Abstract
Genetic variants in MYBPC3 are one of the most common causes of hypertrophic cardiomyopathy (HCM). While variants in MYBPC3 affecting canonical splice site dinucleotides are a well-characterised cause of HCM, only recently has work begun to investigate the pathogenicity of more deeply intronic variants. Here, we present three patients with HCM and intronic splice-affecting MYBPC3 variants and analyse the impact of variants on splicing using in vitro minigene assays. We show that the three variants, a novel c.927-8G>A variant and the previously reported c.1624+4A>T and c.3815-10T>G variants, result in MYBPC3 splicing errors. Analysis of blood-derived patient RNA for the c.3815-10T>G variant revealed only wild type spliced product, indicating that mis-spliced transcripts from the mutant allele are degraded. These data indicate that the c.927-8G>A variant of uncertain significance and likely benign c.3815-10T>G should be reclassified as likely pathogenic. Furthermore, we find shortcomings in commonly applied bioinformatics strategies to prioritise variants impacting MYBPC3 splicing and re-emphasise the need for functional assessment of variants of uncertain significance in diagnostic testing.
1. Introduction
Hypertrophic cardiomyopathy (HCM) is a relatively common genetic disorder (with a prevalence of 1:500) characterised by left ventricular cardiomyopathy, a non-dilated left ventricle and normal, or increased, ejection fraction, and is associated with myocardial fibre disarray [1,2,3]. HCM is usually asymmetrical and develops in the absence of an identifiable secondary cause such as hypertension or aortic valvular stenosis. Patients with HCM usually have a relatively benign clinical course, although HCM can cause sudden cardiac death particularly in adolescents and young adults [3]. Risk factors such as severe cardiac hypertrophy, family history of sudden cardiac death, syncope and non-sustained ventricular tachycardia have been associated with sudden cardiac death in HCM patients, and high-risk patients may be offered implantable cardioverter-defibrillators (ICDs) to ameliorate their risk [2,3,4,5,6,7].
HCM usually shows an autosomal dominant mode of inheritance, although very rare cases of autosomal recessive and X-linked modes of inheritance have been described [3,8,9,10]. Incomplete penetrance and variable clinical expression are common confounding phenomena. The phenotypic variability is thought to arise, at least in part, from interactions between pathogenic variants and genetic and non-genetic modifiers [3,11,12,13]. Variants in at least 12 genes encoding sarcomere and sarcomere-associated proteins cause HCM, the two most common of which (accounting for approximately 50% of families with HCM) are β-myosin heavy chain (MYH7) and myosin-binding protein C3 (MYBPC3) [2,3,14,15,16,17,18,19]. The β-myosin heavy chain has the ATPase enzymatic activity in the myosin head required for force generation in muscle fibres [20]. The myosin-binding protein C3 interacts with both actin and myosin, as well as titin, and regulates cardiac contraction [21,22]. While the majority of variants causing HCM in MYH7 and other less common causative genes are missense variants, pathogenic variants in MYBPC3 tend to be frameshift, nonsense or splice site variants, which result in premature termination codons (PTCs) and predicted loss of function [3,14,16,23]. PTC-containing MYBPC3 transcripts may result in truncated MYBPC3 protein upon translation; alternatively, mutant transcripts may be degraded by the nonsense-mediated decay pathway, causing allelic loss-of-function and reduced expression [23,24]. Approximately 15% of MYBPC3 variants in HCM are non-truncating, including missense and short in-frame insertion/deletion variants. The pathogenic mechanisms of these non-truncating variants are largely unknown, although it has been shown that disease severity and clinical outcome, while highly variable, is largely independent of whether the pathogenic MYBPC3 variant is truncating or non-truncating [23,25,26].
While genetic testing is now common in patients with HCM, the diagnostic yield is only 40–50%, and the causative genotype is unknown for at least 40% of patients, even for those with family histories of the disease [27,28]. There are many factors which may explain this low genetic testing yield. For example, phenocopying occurs in other syndromic conditions such as Noonan syndrome and storage disorders, including Anderson–Fabry disease, while in other cases complex and as-yet unknown genetic mechanisms may cause HCM [27,29,30,31,32]. However, conventional genetic testing methods may also fail to identify pathogenic non-coding variants, in particular cryptic splice variants (i.e., intronic variants out-with the canonical splice acceptor and splice donor dinucleotides which alter pre-mRNA splicing) in known HCM genes, which may be a major contributor to the missing heritability in HCM. For example, several recent publications have identified novel intronic cryptic splice variants within MYBPC3 in cohorts of HCM patients, which would not have been detected as pathogenic variants by standard genetic testing [27,33,34,35,36,37]. These intronic variants were shown to alter MYBPC3 splicing and result in allelic loss-of-function and haploinsufficiency.
Here, we present three patients with HCM presenting with intronic MYBPC3 splice variants out-with the invariant splice donor/acceptor dinucleotides. We use in vitro minigene splicing assays in human cell lines, combined with direct analysis of patient RNA in one case, to confirm the aberrant effect of these variants on MYBPC3 splicing. This study supports the importance of sequencing intronic regions in MYBPC3 to increase the detection of pathogenic variants causing HCM
2. Materials and Methods
2.1. Patient Recruitment and Genetic Testing
The three probands were recruited by the Manchester Centre for Genomic Medicine. For proband 1, clinical DNA sequencing by NGS was undertaken of the entire coding sequence of four genes—MYH7, MYBPC3, TNNT2 and TNNI3. For probands 2 and 3, a panel of >20 genes associated with HCM was screened by NGS, as previously described [38].
2.2. Bioinformatics Analysis
In silico pathogenicity prediction with SpliceAI v1.3.1 and Human Splicing Finder v3.1 were used to score cryptic splice-affecting variants in MYBPC3 [39,40]. Population frequency of MYBPC3 variants was analysed using gnomAD v.3.1 (https://gnomad.broadinstitute.org/) (accessed 12 September 2020) [41].
2.3. Human K562 Cell Culture
Human K562 cells were cultured under standard tissue culture conditions in RPMI-1640 medium (Sigma) supplemented with 10% Fetal Bovine Serum (FBS) (Sigma) and incubated in 5% CO2 at 37 °C. Original K562 cell stocks were supplied at passage 10.
2.4. In Vitro Splicing Minigene Assay
Fragments of MYBPC3 (reference transcript NM_000256) were amplified by polymerase chain reaction (PCR) from commercially available reference genomic DNA (Promega) using either wildtype or mutagenic primers (Table S1) using Phusion High Fidelity DNA Polymerase (NEB) according to the manufacturer’s recommendations. For the c.927-8G>A variant, the total MYBPC3 fragment length was 502bp, containing the 164 bp exon 12 plus 170 bp of intron 11 and 168 bp of intron 12 of MYBPC3 (Figure 1(ai)). For the c.1624+4A>T variant, the total MYBPC3 fragment length was 332bp, containing the 167 bp exon 17 plus 65 bp of intron 16 and 100 bp of intron 17 (Figure 1(aii)). For the c.3815-10T>G variant, the MYBPC3 fragment was 657 bp and contained the 187 bp exon 33 and 37 bp exon 34, plus 120 bp of intron 32, the 190 bp intron 33, and 123 bp of intron 34 (Figure 1(aiii)). The wildtype and mutagenic MYBPC3 fragments were cloned into the SK3 plasmid (a derivative of the pSpliceExpress minigene splice reporter vector, gifted from Stefan Stamm, Addgene #32485) using the Gibson method [42,43]. Constructs were transformed into competent bacteria and candidate colonies were cultured and vector DNA isolated using the GenElute Plasmid Miniprep kit (Sigma, St. Louis, MO, USA). Sequences of the minigene vector constructs were verified by Sanger sequencing (performed by Eurofins genomics).
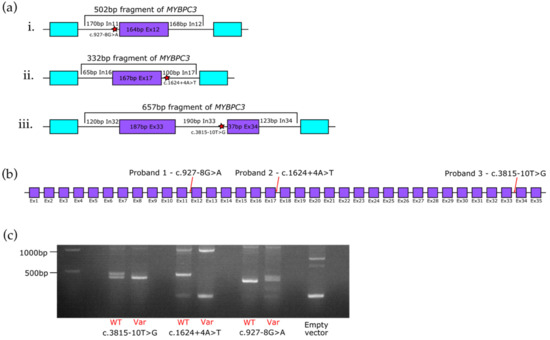
Figure 1.
MYBPC3 intronic splice variants in three probands with hypertrophic cardiomyopathy. (a) Schematic of the minigene constructs used to test the pathogenicity of MYBPC3 splice variants i. c.927-8G>A, ii. c.1624+4A>T and iii. c.3815-10T>G. MYBPC3 exons are shown in purple. Endogenous minigene exons are shown in light blue. The locations of the splice site variants are indicated by red stars. Ex = exon, In = intron. (b) Location of MYBPC3 splice variants analysed in this paper and the probands in which the variants were identified, using the reference transcript NM_000256. Purple boxes refer to exons (Ex), black lines refer to introns. (c) Representative RT-PCR results showing the outcome of the minigene assays. Identities of RT-PCR products were confirmed by Sanger sequencing. High molecular weight bands above 500 bp correspond to unspliced vector DNA contamination. WT = wild type, Var = variant. n = 2.
Human K562 cells were plated in 2 mL RPMI-1640 medium (Sigma) supplemented with 10% Fetal Bovine Serum (FBS) (Sigma) at approximately 80% confluency in 6-well tissue culture plates (Corning) on the day of transfection. Cells were transiently transfected with 2.5 µg plasmid DNA using Lipofectamine LTX (Invitrogen) following the manufacturer’s protocol and incubated for 18–24 h at 37 °C with 5% CO2. The splicing minigene assays were performed twice independently.
2.5. Reverse-Transcription PCR (RT-PCR)
For the splicing minigene assay, total RNA was extracted from K562 cells using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. The resulting RNA was cleaned up using the RNeasy Mini RNA Isolation Kit (Qiagen, Hilden, Germany), including an on-column DNA digestion step. An equal amount of RNA for each sample was converted to cDNA using Superscript IV (Invitrogen, Carlsbad, CA, USA) with random hexamers (Invitrogen) according to the supplier’s recommendations. cDNA was amplified using Phusion polymerase and primers listed in Table S2. PCR products were separated by agarose gel electrophoresis supplemented with SafeView nucleic acid stain (NBS Biologicals, Huntingdon, UK) and visualised under a blue-light transilluminator. Products were extracted using QIAquick gel extraction kit (Qiagen) and sequenced by Sanger sequencing to confirm their identity.
For proband 3 with the MYBPC3 c.3815-10T>G variant, an RNA sample (Paxgene) was derived from blood following the manufacturer’s instructions. A cDNA synthesis was performed as above using patient RNA as a template and the cDNA amplified using Phusion polymerase and primers listed in Table S2. PCR products were separated, gel extracted and sequenced, as described above.
3. Results
Three patients with a clinical diagnosis of HCM, but no known genetic cause underwent genetic and molecular screening by next-generation sequencing (NGS) to establish the genetic basis for their specific cardiac phenotype.
3.1. Clinical Characteristics Associated with Three Intronic MYBPC3 Variants
The proband in family 1 (Figure 2a) was a 77-year-old woman with a history of persistent atrial fibrillation. An echocardiogram revealed severe asymmetrical left ventricular hypertrophy with a septum thickness of 2 cm. The proband’s father was reported to have died suddenly at the age of 53; however, there was no other family history consistent with a diagnosis of cardiomyopathy. Genetic testing revealed a heterozygous c.927-8G>A variant in intron 11 of MYBPC3 (Figure 1b). The affected nucleotide is 8 base pairs (bp) upstream of the canonical 3′ splice site in intron 11, and potentially creates a new cryptic 3′ splice site AG. This c.927-8G>A variant has not been described previously in HCM patients in the literature, is not present in the gnomAD population genomic sequence database and is classified as a variant of uncertain significance. However, this variant has been identified in seven additional unrelated individuals with HCM in diagnostic laboratories performing cardiomyopathy testing in the United Kingdom. Additionally, a variant in the adjacent residue (c.927-9G>A) has been reported as likely pathogenic, although analysis of patient RNA derived from blood for this adjacent variant did not confirm any splicing errors caused by the variant [37,44]. The c.927-8G>A variant was not predicted to have any significant impact on splicing signals by the Human Splicing Finder. In agreement with the Human Splicing Finder, the SpliceAI score for this variant was 0.11 (acceptor loss). Commonly applied SpliceAI scores of 0.2 as potentially splice-altering and 0.5 as likely splice-altering for variant prioritisation would (falsely) exclude this variant from prioritisation [39,45].

Figure 2.
Family pedigrees for the three probands with HCM presented in this study. Pedigrees for (a) family 1, (b) family 2, and (c) family 3. Arrow indicates the probands subject to genetic testing in this study. Filled shapes indicate clinically affected individuals.
The proband in family 2 (Figure 2b) was a 46-year-old man who presented with palpitations and shortness of breath on exertion. A cardiac MRI scan showed moderate to severe concentric left ventricular hypertrophy with septal predominance (basal anteroseptum measured 2.4 cm). There was also evidence of mid-layer fibrosis involving the hypertrophied regions. The proband had type 1 diabetes mellitus but no history of hypertension or other significant medical issues. There was no known family history of cardiomyopathy or sudden cardiac death. The proband was identified as having a heterozygous c.1624+4A>T variant in intron 17 of MYBPC3 (Figure 1b). The affected nucleotide is 4 bp downstream of the 5′ splice site but still within the splice donor consensus sequence. This c.1624+4A>T variant is present in three of 224,880 alleles in gnomAD and is predicted by the Human Splicing Finder to affect splicing by abrogating the splice donor site at the 5′ end of intron 17, although the SpliceAI score for the variant is only 0.16 (donor loss) again suggesting no impact on splicing. The c.1624+4A>T variant is classified as pathogenic, having been reported in a number of HCM patients, both within and outside of the UK, and previous blood-derived RNA analysis from these patients revealed the variant causes exon 17 skipping, which would result in a PTC and loss of the C-terminal region of the protein upon translation removing the major myosin and titin binding sites [24,37,46,47,48,49].
The proband in family 3 (Figure 2c) was diagnosed with HCM after presenting with chest pain whilst walking in his fifth decade. He had a history of paroxysmal atrial fibrillation and had a stroke at the age of 58 years. A cardiac MRI scan performed shortly after his stroke revealed a maximal wall thickness of 2.6 cm and left ventricular outflow tract obstruction, consistent with HCM. On gadolinium imaging, there was evidence of very extensive enhancement involving most of the left ventricle. One of the proband’s brothers was reported to have died in his sleep at the age of 66 years and one of the proband’s sisters was identified as having mild HCM on screening shortly after his diagnosis. Genetic testing in the proband revealed a heterozygous c.3815-10T>G variant in intron 33 of MYBPC3 (Figure 1b). The variant is 10 nucleotides upstream of the 3′ splice site. This c.3815-10T>G variant has been reported once in 242,620 alleles in gnomAD and has been reported previously in one HCM patient in ClinVar, although it is currently classified as a likely benign variant [33]. In silico pathogenicity prediction by Human Splicing Finder suggested no impact of this variant on splicing signals, while the variant had a SpliceAI score of 0.37 (acceptor loss), indicating possible impact on splicing. Genetic testing also revealed a heterozygous MYL2 c.37G>A, p.Ala13Thr variant in this proband.
3.2. In Vitro Minigene Assays to Assess the Impact of Intronic MYBPC3 Splice-Affecting Variants
To test the effects of the identified MYBPC3 intronic variants on MYBPC3 pre-mRNA splicing, we conducted in vitro minigene splicing assays in human cell lines. The assays compared spliced RNA transcripts extracted from human K562 cells following transfection with pairs of wild type and variant-containing minigene constructs containing fragments of MYBPC3 (Figure 1a).
For the c.927-8G>A variant, we found that while the wild type sequence led to canonical splicing of MYBPC3 only, the variant resulted in loss of the wild type splice acceptor and the use of an upstream cryptic splice acceptor site instead, resulting in the inclusion of an additional 19 bp at the 5′ end of exon 12 (Figure 1c). This 19 bp exon extension would result in a frameshift and the creation of a PTC in exon 13.
For the previously characterised c.1624+4A>T variant, the wild type sequence resulted in a primary product containing canonically spliced exon 17, and a small fraction of product with exon 17 skipping. However, the mutant sequence resulted in complete exon 17 skipping (due to mutation of the wild type splice donor consensus sequence) and no canonically spliced product, in agreement with previous reports (Figure 1c and Figure S1) [33,37].
Finally, for the c.3815-10T>G variant, the wild type sequence resulted in two spliced products, one containing exon 34 and one with exon 34 skipped. However, for the mutant construct, there was complete skipping of exon 34 and no exon 34 retention, suggesting the variant led to complete loss of splice acceptor recognition at the 3′ end of intron 33 (Figure 1c). Interestingly, exon 34 (of 35 total exons for the NM_000256 reference transcript used in this study) is the final coding exon of MYBPC3, and so the skipping event observed here would result in loss of the normal stop codon. RNA from blood was available for this patient, and direct analysis of the spliced products in blood-derived patient RNA revealed only a wild type product, suggesting that mutant transcripts produced from the variant-containing allele were degraded (data not shown). This finding suggests the c.3815-10T>G variant results in decreased MYBPC3 expression, in agreement with the known disease mechanism of haploinsufficiency.
4. Discussion
In this study, we present three patients with MYBPC3 intronic variants and assess the effects of these variants on MYBPC3 splicing in minigene assays in vitro. Confirming the pathogenicity of these intronic splice variants makes it possible to utilise the findings clinically and offer targeted genetic testing to at-risk family members. For the c.927-8G>A and c.3815-10T>G variants, we can now apply PS3 from the American College of Medical Genetics (ACMG) guidelines for assigning pathogenicity to sequence variants as we have shown, using a well-established in vitro functional study, the damaging effect on splicing of these two variants [50]. Patient-derived RNA is the ideal analyte to determine the effect of sequence variants on splicing, but where this is not available, we propose that in vitro minigene splicing assays can generate supportive evidence, which can be used as part of the variant classification [51].
While the MYBPC3 c.1624+4A>T variant has been well-characterised and previously classified as pathogenic, we included it in our minigene assays as a positive control to validate our methodology and confirm the sensitivity of our in vitro assay for detecting splice-altering variant effects [24,37,46,47,48,49]. In our system, this variant resulted in complete MYBPC3 exon 17 skipping, in agreement with these previous analyses.
The novel c.927-8G>A variant, described for the first time in proband 1 of our study, had a clear impact on MYBPC3 splicing, activates a cryptic splice site upstream of the canonical 3′ splice site at intron 11, resulting in the extension of exon 12 by 19 bp at the 5′ end, leading to a frameshift and a PTC. It is interesting that the report of a variant in the adjacent nucleotide (c.927-9G>A) could not confirm aberrant splicing in patient RNA samples by RT-PCR; the authors only observed the wild type spliced product [37]. We postulate that the adjacent variant may have the same effect on MYBPC3 splicing as our c.927-8G>A variant as both variants affect the same canonical splice site in intron 11 and likely activate the same cryptic upstream 3′ splice site. It is likely that transcripts from the mutant allele are degraded by nonsense-mediated decay (NMD), such that they are not observed by RT-PCR of patient RNA. Quantitative analysis of the MYBPC3 expression by qPCR and/or heterozygous variant ratios to assess whether transcripts from the mutant allele are present in patient RNA samples could be used to confirm this mechanism, as well as assessing the variant in a minigene assay.
Finally, the previously described c.3815-10T>G variant in proband 3 of our study had a clear effect on splicing in our minigene assay, resulting in total loss of the splice donor recognition at the 3′ end of intron 33 leading to complete exon 34 skipping and loss of the normal stop codon. The nucleotide sequence of exon 35 codes for 46 amino acids in-frame with exon 33 with an in-frame stop codon and so the mis-splicing event associated with the variant could result in a change to the 3′ end of the encoded protein. This event would not be detected from our minigene assay because the construct only contained MYBPC3 exons 33 and 34 and not exon 35. However, RT-PCR analysis of patient RNA indicated that this event causes NMD of transcripts from the mutant allele, as opposed to production of a protein with an altered C-terminal. Two points of interest are worth noting here. Firstly, even for our wild type minigene construct, there is a relatively strong RT-PCR product (approximately the same intensity of band on the gel as the canonically spliced RT-PCR product) corresponding to the exon 34-skipped splice product, and so the variant appears to shift the ratio of splice isoforms towards an exon 34-skipped product, which is already normally observed. In contrast, while a very faint band corresponding to the exon 17-skipped splice product is also present for the wild type construct for the c.1624+4A>T variant, this band is much weaker than the canonically spliced product. MYBPC3 exon 34 is a very short exon (37bp), and previous studies have indicated shorter exon length is associated with more frequently skipped exons, which may explain the relatively high prevalence of the shorter splice isoform even for the wild type minigene constructs [52,53]. The second point of interest with the c.3815-10T>G variant is that this individual also carries a heterozygous MYL2 c.37G>A, p.Ala13Thr variant. This amino acid substitution has been shown to have an important effect on calcium binding and subsequent myosin binding of the encoded protein and has been previously reported in patients with HCM, particularly associated with left ventricular obstruction. This observation is consistent with the phenotype of left ventricular outflow tract obstruction in our patient, but the high minor allele frequency means that the clinical significance of this variant is unclear [54,55,56,57,58]. It may be that because the MYBPC3 c.3815-10T>G splice variant enhances an exon skipping event, which is normally observed at a relatively high frequency, this variant must be combined with a second variant in another sarcomere-associated gene and/or must occur in the presence of a background of particular genetic modifiers to be fully penetrant. Another non-mutually exclusive possibility is that the common MYL2 variant in this patient is only pathogenic in the presence of a second variant in a sarcomere-associated gene (in this case, the MYBPC3 c.3815-10T>G variant), explaining the link between the variant and specific cardiac phenotypes even though its population frequency is relatively high. It is not clear whether the previously reported individuals with the MYBPC3 c.3815-10T>G variant also carried the MYL2 c.37G>A variant [33]. One of this proband’s sisters was identified as having a mild HCM phenotype. Genetic testing will now be extended to the affected sibling.
A further point of interest is that none of the intronic splice-affecting variants presented in this study had particularly high SpliceAI scores and only the c.1624+4A>T variant was predicted by Human Splicing Finder to affect MYBPC3 splicing. Indeed, for this variant, the SpliceAI score was only 0.16, and yet the variant resulted in total loss of the intron 17 splice donor and complete exon 17 skipping and is already classified as pathogenic on the basis of previous RNA analysis. Similarly, the c.927-8G>A variant had a SpliceAI score of just 0.11, and yet had a very clear impact on the MYBPC3 exon 12 splice acceptor site, while the c.3815-10T>G had a SpliceAI score of 0.37 but had a major impact in shifting MYBPC3 splicing towards exon 34 skipping. In general, our laboratory policy is to consider variants with SpliceAI scores >0.2 as potentially splice-altering, while scores >0.5 are considered as very likely to be splice-altering, which is similar to previously described SpliceAI cut-offs used for variant prioritisation [39,45]. However, some reports investigating MYBPC3 intronic splice variants have used very stringent SpliceAI cut-offs of ≥0.9 to initially prioritise variants [27]. Nonetheless, when these studies expanded their inclusion criteria to include variants with lower SpliceAI scores, an MYBPC3 c.1898-23A>G variant with a SpliceAI score of just 0.04 was identified; assessment of patient RNA using RT-PCR showed the inclusion of intron 19, leading to a premature stop codon and presumed NMD [27]. In our study, we further demonstrate that intronic variants beneath commonly applied SpliceAI filters do have a significant impact on splicing, and these data re-emphasise that functional investigation alongside in silico assessments remains an important consideration for diagnostic testing in the context of splicing.
5. Conclusions
Intronic MYBPC3 cryptic splice variants are an important genetic cause of HCM and may not be accurately predicted by in silico bioinformatic methods. Analysis of intronic MYBPC3 splice variants increases the diagnostic yield in HCM patients.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/cardiogenetics11020009/s1. Table S1: Primers used for generating MYBPC3 minigene constructs. Table S2: Primers used for reverse-transcription polymerase chain reaction (RT-PCR) experiments. Figure S1: Sequence traces showing the 19 bp insertion at the 5′ end of MYBPC3 exon 12 for the c.927-8G>A variant in the minigene assay.
Author Contributions
Conceptualization, W.G.N.; methodology, H.B.T. and K.A.W.; software, J.M.E.; validation, K.A.W., H.B.T. and J.M.E.; formal Analysis, K.A.W., H.B.T. and J.M.E.; investigation, K.A.W. and J.M.E.; resources, W.G.N. and R.T.O.; data curation, K.A.W., J.M.E. and C.H.; writing—original draft preparation, K.A.W. and C.H.; writing—review and editing, H.B.T., J.M.E., J.E., R.T.O. and W.G.N.; visualization, K.A.W.; supervision, W.G.N. and R.T.O.; project administration, W.G.N.; funding acquisition, W.G.N., R.T.O. and J.M.E. All authors have read and agreed to the published version of the manuscript.
Funding
K.A.W. is funded by a Medical Research PhD Council studentship (1916606). W.G.N. is supported by the Manchester NIHR BRC (IS-BRC-1215-20007). C.H. is a clinical lecturer supported by the NIHR. R.T.O. and H.B.T. are supported by the BBSRC (BB/N000258/1). J.M.E. is supported by the Health Education England Genomics Education Programme.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the South Manchester Research Ethics Committee (11/H10003/3).
Informed Consent Statement
Informed consent was obtained from all subjects involved in this study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to the sensitivity of patient information.
Acknowledgments
The authors acknowledge the NW Genomic Laboratory Hub for genetic screening by NGS of patient samples.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Towbin, J.A. Hypertrophic Cardiomyopathy. Pacing Clin. Electrophysiol. 2009, 32, S23–S31. [Google Scholar] [CrossRef] [PubMed]
- Hensley, N.; Dietrich, J.; Nyhan, D.; Mitter, N.; Yee, M.-S.; Brady, M. Hypertrophic Cardiomyopathy. Anesthesia Analg. 2015, 120, 554–569. [Google Scholar] [CrossRef]
- Marian, A.J.; Braunwald, E. Hypertrophic Cardiomyopathy. Circ. Res. 2017, 121, 749–770. [Google Scholar] [CrossRef] [PubMed]
- Elliott, P.M.; Poloniecki, J.; Dickie, S.; Sharma, S.; Monserrat, L.; Varnava, A.; Mahon, N.G.; McKenna, W.J. Sudden death in hypertrophic cardiomyopathy: Identification of high risk patients. J. Am. Coll. Cardiol. 2000, 36, 2212–2218. [Google Scholar] [CrossRef]
- Frenneaux, M.P.; Counihan, P.J.; Caforio, A.L.; Chikamori, T.; McKenna, W.J. Abnormal blood pressure response during exercise in hypertrophic cardiomyopathy. Circulation 1990, 82, 1995–2002. [Google Scholar] [CrossRef]
- Spirito, P.; Bellone, P.; Harris, K.M.; Bernabò, P.; Bruzzi, P.; Maron, B.J. Magnitude of Left Ventricular Hypertrophy and Risk of Sudden Death in Hypertrophic Cardiomyopathy. N. Engl. J. Med. 2000, 342, 1778–1785. [Google Scholar] [CrossRef]
- McKenna, W.J.; Franklin, R.C.; Nihoyannopoulos, P.; Robinson, K.C.; Deanfield, J.E.; Dickie, S.; Krikler, S.J. Arrhythmia and prognosis in infants, children and adolescents with hypertrophic cardiomyopathy. J. Am. Coll. Cardiol. 1988, 11, 147–153. [Google Scholar] [CrossRef]
- Greaves, S.C.; Roche, A.H.; Neutze, J.M.; Whitlock, R.M.; Veale, A.M. Inheritance of hypertrophic cardiomyopathy: A cross sectional and M mode echocardiographic study of 50 families. Heart 1987, 58, 259–266. [Google Scholar] [CrossRef]
- Branzi, A.; Romeo, G.; Specchia, S.; Lolli, C.; Binetti, G.; Devoto, M.; Bacchi, M.; Magnani, B. Genetic heterogeneity of hypertrophic cardiomyopathy. Int. J. Cardiol. 1985, 7, 129–133. [Google Scholar] [CrossRef]
- Hartmannova, H.; Kubanek, M.; Sramko, M.; Piherova, L.; Noskova, L.; Hodanova, K.; Stranecky, V.; Pristoupilova, A.; Sovova, J.; Marek, T.; et al. Isolated X-Linked Hypertrophic Cardiomyopathy Caused by a Novel Mutation of the Four-and-a-Half LIM Domain 1 Gene. Circ. Cardiovasc. Genet. 2013, 6, 543–551. [Google Scholar] [CrossRef][Green Version]
- Marian, A.J. Modifier genes for hypertrophic cardiomyopathy. Curr. Opin. Cardiol. 2002, 17, 242–252. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, X.; Xie, J.; Zhu, S.; Chen, Y.; Wang, L.; Xu, B. Next-generation sequencing identifies pathogenic and modifier mutations in a consanguineous Chinese family with hypertrophic cardiomyopathy. Medicine 2017, 96, e7010. [Google Scholar] [CrossRef]
- Chen, Y.; Xu, F.; Munkhsaikhan, U.; Boyle, C.; Borcky, T.; Zhao, W.; Purevjav, E.; Towbin, J.A.; Liao, F.; Williams, R.W.; et al. Identifying modifier genes for hypertrophic cardiomyopathy. J. Mol. Cell. Cardiol. 2020, 144, 119–126. [Google Scholar] [CrossRef]
- Millat, G.; Bouvagnet, P.; Chevalier, P.; Dauphin, C.; Jouk, P.S.; Da Costa, A.; Prieur, F.; Bresson, J.-L.; Faivre, L.; Eicher, J.-C.; et al. Prevalence and spectrum of mutations in a cohort of 192 unrelated patients with hypertrophic cardiomyopathy. Eur. J. Med. Genet. 2010, 53, 261–267. [Google Scholar] [CrossRef]
- Kaski, J.P.; Syrris, P.; Esteban, M.T.T.; Jenkins, S.; Pantazis, A.; Deanfield, J.; McKenna, W.J.; Elliott, P.M. Prevalence of Sarcomere Protein Gene Mutations in Preadolescent Children with Hypertrophic Cardiomyopathy. Circ. Cardiovasc. Genet. 2009, 2, 436–441. [Google Scholar] [CrossRef]
- Erdmann, J.; Daehmlow, S.; Wischke, S.; Senyuva, M.; Werner, U.; Raible, J.; Tanis, N.; Dyachenko, S.; Hummel, M.; Hetzer, R.; et al. Mutation spectrum in a large cohort of unrelated consecutive patients with hypertrophic cardiomyopathy. Clin. Genet. 2003, 64, 339–349. [Google Scholar] [CrossRef]
- Richard, P.; Charron, P.; Carrier, L.; Ledeuil, C.; Cheav, T.; Pichereau, C.; Benaiche, A.; Isnard, R.; Dubourg, O.; Burban, M.; et al. Hypertrophic Cardiomyopathy. Circulation 2003, 107, 2227–2232. [Google Scholar] [CrossRef]
- Wigle, E.D.; Rakowski, H.; Kimball, B.P.; Williams, W.G. Hypertrophic Cardiomyopathy. Circulation 1995, 92, 1680–1692. [Google Scholar] [CrossRef] [PubMed]
- Maron, B.J.; Maron, M.S. Hypertrophic cardiomyopathy. Lancet 2013, 381, 242–255. [Google Scholar] [CrossRef]
- Colegrave, M.; Peckham, M. Structural Implications of β-Cardiac Myosin Heavy Chain Mutations in Human Disease. Anat. Rec. Adv. Integr. Anat. Evol. Biol. 2014, 297, 1670–1680. [Google Scholar] [CrossRef] [PubMed]
- Previs, M.J.; Previs, S.B.; Gulick, J.; Robbins, J.; Warshaw, D.M. Molecular Mechanics of Cardiac Myosin-Binding Protein C in Native Thick Filaments. Science 2012, 337, 1215–1218. [Google Scholar] [CrossRef]
- Previs, M.J.; Prosser, B.L.; Mun, J.Y.; Previs, S.B.; Gulick, J.; Lee, K.; Robbins, J.; Craig, R.; Lederer, W.J.; Warshaw, D.M. Myosin-binding protein C corrects an intrinsic inhomogeneity in cardiac excitation-contraction coupling. Sci. Adv. 2015, 1, e1400205. [Google Scholar] [CrossRef]
- Helms, A.S.; Thompson, A.D.; Glazier, A.A.; Hafeez, N.; Kabani, S.; Rodriguez, J.; Yob, J.M.; Woolcock, H.; Mazzarotto, F.; Lakdawala, N.K.; et al. Spatial and Functional Distribution of MYBPC3 Pathogenic Variants and Clinical Outcomes in Patients with Hypertrophic Cardiomyopathy. Circ. Genom. Precis. Med. 2020, 13, 396–405. [Google Scholar] [CrossRef]
- Marston, S.; Copeland, O.; Jacques, A.; Livesey, K.; Tsang, V.; McKenna, W.J.; Jalilzadeh, S.; Carballo, S.; Redwood, C.; Watkins, H. Evidence from Human Myectomy Samples That MYBPC3 Mutations Cause Hypertrophic Cardiomyopathy Through Haploinsufficiency. Circ. Res. 2009, 105, 219–222. [Google Scholar] [CrossRef] [PubMed]
- Erdmann, J.; Raible, J.; Maki-Abadi, J.; Hammann, J.; Wollnik, B.; Frantz, E.; Fleck, E.; Regitz-Zagrosek, V.; Hummel, M.; Hetzer, R. Spectrum of clinical phenotypes and gene variants in cardiac myosin-binding protein C mutation carriers with hypertrophic cardiomyopathy. J. Am. Coll. Cardiol. 2001, 38, 322–330. [Google Scholar] [CrossRef]
- Page, S.P.; Kounas, S.; Syrris, P.; Christiansen, M.; Frank-Hansen, R.; Andersen, P.S.; Elliott, P.M.; McKenna, W.J. Cardiac Myosin Binding Protein-C Mutations in Families with Hypertrophic Cardiomyopathy. Circ. Cardiovasc. Genet. 2012, 5, 156–166. [Google Scholar] [CrossRef]
- Lopes, L.R.; Barbosa, P.; Torrado, M.; Quinn, E.; Merino, A.; Ochoa, J.P.; Jager, J.; Futema, M.; Carmo-Fonseca, M.; Monserrat, L.; et al. Cryptic Splice-Altering Variants in MYBPC3 Are a Prevalent Cause of Hypertrophic Cardiomyopathy. Circ. Genom. Precis. Med. 2020, 13. [Google Scholar] [CrossRef] [PubMed]
- Ko, C.; Arscott, P.; Concannon, M.; Saberi, S.; Day, S.M.; Yashar, B.M.; Helms, A.S. Genetic testing impacts the utility of prospective familial screening in hypertrophic cardiomyopathy through identification of a nonfamilial subgroup. Genet. Med. 2017, 20, 69–75. [Google Scholar] [CrossRef]
- Marian, A.J. Challenges in the Diagnosis of Anderson-Fabry Disease. J. Am. Coll. Cardiol. 2016, 68, 1051–1053. [Google Scholar] [CrossRef]
- Gelb, B.D.; Roberts, A.E.; Tartaglia, M. Cardiomyopathies in Noonan syndrome and the other RASopathies. Prog. Pediatr. Cardiol. 2015, 39, 13–19. [Google Scholar] [CrossRef]
- Elliott, P.; Baker, R.; Pasquale, F.; Quarta, G.; Ebrahim, H.; Mehta, A.B.; Hughes, D.; on Behalf of the ACES Study Group. Prevalence of Anderson-Fabry disease in patients with hypertrophic cardiomyopathy: The European Anderson-Fabry Disease Survey. Heart 2011, 97, 1957–1960. [Google Scholar] [CrossRef]
- Sankaranarayanan, R.; Fleming, E.J.; Garratt, C.J. Mimics of Hypertrophic Cardiomyopathy—Diagnostic Clues to Aid Early Identification of Phenocopies. Arrhythmia Electrophysiol. Rev. 2013, 2, 36–40. [Google Scholar] [CrossRef]
- Ito, K.; Patel, P.N.; Gorham, J.M.; McDonough, B.; DePalma, S.R.; Adler, E.E.; Lam, L.; MacRae, C.A.; Mohiuddin, S.M.; Fatkin, D.; et al. Identification of pathogenic gene mutations in LMNA and MYBPC3 that alter RNA splicing. Proc. Natl. Acad. Sci. USA 2017, 114, 7689–7694. [Google Scholar] [CrossRef] [PubMed]
- Bagnall, R.D.; Ingles, J.; Dinger, M.E.; Cowley, M.J.; Ross, S.B.; Minoche, A.E.; Lal, S.; Turner, C.; Colley, A.; Rajagopalan, S.; et al. Whole Genome Sequencing Improves Outcomes of Genetic Testing in Patients with Hypertrophic Cardiomyopathy. J. Am. Coll. Cardiol. 2018, 72, 419–429. [Google Scholar] [CrossRef]
- Janin, A.; Chanavat, V.; Rollat-Farnier, P.; Bardel, C.; Nguyen, K.; Chevalier, P.; Eicher, J.; Faivre, L.; Piard, J.; Albert, E.; et al. Whole MYBPC3 NGS sequencing as a molecular strategy to improve the efficiency of molecular diagnosis of patients with hypertrophic cardiomyopathy. Hum. Mutat. 2019, 41, 465–475. [Google Scholar] [CrossRef]
- Frank-Hansen, R.; Page, S.P.; Syrris, P.; McKenna, W.J.; Christiansen, M.; Andersen, P.S. Micro-exons of the cardiac myosin binding protein C gene: Flanking introns contain a disproportionately large number of hypertrophic cardiomyopathy mutations. Eur. J. Hum. Genet. 2008, 16, 1062–1069. [Google Scholar] [CrossRef] [PubMed]
- Singer, E.S.; Ingles, J.; Semsarian, C.; Bagnall, R.D. Key Value of RNA Analysis of MYBPC3 Splice-Site Variants in Hypertrophic Cardiomyopathy. Circ. Genom. Precis. Med. 2019, 12, e002368. [Google Scholar] [CrossRef]
- Xu, M.; Orsborne, C.; Eden, J.; Wallace, A.; Church, H.J.; Tylee, K.; Deepak, S.; Cassidy, C.; Woolfson, P.; Miller, C.; et al. Mosaic Fabry Disease in a Male Presenting as Hypertrophic Cardiomyopathy. Cardiogenetics 2020, 11, 1–9. [Google Scholar] [CrossRef]
- Jaganathan, K.; Panagiotopoulou, S.K.; McRae, J.F.; Darbandi, S.F.; Knowles, D.; Li, Y.I.; Kosmicki, J.A.; Arbelaez, J.; Cui, W.; Schwartz, G.B.; et al. Predicting Splicing from Primary Sequence with Deep Learning. Cell 2019, 176, 535–548.e24. [Google Scholar] [CrossRef]
- Desmet, F.O.; Hamroun, D.; Lalande, M.; Collod-Béroud, G.; Claustres, M.; Béroud, C. Human splicing finder: An online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009, 37, e67. [Google Scholar] [CrossRef]
- Koch, L. Exploring human genomic diversity with gnomAD. Nat. Rev. Genet. 2020, 21, 448. [Google Scholar] [CrossRef] [PubMed]
- Kishore, S.; Khanna, A.; Stamm, S. Rapid generation of splicing reporters with pSpliceExpress. Gene 2008, 427, 104–110. [Google Scholar] [CrossRef]
- Thomas, H.B.; Wood, K.A.; Buczek, W.A.; Gordon, C.T.; Pingault, V.; Attié-Bitach, T.; Hentges, K.E.; Varghese, V.C.; Amiel, J.; Newman, W.G.; et al. EFTUD2 missense variants disrupt protein function and splicing in mandibulofacial dysostosis Guion-Almeida type. Hum. Mutat. 2020, 41, 1372–1382. [Google Scholar] [CrossRef]
- Rodríguez-García, M.I.; Monserrat, L.; Ortiz, M.; Fernández, X.; Cazón, L.; Núñez, L.; Barriales-Villa, R.; Maneiro, E.; Veira, E.; Castro-Beiras, A.; et al. Screening mutations in myosin binding protein C3 gene in a cohort of patients with Hypertrophic Cardiomyopathy. BMC Med. Genet. 2010, 11, 67. [Google Scholar] [CrossRef] [PubMed]
- Ellingford, J.M.; Thomas, H.B.; Rowlands, C.; Arno, G.; Beaman, G.; Gomes-Silva, B.; Campbell, C.; Gossan, N.; Hardcastle, C.; Webb, K.; et al. Functional and in-silico interrogation of rare genomic variants impacting RNA splicing for the diagnosis of genomic disorders. bioRxiv 2019, 781088. [Google Scholar] [CrossRef]
- Ingles, J.; Doolan, A.; Chiu, C.; Seidman, J.; Seidman, C.; Semsarian, C. Compound and double mutations in patients with hypertrophic cardiomyopathy: Implications for genetic testing and counselling. J. Med. Genet. 2005, 42, e59. [Google Scholar] [CrossRef]
- Helms, A.S.; Davis, F.M.; Coleman, D.; Bartolone, S.; Glazier, A.A.; Pagani, F.; Yob, J.M.; Sadayappan, S.; Pedersen, E.; Lyons, R.; et al. Sarcomere Mutation-Specific Expression Patterns in Human Hypertrophic Cardiomyopathy. Circ. Cardiovasc. Genet. 2014, 7, 434–443. [Google Scholar] [CrossRef]
- Carrier, L.; Bonne, G.; Bahrend, E.; Yu, B.; Richard, P.; Niel, F.; Hainque, B.; Cruaud, C.; Gary, F.; Labeit, S.; et al. Organization and Sequence of Human Cardiac Myosin Binding Protein C Gene (MYBPC3) and Identification of Mutations Predicted to Produce Truncated Proteins in Familial Hypertrophic Cardiomyopathy. Circ. Res. 1997, 80, 427–434. [Google Scholar] [CrossRef]
- Flavigny, J.; Souchet, M.; Sébillon, P.; Berrebi-Bertrand, I.; Hainque, B.; Mallet, A.; Bril, A.; Schwartz, K.; Carrier, L. COOH-terminal truncated cardiac myosin-binding protein C mutants resulting from familial hypertrophic cardiomyopathy mutations exhibit altered expression and/or incorporation in fetal rat cardiomyocytes. J. Mol. Biol. 1999, 294, 443–456. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–423. [Google Scholar] [CrossRef]
- Kanavy, D.M.; McNulty, S.M.; Jairath, M.K.; Brnich, S.E.; Bizon, C.; Powell, B.C.; Berg, J.S. Comparative analysis of functional assay evidence use by ClinGen Variant Curation Expert Panels. Genome Med. 2019, 11, 77. [Google Scholar] [CrossRef]
- Zheng, C.L.; Fu, X.-D.; Gribskov, M. Characteristics and regulatory elements defining constitutive splicing and different modes of alternative splicing in human and mouse. RNA 2005, 11, 1777–1787. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, J.; Huang, B.O.; Xu, Y.-M.; Huang, L.-F.; Lin, J.; Zhang, J.; Min, Q.-H.; Yang, W.-M.; Wang, X.-Z. Mechanism of alternative splicing and its regulation. Biomed. Rep. 2015, 3, 152–158. [Google Scholar] [CrossRef]
- Walsh, R.; Thomson, K.L.; Ware, J.S.; Funke, B.H.; Woodley, J.; McGuire, K.J.; Mazzarotto, F.; Blair, E.; Seller, A.; Exome Aggregation Consortium; et al. Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples. Genet. Med. 2017, 19, 192–203. [Google Scholar] [CrossRef]
- Poetter, K.; Jiang, H.; Hassanzadeh, S.; Master, S.R.; Chang, A.; Dalakas, M.C.; Rayment, I.; Sellers, J.R.; Fananapazir, L.; Epstein, N.D. Mutations in either the essential or regulatory light chains of myosin are associated with a rare myopathy in human heart and skeletal muscle. Nat. Genet. 1996, 13, 63–69. [Google Scholar] [CrossRef]
- Szczesna, D.; Ghosh, D.; Li, Q.; Gomes, A.V.; Guzman, G.; Arana, C.; Zhi, G.; Stull, J.T.; Potter, J.D. Familial Hypertrophic Cardiomyopathy Mutations in the Regulatory Light Chains of Myosin Affect Their Structure, Ca2+ Binding, and Phosphorylation. J. Biol. Chem. 2001, 276, 7086–7092. [Google Scholar] [CrossRef] [PubMed]
- Kaźmierczak, K.; Muthu, P.; Huang, W.; Jones, M.; Wang, Y.; Szczesna-Cordary, D. Myosin regulatory light chain mutation found in hypertrophic cardiomyopathy patients increases isometric force production in transgenic mice. Biochem. J. 2012, 442, 95–103. [Google Scholar] [CrossRef]
- Farman, G.P.; Muthu, P.; Kazmierczak, K.; Szczesna-Cordary, D.; Moore, J.R. Impact of familial hypertrophic cardiomyopathy-linked mutations in the NH2 terminus of the RLC on β-myosin cross-bridge mechanics. J. Appl. Physiol. 2014, 117, 1471–1477. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).