besides, the average size of haplotype blocks in the European population is higher than in the African and Asian populations, which is beneficial in genetic studies. Nonetheless, our pipeline is flexible and if a researcher wishes to use a different definition of a block or use data from another population, this can be done by simply replacing the input file for haplotype blocks with another file.
It is also important to note that not all of the human genome is within haplotype blocks; therefore, in situations where a variant cannot be assigned to a haplotype block, our algorithm searches for nearby SNPs that are within 5 Kbp of the mutation (locus), and if the search result is null, it increments its search window by 10 Kbp and stops until it reaches the search window of 45 Kbp (the largest size of an average haplotype block across human populations) [2].
Finding functional features: The list of SNPs obtained from the previous step then is used to identify the functional features (probes) associated (p < 5 × 10−8) with them. We use the SMR software (version 1.03) [3] for this step because it stores quantitative trait locus (QTL) data for functional features in binary format; moreover, it provides flexible options to query the data. QTL data can be obtained from previous studies; we also provide a collection of QTL data for different types of functional features (see the Data/Code Availability section).
Statistical analysis: The aim of this stage is to test whether any of the functional features obtained from the previous step are associated with the studied phenotype. For this purpose, we use Mendelian randomization, which can infer causality between a functional feature and a trait. QTLs included in the instrument for the MR must be non-pleiotropic (p < 0.01), be associated with the functional feature (p < 5 × 10−8) and be in linkage equilibrium (r2 < 0.05). MR analysis was performed using the GSMR algorithm implemented in GCTA software (version 1.92) [4]. During this stage, if we find a significant association between a functional feature and a trait, we can conclude that our queried variant/locus impacts the trait through the functional feature.
MR analysis requires GWAS summary input files in GSMR format. For functional features, our workflow automatically generates such files from the SMR binary files; however, for a phenotype, this file must be obtained from a previous study. Of note, we provided a script that can generate the required input file for a trait by specifying its identifier from the OpenGWAS website (https://gwas.mrcieu.ac.uk/ (accessed on 6 December 2021)), which is a repertoire of GWAS summary datasets [5].
Test of interaction: The purpose of this step is to obtain additional functional insight by testing whether the identified probes contribute to the trait independently or not. The underlying script uses MR to carry out pairwise interaction analysis between the probes; however, in addition to causality (SNPs → Probe A → Probe B), it also examines the presence of pleiotropy (Probe A ← SNPs → Probe B) between the probes by keeping the pleiotropic SNPs in the instrument. This helps to identify probes that are under the regulatory impact of the same set of SNPs.
Summary report: If the algorithm finds a significant association, it reports the findings in a comma-delimited file that contains statistics that describe the nature of the association between the outcome and exposure and the magnitude of association. The columns in this file are described in Table 1. In Table S1, we provide the output for the search examples underlined in the Section 3. The estimated effect size (β) is interpreted in standard deviation (SD) units. Namely, for one SD change in the level of the exposure, the outcome changes by β (assuming that the data are unadjusted). The sign of β indicates the direction of association, i.e., a positive β indicates that a higher value (level) of the exposure is associated with a higher value of the outcome and a negative β indicates an inverse association.

Table 1.
Description of the columns provided in the report file.
In the following section, we explain the performance of our approach through examples of well-studied loci.
3. Results
3.1. PCSK1
PCSK1 was one of the first genes linked to monogenic early-onset obesity. It encodes the enzyme proprotein convertase 1. Substrates of PCSK1 enzyme, such as POMC, insulin, NPY, ghrelin and GLP-1, are involved in the regulation of energy homeostasis and food behavior. Patients with mutations in PCSK1 develop a profound appetite that results in significant weight gain and eventually obesity early in life [6]. PCSK1 is located in chromosome 5q15 and rare variants within this locus are reported for obesity in the ClinVar database (Table S2). In this study, we tested whether our algorithm could link the rare variants to this gene and this gene to obesity. We passed the location of a rare variant within this locus and the name of the relevant phenotype (BMI) to the wrapper script and executed it as:
Bash wrapper.sh chr5:95734724 BMI_PMID30239722
The first argument represents the coordinate of the variant in the human reference genome (build GRCh37). The second argument indicates the phenotype name and its study identifier (represented by PMID). Our algorithm performed haplotype mapping and retrieved the list of biomarkers (functional features) tagged by the SNPs surrounding the rare variant (chr5:95734724). It then tested the association between the biomarkers and BMI using MR and reported the result. The report indicated that PCSK1.13388.57.3, which is a biomarker that measures the level of PCSK1 in the blood, is associated with obesity (B = −0.02, p = 4 × 10−19). This finding indicates that the rare variant is among the pQTLs for PCSK1 (Figure 2a) and, as such, contributes to obesity by lowering the level of this protein in the blood (Figure 2b).
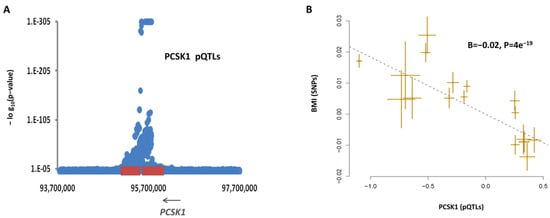
Figure 2.
Impact of rare variants within 5q15 on obesity. Rare variants within 5q15 chromosome band are reported for obesity in ClinVar database. In this region, PCSK1 deficiency is known to cause obesity. We investigate whether our workflow can link variants within this locus to PCSK1 and PCSK1 protein level to obesity. The haplotype mapping indicates that the reported variants are within the coordinate of pQTLs of PCSK1 (A). MR analysis indicates subjects that are genetically susceptible to lower levels of PCSK1 in blood tend to have higher risk of obesity (B). MR was done using non-pleiotropic SNPs (p > 0.01) that are independently (r2 < 0.05) and significantly associated (p < 5 × 10−8) with PCSK1 protein level. Each point on the MR plot represents a SNP; the x-value of a SNP is its beta effect size on PCSK1 protein level, and the horizontal error bar represents the standard error around the beta. The y-value of the SNP is its beta effect size on BMI and the vertical error bar represents the standard error around its beta. The dashed line represents the line of best fit (a line with the intercept of 0 and the slope of β from the MR test). pQTL summary statistics (beta and SE) were obtained from PMID: 29875488. For BMI, we obtained these data from PMID: 30239722.
3.2. APOE
APOE is involved in the metabolism of lipids, and mutations in this gene cause lipid abnormalities. We tested whether our approach could link this gene to lipid phenotypes. For this purpose, we passed the genomic coordinates of the locus (chr19:45409039-45412650) and the name of the lipid phenotype to the wrapper script to initiate the search, e.g.,
bash wrapper.sh chr19:45409039-45412650 LDL_PMID24097068.gz
The result (Figure 3) indicates that a higher protein level of APOE in the blood is associated with higher LDL (B = 0.8, p = 1.2 × 10−83) and total cholesterol (B = 0.5, p = 1.3 × 10−56) and lower HDL (B = −0.14, p = 9 × 10−23). The report generated by the interaction test indicates that the APOE protein level is under the impact of the cg13375295 site. Higher methylation at this site was associated (B = −3.5, p = 4.2 × 10−9) with a lower level of APOE in the blood (Figure 3).
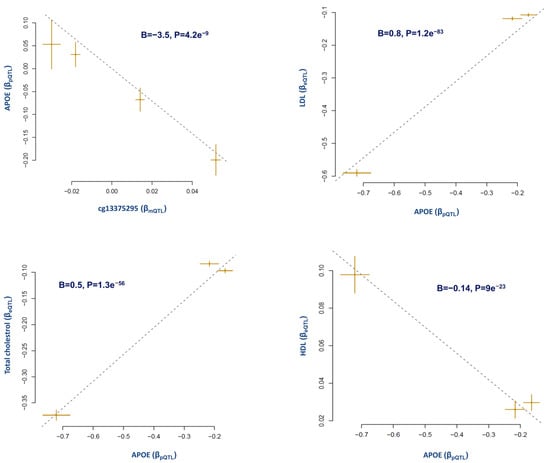
Figure 3.
Impact of APOE blood level on lipid phenotype. In this example, we tested whether our workflow could detect the association of APOE protein level with lipid phenotypes. The results indicate that the APOE level in blood is under the regulatory impact of the cg13375295 site upstream of this gene. Furthermore, we detected that a higher level of APOE is associated with higher LDL and higher total cholesterol but lower HDL. Summary statistics provided in parentheses are from the Mendelian randomization analysis using non-pleiotropic SNPs (p > 0.01) that were independently (r2 < 0.05) and significantly associated (p < 5 × 10−8) with the exposure. Each point on the plots represents a SNP, the x-value of a SNP is its beta effect size on the exposure (cg13375295), and the horizontal error bar represents the standard error around the beta. The y-value of the SNP is its beta effect size on the outcome and the vertical error bar represents the standard error around its beta. The dashed line represents the line of best fit (a line with the intercept of 0 and the slope of β from the MR test). pQTL summary statistics (beta and SE) are from PMID: 29875488; mQTL summary statistics are from PMID: 30401456 and lipid summary statistics are from PMID: 24097068.
4. Discussion
In this study, we devised a workflow to understand the function of a locus using knowledge available at the SNP level and we demonstrated its application through examples for the PCSK1 and APOE locus. The underlying algorithm that carries out the task is written in the shell scripting language. This allows the use of parallel computing and therefore the possibility to conduct screening at phenome-/genome-wide scales. Considering the volume of existing and upcoming functional data, parallel computing will become a necessity to integrate/relate various layers of omics data in a time-efficient manner.
In this study, we used Mendelian randomization to quantify the association between two entities. MR allows the incorporation of summary association statistics from large GWAS consortia and therefore provides higher statistical power as compared to traditional association studies conducted in a sample of individuals. The MR design also provides a shield against the confounding effect of environmental factors because it uses a set of independent SNPs (an instrument) to gauge the relationship between two entities and alleles of independent SNPs are allocated to offspring randomly. Of note, the use of SNPs as an instrument also brings the caveat of weak instrument bias when testing the association between entities that are highly polygenic (e.g., complex traits); however, this is a lesser issue for a functional feature that is under the regulatory impact of fewer SNPs.
Rare variant characterization is a pending problem in molecular biology. Overall, there are two approaches to infer the causal impact of a rare variant on a phenotype. Bioinformatics approaches use functional data to prioritize variants and identify those that carry more functional weight and, as such, are more likely to be the causal variant. Functional data that are used in this approach were previously generated by considering the genome itself, i.e., without taking the studied outcome (phenotype) into account. In contrast, statistics approaches directly test the effect of a rare variant on the phenotype. Hence, they require a sample of subjects with available genotype and phenotype data (in order to detect a significant association), which are not always readily available. Here, we present a workflow that exploits the benefits of both approaches while addressing the shortcomings. Our approach does not require a study sample; it takes functional data into account and carries out the analysis with regard to the studied phenotype. The underlying algorithm is provided as a series of shell scripts. This gives the user the flexibility to adjust them according to the research requirements. For example, by providing a list of phenotypes, it is possible to comprehensively investigate the impact of a rare variant on the entire phenome.
By using SNPs as connectors, we demonstrate that it is possible to merge various omics data and repurpose them for new applications. Such an initiative also has the potential to unite findings from different fields of omics into a single network of knowledge. This would allow biologists to better investigate the mechanism that regulates a biological entity, and to track its impact on other components of the network.
Our workflow relies on the power of GWAS data to provide molecular insights; however, downloading and reformatting these data could be cumbersome for a user. This is a limitation of our approach. One solution would be to provide an online platform where data are collected and centralized and a user can obtain the results by simply entering a search term. Unfortunately, we did not receive support to set up such a platform; however, this is a direction of work for future studies that are interested in this subject. Another limitation of our approach is that QTL data are mainly available at the transcriptome, proteome and methylome levels and, to a lesser extent, at other levels of functional annotation. Therefore, it would be valuable if the practice of reporting the GWAS summary statistics for various functional elements becomes frequent. Another issue in this field of research is inconsistencies in SNP density. New studies are using denser genotyping panels and improving their coverage through genotype imputation; however, this is not always the case in older studies. This impacts the power of analyses; therefore, the development of convenient tools that make the GWAS data uniform by imputing the missing SNPs is valuable.
In summary, this study provides a solution to navigate through various layers of omics data in order to understand the function of a locus. We show its application in finding the mechanism whereby a mutation causes a disease and in revealing the path whereby a gene is being regulated and impacts phenotypes. We provide freely available shell scripts that perform the analyses.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/cardiogenetics11040024/s1, Table S1. The output of the analyses based on search examples presented in the manuscript; Table S2. List of genomic variants within 5q15 chromosome band from ClinVar database that are reported to cause obesity/proprotein convertase 1/3 deficiency.
Author Contributions
Conceptualization, M.N., R.M.; formal analysis, M.N.; investigation, S.R.; data resources, R.M. and R.D.; draft preparation, M.N.; editing, R.M. and S.R. All authors have read and agreed to the published version of the manuscript.
Funding
Supported by the Canadian Institutes of Health Research # FDN-154308 (R.M.).
Institutional Review Board Statement
Not applicable, this study was done using publicly available data.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data, instructions and shell scripts to carry out the analyses are available from: https://github.com/mnikpay/locus-annotator.git (accessed on 6 December 2021).
Acknowledgments
This research was enabled in part by computational resources and support provided by the Compute Ontario and the Compute Canada.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. GigaScience 2015, 4, 7. [Google Scholar] [CrossRef] [PubMed]
- Gabriel, S.; Schaffner, S.; Nguyen, H.; Moore, J.; Roy, J.; Blumenstiel, B.; Higgins, J.; DeFelice, M.; Lochner, A.; Faggart, M.; et al. The Structure of Haplotype Blocks in the Human Genome. Science 2002, 296, 2225–2229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Z.; Zhang, F.; Hu, H.; Bakshi, A.; Robinson, M.R.; Powell, J.E.; Montgomery, G.W.; Goddard, M.E.; Wray, N.R.; Visscher, P.M.; et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 2016, 48, 481–487. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Zheng, Z.; Zhang, F.; Wu, Y.; Trzaskowski, M.; Maier, R.; Robinson, M.R.; McGrath, J.J.; Visscher, P.M.; Wray, N.R.; et al. Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat. Commun. 2018, 9, 224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elsworth, B.; Lyon, M.; Alexander, T.; Liu, Y.; Matthews, P.; Hallett, J.; Bates, P.; Palmer, T.; Haberland, V.; Smith, G.D.; et al. The MRC IEU OpenGWAS data infrastructure. bioRxiv 2020. [Google Scholar] [CrossRef]
- Ramos-Molina, B.; Martin, M.G.; Lindberg, I. PCSK1 Variants and Human Obesity. Prog. Mol. Biol. Transl. Sci. 2016, 140, 47–74. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).