The Emerging Role of Omics-Based Approaches in Plant Virology
Abstract
1. Introduction
1.1. Plant–Virus Interactions
1.2. Introduction to Omics: Basics
2. Application of Genomics in Plant Virus Disease Control
3. Application of Transcriptomics in Plant Virus Disease Control
4. Application of Proteomics in Plant Virus Disease Control
5. Application of Metabolomics in Plant Virus Disease Control
6. Challenges and Pitfalls of Individual Omics Technologies
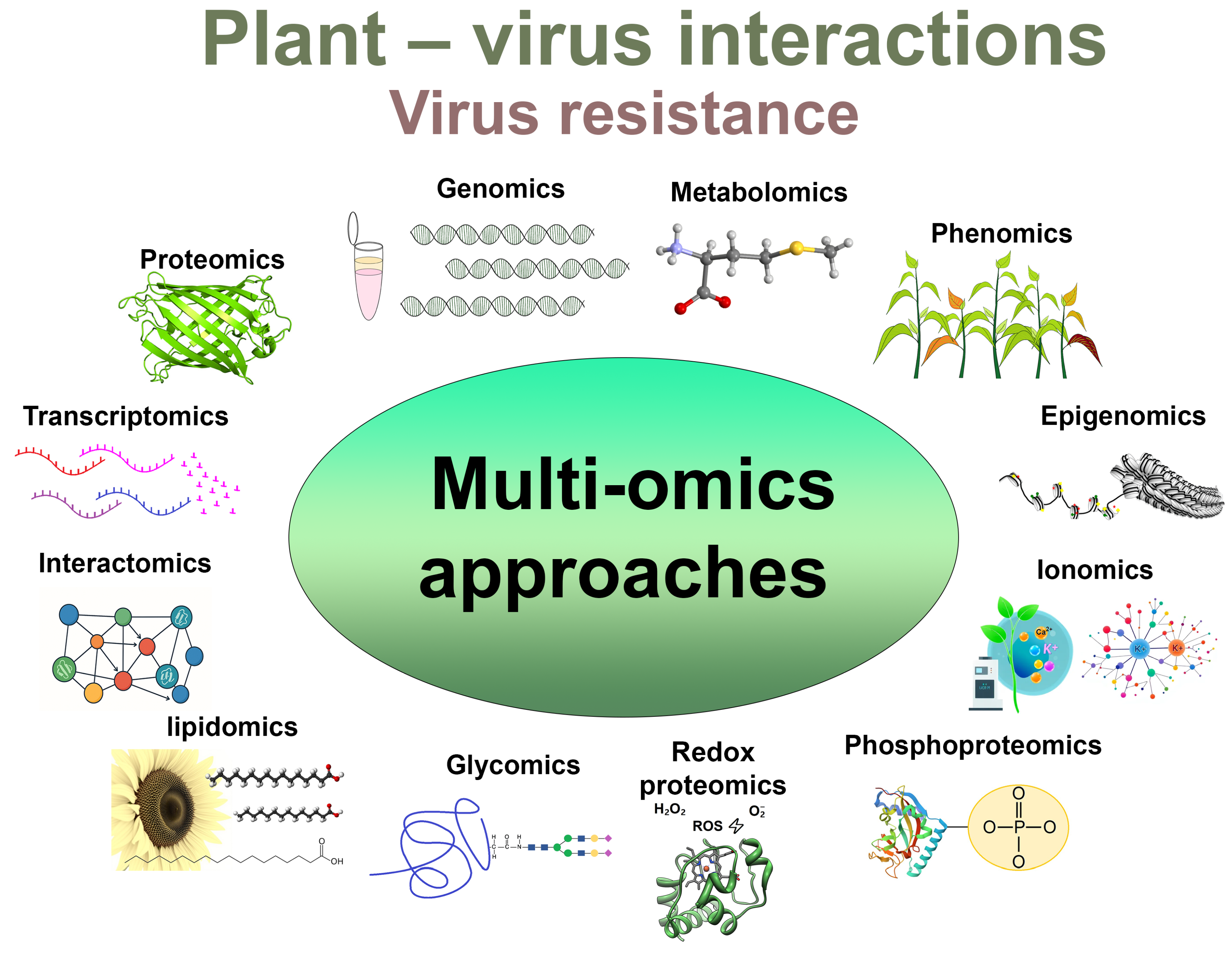
7. Multi-Omics
8. Future Directions
8.1. Single-Cell and Spatial Omics
8.2. Artificial Intelligence and Machine Learning
8.3. Experimental Verification
8.4. Localisation Studies: Relocalisation, Retention, Sequestration—New Omics?
8.5. Biological Pitfalls
- Some additional biological aspects should be considered when planning omics related research on plant–virus interactions.
- Effect of viral tropism. Plant viral tropism refers to the bias a virus has for infecting and replicating within specific cell types or tissues [183]. Certain viruses may preferentially invade reproductive organs, meristems, and seeds, overcoming antiviral barriers to establish persistent infections [183]. In contrast, other viruses may be strictly limited to specific tissues, e.g., the phloem, the plant′s vascular tissue responsible for transporting organic nutrients [184]. It is also well known that viral replication sites vary depending on the virus type. Most RNA viruses replicate in the cytoplasm, while many DNA viruses replicate in the nucleus [185]. Some viruses, regardless of their genome type, establish specialised compartments within the cell to facilitate their replication [19,185]. These compartments may concentrate viral proteins and RNA to spatially organise the replication events.
- Mixed infections. In the field, mixed infections consist of two or more plant viruses, which often culminate in very complex multifaceted interactions between the host and viruses which may differentially modulate plant responses to environmental conditions, virus loads, virus tissue tropisms and light period [186]. Mixed infections very commonly occur and therefore omics-based research should recognise the complexities of mixed infections so that important knowledge can be obtained for effective control of plant viruses.
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Correction Statement
Abbreviations
| PAMP | Pathogen-associated molecular pattern |
| PTI | PAMP-triggered immunity |
| ETI | Effector-triggered immunity |
| PRRs | Pattern recognition receptors |
| ROS | Reactive oxygen species |
| SA | Salicylic acid |
| SAR | Systemic acquired resistance |
| RNAi | RNA interference |
| TGS | Transcriptional gene silencing |
| PTGS | Post-transcriptional gene silencing |
| siRNAs | Small interfering RNAs |
| NGS | Next generation sequencing |
| RNAseq | RNA sequencing |
| MS | Mass spectrometry |
| NMR | Nuclear magnetic resonance |
| SNPs | Single nucleotide polymorphisms |
| GWAS | Genome-wide association studies |
| lncRNAs | Long non-coding RNAs |
| smRNAs | Small RNAs |
| DEGs | Differentially expressed genes |
| vsiRNAs | Viral small interfering RNAs |
| MTC | Methionine cycle |
| PARylation | Poly ADP-ribosylation |
| mROS | Mitochondrial reactive oxidative species |
| AI | Artificial intelligence |
| ML | Machine learning |
| SG | Stress granules |
| PB | Processing bodies |
References
- Rybicki, E.P. A Top Ten List for Economically Important Plant Viruses. Arch. Virol. 2015, 160, 17–20. [Google Scholar] [CrossRef] [PubMed]
- Tatineni, S.; Hein, G.L. Plant Viruses of Agricultural Importance: Current and Future Perspectives of Virus Disease Management Strategies. Phytopathology 2023, 113, 117–141. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhang, X.; Hong, Y.; Liu, Y. Chloroplast in Plant-Virus Interaction. Front. Microbiol. 2016, 7, 1565. [Google Scholar] [CrossRef]
- García-Arenal, F.; Fraile, A. Trade-offs in Host Range Evolution of Plant Viruses. Plant Pathol. 2013, 62, 2–9. [Google Scholar] [CrossRef]
- Ramlal, A.; Sarma, R.; Rani, A.; Nautiyal, A.; Kumar, J.; Mishra, V. Chapter 15—Plant-Virus Interactions in Plant Innate Immunity. In Plant RNA Viruses; Gaur, R.K., Patil, B.L., Selvarajan, R., Eds.; Academic Press: Cambridge, MA, USA, 2023; pp. 297–310. ISBN 978-0-323-95339-9. [Google Scholar]
- Niehl, A.; Wyrsch, I.; Boller, T.; Heinlein, M. Double-Stranded RNAs Induce a Pattern-Triggered Immune Signaling Pathway in Plants. New Phytol. 2016, 211, 1008–1019. [Google Scholar] [CrossRef]
- Amari, K.; Niehl, A. Nucleic Acid-Mediated PAMP-Triggered Immunity in Plants. Curr. Opin. Virol. 2020, 42, 32–39. [Google Scholar] [CrossRef]
- Kim, T.-J.; Lim, G.-H. Salicylic Acid and Mobile Regulators of Systemic Immunity in Plants: Transport and Metabolism. Plants 2023, 12, 1013. [Google Scholar] [CrossRef]
- Yang, Z.; Li, Y. Dissection of RNAi-Based Antiviral Immunity in Plants. Curr. Opin. Virol. 2018, 32, 88–99. [Google Scholar] [CrossRef]
- Guo, Z.; Li, Y.; Ding, S.-W. Small RNA-Based Antimicrobial Immunity. Nat. Rev. Immunol. 2019, 19, 31–44. [Google Scholar] [CrossRef]
- Baulcombe, D. RNA Silencing. Trends Biochem. Sci. 2005, 30, 290–293. [Google Scholar] [CrossRef]
- Alvarado, V.; Scholthof, H.B. Plant Responses against Invasive Nucleic Acids: RNA Silencing and Its Suppression by Plant Viral Pathogens. Semin. Cell Dev. Biol. 2009, 20, 1032–1040. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Ruiz, H.; Takeda, A.; Chapman, E.J.; Sullivan, C.M.; Fahlgren, N.; Brempelis, K.J.; Carrington, J.C. Arabidopsis RNA-Dependent RNA Polymerases and Dicer-like Proteins in Antiviral Defense and Small Interfering RNA Biogenesis during Turnip Mosaic Virus Infection. Plant Cell 2010, 22, 481–496. [Google Scholar] [CrossRef] [PubMed]
- Sanfaçon, H. Plant Translation Factors and Virus Resistance. Viruses 2015, 7, 3392–3419. [Google Scholar] [CrossRef]
- Wang, X.; Cao, X.; Liu, M.; Zhang, R.; Zhang, X.; Gao, Z.; Zhao, X.; Xu, K.; Li, D.; Zhang, Y. Hsc70-2 Is Required for Beet Black Scorch Virus Infection through Interaction with Replication and Capsid Proteins. Sci. Rep. 2018, 8, 4526. [Google Scholar] [CrossRef]
- Kim, S.H.; MacFarlane, S.; Kalinina, N.O.; Rakitina, D.V.; Ryabov, E.V.; Gillespie, T.; Haupt, S.; Brown, J.W.S.; Taliansky, M. Interaction of a Plant Virus-Encoded Protein with the Major Nucleolar Protein Fibrillarin Is Required for Systemic Virus Infection. Proc. Natl. Acad. Sci. USA 2007, 104, 11115–11120. [Google Scholar] [CrossRef]
- Ding, Y.; Lozano-Durán, R. The Cajal Body in Plant-Virus Interactions. Viruses 2020, 12, 250. [Google Scholar] [CrossRef]
- Shaw, J.; Love, A.J.; Makarova, S.S.; Kalinina, N.O.; Harrison, B.D.; Taliansky, M.E. Coilin, the Signature Protein of Cajal Bodies, Differentially Modulates the Interactions of Plants with Viruses in Widely Different Taxa. Nucleus 2014, 5, 85–94. [Google Scholar] [CrossRef]
- Lin, W.; Nagy, P.D. Co-Opted Cytosolic Proteins Form Condensate Substructures within Membranous Replication Organelles of a Positive-Strand RNA Virus. New Phytol. 2024, 243, 1917–1935. [Google Scholar] [CrossRef]
- Jiang, T.; Hao, T.; Chen, W.; Li, C.; Pang, S.; Fu, C.; Cheng, J.; Zhang, C.; Ghorbanpour, M.; Miao, S. Reprogrammed Plant Metabolism During Viral Infections: Mechanisms, Pathways and Implications. Mol. Plant Pathol. 2025, 26, e70066. [Google Scholar] [CrossRef]
- Yang, Y. The Application of Genomics in Agriculture. Agric. Sci. Food Process. 2025, 2, 26–46. Available online: https://www.icck.org/article/html/asfp.2025.111574 (accessed on 23 May 2025).
- Zanardo, L.G.; de Souza, G.B.; Alves, M.S. Transcriptomics of Plant–Virus Interactions: A Review. Theor. Exp. Plant Physiol. 2019, 31, 103–125. [Google Scholar] [CrossRef]
- Rurek, M.; Smolibowski, M. Variability of Plant Transcriptomic Responses under Stress Acclimation: A Review from High Throughput Studies. Acta Biochim. Pol. 2024, 71, 13585. [Google Scholar] [CrossRef]
- Souza, P.F.N.; Garcia-Ruiz, H.; Carvalho, F.E.L. What Proteomics Can Reveal about Plant–Virus Interactions? Photosynthesis-Related Proteins on the Spotlight. Theor. Exp. Plant Physiol. 2019, 31, 227–248. [Google Scholar] [CrossRef] [PubMed]
- Saleem, M.H.; Noreen, S.; Ishaq, I.; Saleem, A.; Khan, K.A.; Ercisli, S.; Anas, M.; Khalid, A.; Ahmed, T.; Hassan, A.; et al. Omics Technologies: Unraveling Abiotic Stress Tolerance Mechanisms for Sustainable Crop Improvement. J. Plant Growth Regul. 2025, 1–23. [Google Scholar] [CrossRef]
- Shalini, S.; Singla, A.; Goyal, M.; Kaur, V.; Kumar, P. Omics in Agriculture: Applications, Challenges and Future Perspectives. In Crop Improvement for Sustainability; Astral International (P) Ltd.: New Delhi, India, 2018; Volume 1, pp. 343–356. ISBN 978-93-5124-942-9. [Google Scholar]
- Raza, Q.; Azhar, M.T.; Rana, I.A.; Ahmad, M.Q.; Atif, R.M. 1—Biofortification of Crops to Achieve Food and Nutritional Security. In Biofortification of Grain and Vegetable Crops; Azhar, M.T., Ahmad, M.Q., Rana, I.A., Atif, R.M., Eds.; Academic Press: Cambridge, MA, USA, 2024; pp. 1–17. ISBN 978-0-323-91735-3. [Google Scholar]
- Zhang, C.; Wang, D.; Li, Y.; Wang, Z.; Wu, Z.; Zhang, Q.; Jia, H.; Dong, X.; Qi, L.; Shi, J.; et al. Gibberellin Positively Regulates Tomato Resistance to Tomato Yellow Leaf Curl Virus (TYLCV). Plants 2024, 13, 1277. [Google Scholar] [CrossRef]
- Zhang, K.; Zhuang, X.; Dong, Z.; Xu, K.; Chen, X.; Liu, F.; He, Z. The Dynamics of N6-Methyladenine RNA Modification in Interactions between Rice and Plant Viruses. Genome Biol. 2021, 22, 189. [Google Scholar] [CrossRef]
- He, Y.; Li, L.; Yao, Y.; Li, Y.; Zhang, H.; Fan, M. Transcriptome-Wide N6-Methyladenosine (m6A) Methylation in Watermelon under CGMMV Infection. BMC Plant Biol. 2021, 21, 516. [Google Scholar] [CrossRef]
- Yang, T.; Zhao, X.; Bai, J.; Lv, W.; Chen, Q.; Hu, J.; Liu, G.; Zhao, Y.; Zhou, H.; Zhao, M.; et al. Transcriptome Analysis of Genes Involved in the Pathogenesis Mechanism of Potato Virus Y in Potato Cultivar YouJin. Front. Microbiol. 2024, 15, 1353814. [Google Scholar] [CrossRef]
- Chen, T.; Lv, Y.; Zhao, T.; Li, N.; Yang, Y.; Yu, W.; He, X.; Liu, T.; Zhang, B. Comparative Transcriptome Profiling of a Resistant vs. Susceptible Tomato (Solanum lycopersicum) Cultivar in Response to Infection by Tomato Yellow Leaf Curl Virus. PLoS ONE 2013, 8, e80816. [Google Scholar] [CrossRef]
- Goyer, A.; Hamlin, L.; Crosslin, J.M.; Buchanan, A.; Chang, J.H. RNA-Seq Analysis of Resistant and Susceptible Potato Varieties during the Early Stages of Potato Virus Y Infection. BMC Genom. 2015, 16, 472. [Google Scholar] [CrossRef]
- Cho, W.K.; Lian, S.; Kim, S.-M.; Seo, B.Y.; Jung, J.K.; Kim, K.-H. Time-Course RNA-Seq Analysis Reveals Transcriptional Changes in Rice Plants Triggered by Rice Stripe Virus Infection. PLoS ONE 2015, 10, e0136736. [Google Scholar] [CrossRef] [PubMed]
- Zorzatto, C.; Machado, J.P.B.; Lopes, K.V.G.; Nascimento, K.J.T.; Pereira, W.A.; Brustolini, O.J.B.; Reis, P.A.B.; Calil, I.P.; Deguchi, M.; Sachetto-Martins, G.; et al. NIK1-Mediated Translation Suppression Functions as a Plant Antiviral Immunity Mechanism. Nature 2015, 520, 679–682. [Google Scholar] [CrossRef] [PubMed]
- Kamitani, M.; Nagano, A.J.; Honjo, M.N.; Kudoh, H. RNA-Seq Reveals Virus-Virus and Virus-Plant Interactions in Nature. FEMS Microbiol. Ecol. 2016, 92, fiw176. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, H.; Feng, M.; Zuo, D.; Hu, Y.; Jiang, T. Transcriptome Analysis of Woodland Strawberry (Fragaria vesca) Response to the Infection by Strawberry Vein Banding Virus (SVBV). Virol. J. 2016, 13, 128. [Google Scholar] [CrossRef]
- Collum, T.D.; Padmanabhan, M.S.; Hsieh, Y.-C.; Culver, J.N. Tobacco Mosaic Virus-Directed Reprogramming of Auxin/Indole Acetic Acid Protein Transcriptional Responses Enhances Virus Phloem Loading. Proc. Natl. Acad. Sci. USA 2016, 113, E2740–E2749. [Google Scholar] [CrossRef]
- Chen, L.; Fei, C.; Zhu, L.; Xu, Z.; Zou, W.; Yang, T.; Lin, H.; Xi, D. RNA-Seq Approach to Analysis of Gene Expression Profiles in Dark Green Islands and Light Green Tissues of Cucumber Mosaic Virus-Infected Nicotiana tabacum. PLoS ONE 2017, 12, e0175391. [Google Scholar] [CrossRef]
- Sun, X.; Wang, Z.; Gu, Q.; Li, H.; Han, W.; Shi, Y. Transcriptome Analysis of Cucumis sativus Infected by Cucurbit Chlorotic Yellows Virus. Virol. J. 2017, 14, 18. [Google Scholar] [CrossRef]
- Ghorbani, A.; Izadpanah, K.; Dietzgen, R.G. Changes in Maize Transcriptome in Response to Maize Iranian Mosaic Virus Infection. PLoS ONE 2018, 13, e0194592. [Google Scholar] [CrossRef]
- Anjanappa, R.B.; Mehta, D.; Okoniewski, M.J.; Szabelska-Berȩsewicz, A.; Gruissem, W.; Vanderschuren, H. Molecular Insights into Cassava brown streak virus Susceptibility and Resistance by Profiling of the Early Host Response. Mol. Plant Pathol. 2018, 19, 476–489. [Google Scholar] [CrossRef]
- Wang, J.; Yang, Y.; Jin, L.; Ling, X.; Liu, T.; Chen, T.; Ji, Y.; Yu, W.; Zhang, B. Re-Analysis of Long Non-Coding RNAs and Prediction of circRNAs Reveal Their Novel Roles in Susceptible Tomato Following TYLCV Infection. BMC Plant Biol. 2018, 18, 104. [Google Scholar] [CrossRef]
- Zhu, C.; Li, X.; Zheng, J. Transcriptome Profiling Using Illumina- and SMRT-Based RNA-Seq of Hot Pepper for in-Depth Understanding of Genes Involved in CMV Infection. Gene 2018, 666, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Withycombe, J.; Han, J.; MacWilliams, J.; Dorn, K.M.; Nalam, V.J.; Nachappa, P. Transcriptomic Profiling Reveals Distinct Responses to Beet Curly Top Virus (BCTV) Infection in Resistant and Susceptible Sugar Beet Genotypes. BMC Genom. 2024, 25, 1237. [Google Scholar] [CrossRef] [PubMed]
- He, W.-Z.; Zhao, L.; Sun, K.; Feng, Z.; Zhou, G.; Rao, Q. Transcriptomic Profiling Reveals the Complex Interaction between a Bipartite Begomovirus and a Cucurbitaceous Host Plant. BMC Genom. 2024, 25, 876. [Google Scholar] [CrossRef]
- Glushkevich, A.; Spechenkova, N.; Fesenko, I.; Knyazev, A.; Samarskaya, V.; Kalinina, N.O.; Taliansky, M.; Love, A.J. Transcriptomic Reprogramming, Alternative Splicing and RNA Methylation in Potato (Solanum tuberosum L.) Plants in Response to Potato Virus Y Infection. Plants 2022, 11, 635. [Google Scholar] [CrossRef]
- He, L.; Jin, P.; Chen, X.; Zhang, T.-Y.; Zhong, K.-L.; Liu, P.; Chen, J.-P.; Yang, J. Comparative Proteomic Analysis of Nicotiana Benthamiana Plants under Chinese Wheat Mosaic Virus Infection. BMC Plant Biol. 2021, 21, 51. [Google Scholar] [CrossRef]
- Niehl, A.; Zhang, Z.J.; Kuiper, M.; Peck, S.C.; Heinlein, M. Label-Free Quantitative Proteomic Analysis of Systemic Responses to Local Wounding and Virus Infection in Arabidopsis thaliana. J. Proteom. Res. 2013, 12, 2491–2503. [Google Scholar] [CrossRef]
- Serra-Soriano, M.; Navarro, J.A.; Genoves, A.; Pallás, V. Comparative Proteomic Analysis of Melon Phloem Exudates in Response to Viral Infection. J. Proteom. 2015, 124, 11–24. [Google Scholar] [CrossRef]
- Wang, B.; Hajano, J.-U.-D.; Ren, Y.; Lu, C.; Wang, X. iTRAQ-Based Quantitative Proteomics Analysis of Rice Leaves Infected by Rice Stripe Virus Reveals Several Proteins Involved in Symptom Formation. Virol. J. 2015, 12, 99. [Google Scholar] [CrossRef]
- Wang, J.; Wang, X.; Zhou, Q.; Yang, J.; Guo, H.; Yang, L.; Liu, W. iTRAQ Protein Profile Analysis Provides Integrated Insight into Mechanisms of Tolerance to TMV in Tobacco (Nicotiana tabacum). J. Proteom. 2016, 132, 21–30. [Google Scholar] [CrossRef]
- Zhong, X.; Wang, Z.Q.; Xiao, R.; Wang, Y.; Xie, Y.; Zhou, X. iTRAQ Analysis of the Tobacco Leaf Proteome Reveals That RNA-Directed DNA Methylation (RdDM) Has Important Roles in Defense against Geminivirus-Betasatellite Infection. J. Proteom. 2017, 152, 88–101. [Google Scholar] [CrossRef]
- Paiva, A.L.S.; Oliveira, J.T.A.; de Souza, G.A.; Vasconcelos, I.M. Label-Free Proteomic Reveals That Cowpea Severe Mosaic Virus Transiently Suppresses the Host Leaf Protein Accumulation During the Compatible Interaction with Cowpea (Vigna unguiculata [L.] Walp.). J. Proteom. Res. 2016, 15, 4208–4220. [Google Scholar] [CrossRef] [PubMed]
- Cerna, H.; Černý, M.; Habánová, H.; Šafářová, D.; Abushamsiya, K.; Navrátil, M.; Brzobohatý, B. Proteomics Offers Insight to the Mechanism behind Pisum sativum L. Response to Pea Seed-Borne Mosaic Virus (PSbMV). J. Proteom. 2017, 153, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.; Yu, L.; Chen, Z.; Zhang, S.; Shi, J.; Zhao, X.; Yang, Y.; Hu, D.; Song, B. Label-Free Quantitative Proteomic Analysis of Chitosan Oligosaccharide-Treated Rice Infected with Southern Rice Black-Streaked Dwarf Virus. Viruses 2017, 9, 115. [Google Scholar] [CrossRef]
- Varela, A.L.N.; Oliveira, J.T.A.; Komatsu, S.; Silva, R.G.G.; Martins, T.F.; Souza, P.F.N.; Lobo, A.K.M.; Vasconcelos, I.M.; Carvalho, F.E.L.; Silveira, J.A.G. A Resistant Cowpea (Vigna unguiculata [L.] Walp.) Genotype Became Susceptible to Cowpea Severe Mosaic Virus (CPSMV) after Exposure to Salt Stress. J. Proteom. 2019, 194, 200–217. [Google Scholar] [CrossRef]
- Feng, H.; Mon, W.; Su, X.; Li, Y.; Zhang, S.; Zhang, Z.; Zheng, K. Integrated Biological Experiments and Proteomic Analyses of Nicotiana tabacum Xylem Sap Revealed the Host Response to Tomato Spotted Wilt Orthotospovirus Infection. Int. J. Mol. Sci. 2024, 25, 10907. [Google Scholar] [CrossRef]
- Fesenko, I.; Spechenkova, N.; Mamaeva, A.; Makhotenko, A.V.; Love, A.J.; Kalinina, N.O.; Taliansky, M. Role of the Methionine Cycle in the Temperature-Sensitive Responses of Potato Plants to Potato Virus Y. Mol. Plant Pathol. 2021, 22, 77–91. [Google Scholar] [CrossRef]
- Fernández-Calvino, L.; Osorio, S.; Hernández, M.L.; Hamada, I.B.; del Toro, F.J.; Donaire, L.; Yu, A.; Bustos, R.; Fernie, A.R.; Martínez-Rivas, J.M.; et al. Virus-Induced Alterations in Primary Metabolism Modulate Susceptibility to Tobacco rattle virus in Arabidopsis. Plant Physiol. 2014, 166, 1821–1838. [Google Scholar] [CrossRef]
- Wang, S.; Wang, X.; Li, S.; Sun, X.; Xue, M.; Di, D.; Zhang, A.; Zhang, Y.; Xia, Y.; Zhou, T.; et al. Maize Lipid Droplet-Associated Protein 2 Is Recruited by a Virus to Enhance Viral Multiplication and Infection through Regulating Cellular Fatty Acid Metabolism. Plant J. 2024, 119, 2484–2499. [Google Scholar] [CrossRef]
- Zhang, Z.; He, H.; Yan, M.; Zhao, C.; Lei, C.; Li, J.; Yan, F. Widely Targeted Analysis of Metabolomic Changes of Cucumis Sativus Induced by Cucurbit Chlorotic Yellows Virus. BMC Plant Biol. 2022, 22, 158. [Google Scholar] [CrossRef]
- Yuan, W.; Jiang, T.; Du, K.; Chen, H.; Cao, Y.; Xie, J.; Li, M.; Carr, J.P.; Wu, B.; Fan, Z.; et al. Maize Phenylalanine Ammonia-Lyases Contribute to Resistance to Sugarcane Mosaic Virus Infection, Most Likely through Positive Regulation of Salicylic Acid Accumulation. Mol. Plant Pathol. 2019, 20, 1365–1378. [Google Scholar] [CrossRef]
- Montero, R.; Pérez-Bueno, M.L.; Barón, M.; Florez-Sarasa, I.; Tohge, T.; Fernie, A.R.; Ouad, H.E.A.; Flexas, J.; Bota, J. Alterations in Primary and Secondary Metabolism in Vitis vinifera “Malvasía de Banyalbufar” upon Infection with Grapevine Leafroll-Associated Virus 3. Physiol. Plant. 2016, 157, 442–452. [Google Scholar] [CrossRef] [PubMed]
- Pistelli, L.; Ferri, B.; Cioni, P.L.; Koziara, M.; Agacka, M.; Skomra, U. Aroma Profile and Bitter Acid Characterization of Hop Cones (Humulus lupulus L.) of Five Healthy and Infected Polish Cultivars. Ind. Crops Prod. 2018, 124, 653–662. [Google Scholar] [CrossRef]
- Lan, H.; Lai, B.; Zhao, P.; Dong, X.; Wei, W.; Ye, Y.; Wu, Z. Cucumber Mosaic Virus Infection Modulated the Phytochemical Contents of Passiflora edulis. Microb. Pathog. 2020, 138, 103828. [Google Scholar] [CrossRef]
- Parizad, S.; Dizadji, A.; Habibi, M.K.; Winter, S.; Kalantari, S.; Movi, S.; Lorenzo Tendero, C.; Alonso, G.L.; Moratalla-Lopez, N. The Effects of Geographical Origin and Virus Infection on the Saffron (Crocus sativus L.) Quality. Food Chem. 2019, 295, 387–394. [Google Scholar] [CrossRef]
- Rajaprakasam, S.; Shanmugavel, P.; Chockalingam, V.; Jegadeesan, S.; Latha, T.K.S.; Ananthan, S.N.; Muthurajan, R.; Kanagarajan, S. Comparative Metabolomic Profiling of Horse Gram (Macrotyloma uniflorum (Lam.) Verdc.) Genotypes for Horse Gram Yellow Mosaic Virus Resistance. Metabolites 2023, 13, 165. [Google Scholar] [CrossRef]
- Kumar, B.K.P.; Kanakala, S.; Malathi, V.G.; Gopal, P.; Usha, R. Transcriptomic and Proteomic Analysis of Yellow Mosaic Diseased Soybean. J. Plant Biochem. Biotechnol. 2017, 26, 224–234. [Google Scholar] [CrossRef]
- Stare, T.; Stare, K.; Weckwerth, W.; Wienkoop, S.; Gruden, K. Comparison between Proteome and Transcriptome Response in Potato (Solanum tuberosum L.) Leaves Following Potato Virus Y (PVY) Infection. Proteomes 2017, 5, 14. [Google Scholar] [CrossRef]
- Stare, T.; Ramšak, Ž.; Križnik, M.; Gruden, K. Multiomics Analysis of Tolerant Interaction of Potato with Potato Virus Y. Sci. Data 2019, 6, 250. [Google Scholar] [CrossRef]
- Akbar, S.; Yao, W.; Yu, K.; Qin, L.; Ruan, M.; Powell, C.A.; Chen, B.; Zhang, M. Photosynthetic Characterization and Expression Profiles of Sugarcane Infected by Sugarcane Mosaic Virus (SCMV). Photosynth. Res. 2021, 150, 279–294. [Google Scholar] [CrossRef]
- Akbar, S.; Yao, W.; Qin, L.; Yuan, Y.; Powell, C.A.; Chen, B.; Zhang, M. Comparative Analysis of Sugar Metabolites and Their Transporters in Sugarcane Following Sugarcane Mosaic Virus (SCMV) Infection. Int. J. Mol. Sci. 2021, 22, 13574. [Google Scholar] [CrossRef]
- Lyu, S.; Gao, L.; Zhang, R.; Zhang, C.; Hou, X. Correlation Analysis of Expression Profile and Quantitative iTRAQ-LC-MS/MS Proteomics Reveals Resistance Mechanism Against TuMV in Chinese Cabbage (Brassica rapa ssp. Pekinensis). Front. Genet. 2020, 11, 963. [Google Scholar] [CrossRef] [PubMed]
- Roy, B.G.; DeBlasio, S.; Yang, Y.; Thannhauser, T.; Heck, M.; Fuchs, M. Profiling Plant Proteome and Transcriptome Changes during Grapevine Fanleaf Virus Infection. J. Proteom. Res. 2023, 22, 1997–2017. [Google Scholar] [CrossRef] [PubMed]
- Kogovšek, P.; Pompe-Novak, M.; Petek, M.; Fragner, L.; Weckwerth, W.; Gruden, K. Primary Metabolism, Phenylpropanoids and Antioxidant Pathways Are Regulated in Potato as a Response to Potato Virus Y Infection. PLoS ONE 2016, 11, e0146135. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Ulate, B.; Hopfer, H.; Figueroa-Balderas, R.; Ye, Z.; Rivero, R.M.; Albacete, A.; Pérez-Alfocea, F.; Koyama, R.; Anderson, M.M.; Smith, R.J.; et al. Red Blotch Disease Alters Grape Berry Development and Metabolism by Interfering with the Transcriptional and Hormonal Regulation of Ripening. J. Exp. Bot. 2017, 68, 1225–1238. [Google Scholar] [CrossRef]
- Rumbaugh, A.C.; Durbin-Johnson, B.; Padhi, E.; Lerno, L.; Cauduro Girardello, R.; Britton, M.; Slupsky, C.; Sudarshana, M.R.; Oberholster, A. Investigating Grapevine Red Blotch Virus Infection in Vitis vinifera L. Cv. Cabernet Sauvignon Grapes: A Multi-Omics Approach. Int. J. Mol. Sci. 2022, 23, 13248. [Google Scholar] [CrossRef]
- Lu, X.; Zhang, L.; Huang, W.; Zhang, S.; Zhang, S.; Li, F.; Zhang, H.; Sun, R.; Zhao, J.; Li, G. Integrated Volatile Metabolomics and Transcriptomics Analyses Reveal the Influence of Infection TuMV to Volatile Organic Compounds in Brassica rapa. Horticulturae 2022, 8, 57. [Google Scholar] [CrossRef]
- Lv, J.; Deng, M.; Li, Z.; Zhu, H.; Wang, Z.; Yue, Y.; Yang, Z.; Xu, J.; Jiang, S.; Zhao, W.; et al. Integrative Analysis of the Transcriptome and Metabolome Reveals the Response Mechanism to Tomato Spotted Wilt Virus. Hortic. Plant J. 2023, 9, 958–970. [Google Scholar] [CrossRef]
- Siriwan, W.; Vannatim, N.; Chaowongdee, S.; Roytrakul, S.; Charoenlappanit, S.; Pongpamorn, P.; Paemanee, A.; Malichan, S. Integrated Proteomic and Metabolomic Analysis of Cassava Cv. Kasetsart 50 Infected with Sri Lankan Cassava Mosaic Virus. Agronomy 2023, 13, 945. [Google Scholar] [CrossRef]
- Zhai, Y.; Gnanasekaran, P.; Pappu, H.R. Identification and Characterization of Plant-Interacting Targets of Tomato Spotted Wilt Virus Silencing Suppressor. Pathogens 2021, 10, 27. [Google Scholar] [CrossRef]
- Salgotra, R.K.; Gupta, B.B.; Stewart, C.N. From Genomics to Functional Markers in the Era of Next-Generation Sequencing. Biotechnol. Lett. 2014, 36, 417–426. [Google Scholar] [CrossRef]
- Jha, U.C.; Nayyar, H.; Chattopadhyay, A.; Beena, R.; Lone, A.A.; Naik, Y.D.; Thudi, M.; Prasad, P.V.V.; Gupta, S.; Dixit, G.P.; et al. Major Viral Diseases in Grain Legumes: Designing Disease Resistant Legumes from Plant Breeding and OMICS Integration. Front Plant Sci. 2023, 14, 1183505. [Google Scholar] [CrossRef] [PubMed]
- Asano, K.; Endelman, J.B. Development of KASP Markers for the Potato Virus Y Resistance Gene Rychc Using Whole-Genome Resequencing Data. bioRxiv 2023. [Google Scholar] [CrossRef]
- Pérez-de-Castro, A.M.; Vilanova, S.; Cañizares, J.; Pascual, L.; Blanca, J.M.; Díez, M.J.; Prohens, J.; Picó, B. Application of Genomic Tools in Plant Breeding. Curr. Genom. 2012, 13, 179–195. [Google Scholar] [CrossRef] [PubMed]
- Monnot, S.; Desaint, H.; Mary-Huard, T.; Moreau, L.; Schurdi-Levraud, V.; Boissot, N. Deciphering the Genetic Architecture of Plant Virus Resistance by GWAS, State of the Art and Potential Advances. Cells 2021, 10, 3080. [Google Scholar] [CrossRef] [PubMed]
- Kalinina, N.O.; Khromov, A.; Love, A.J.; Taliansky, M.E. CRISPR Applications in Plant Virology: Virus Resistance and Beyond. Phytopathology 2020, 110, 18–28. [Google Scholar] [CrossRef]
- Tamisier, L.; Szadkowski, M.; Nemouchi, G.; Lefebvre, V.; Szadkowski, E.; Duboscq, R.; Santoni, S.; Sarah, G.; Sauvage, C.; Palloix, A.; et al. Genome-Wide Association Mapping of QTLs Implied in Potato Virus Y Population Sizes in Pepper: Evidence for Widespread Resistance QTL Pyramiding. Mol. Plant Pathol. 2020, 21, 3–16. [Google Scholar] [CrossRef]
- Montes, N.; Cobos, A.; Gil-Valle, M.; Caro, E.; Pagán, I. Arabidopsis thaliana Genes Associated with Cucumber mosaic virus Virulence and Their Link to Virus Seed Transmission. Microorganisms 2021, 9, 692. [Google Scholar] [CrossRef]
- Liu, Q.; Lan, G.; Zhu, Y.; Chen, K.; Shen, C.; Zhao, X.; Zhang, F.; Xu, J.; Li, Z. Genome-Wide Association Study on Resistance to Rice Black-Streaked Dwarf Disease Caused by Rice black-streaked dwarf virus. Plant Dis. 2021, 105, 607–615. [Google Scholar] [CrossRef]
- Wolfe, M.D.; Rabbi, I.Y.; Egesi, C.; Hamblin, M.; Kawuki, R.; Kulakow, P.; Lozano, R.; Carpio, D.P.D.; Ramu, P.; Jannink, J.-L. Genome-Wide Association and Prediction Reveals Genetic Architecture of Cassava Mosaic Disease Resistance and Prospects for Rapid Genetic Improvement. Plant Genome 2016, 9, 1–13. [Google Scholar] [CrossRef]
- Li, J.; Tang, Y.; Jacobson, A.L.; Dang, P.M.; Li, X.; Wang, M.L.; Hagan, A.; Chen, C.Y. Population Structure and Association Mapping to Detect QTL Controlling Tomato Spotted Wilt Virus Resistance in Cultivated Peanuts. Crop J. 2018, 6, 516–526. [Google Scholar] [CrossRef]
- Kante, M.; Lindqvist-Kreuze, H.; Portal, L.; David, M.; Gastelo, M. Kompetitive Allele Specific PCR (KASP) Markers for Potato: An Effective Tool for Increased Genetic Gains. Agronomy 2021, 11, 2315. [Google Scholar] [CrossRef]
- Garcia-Ruiz, H. Susceptibility Genes to Plant Viruses. Viruses 2018, 10, 484. [Google Scholar] [CrossRef] [PubMed]
- McLeish, M.J.; Fraile, A.; García-Arenal, F. Population Genomics of Plant Viruses: The Ecology and Evolution of Virus Emergence. Phytopathology 2021, 111, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Villamor, D.E.V.; Ho, T.; Al Rwahnih, M.; Martin, R.R.; Tzanetakis, I.E. High Throughput Sequencing for Plant Virus Detection and Discovery. Phytopathology 2019, 109, 716–725. [Google Scholar] [CrossRef]
- Hrdlickova, R.; Toloue, M.; Tian, B. RNA-Seq Methods for Transcriptome Analysis. Wiley Interdiscip. Rev. RNA 2017, 8, 1364. [Google Scholar] [CrossRef]
- Monzó, C.; Liu, T.; Conesa, A. Transcriptomics in the Era of Long-Read Sequencing. Nat. Rev. Genet. 2025; Online ahead of print. [Google Scholar] [CrossRef]
- Conesa, A.; Madrigal, P.; Tarazona, S.; Gomez-Cabrero, D.; Cervera, A.; McPherson, A.; Szcześniak, M.W.; Gaffney, D.J.; Elo, L.L.; Zhang, X.; et al. A Survey of Best Practices for RNA-Seq Data Analysis. Genome Biol. 2016, 17, 13. [Google Scholar] [CrossRef]
- Babraham Bioinformatics. FastQC a Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 16 May 2025).
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-Based Genome Alignment and Genotyping with HISAT2 and HISAT-Genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De Novo Transcript Sequence Reconstruction from RNA-Seq Using the Trinity Platform for Reference Generation and Analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows–Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Suleski, M.; Sanderford, M.; Sharma, S.; Tamura, K. MEGA12: Molecular Evolutionary Genetic Analysis Version 12 for Adaptive and Green Computing. Mol. Biol. Evol. 2024, 41, msae263. [Google Scholar] [CrossRef]
- Martin, D.P.; Murrell, B.; Golden, M.; Khoosal, A.; Muhire, B. RDP4: Detection and Analysis of Recombination Patterns in Virus Genomes. Virus Evol. 2015, 1, vev003. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Golosova, O.; Fursov, M. Unipro UGENE: A Unified Bioinformatics Toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML-VI-HPC: Maximum Likelihood-Based Phylogenetic Analyses with Thousands of Taxa and Mixed Models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef]
- Zagordi, O.; Bhattacharya, A.; Eriksson, N.; Beerenwinkel, N. ShoRAH: Estimating the Genetic Diversity of a Mixed Sample from next-Generation Sequencing Data. BMC Bioinform. 2011, 12, 119. [Google Scholar] [CrossRef]
- Fritz, A.; Bremges, A.; Deng, Z.-L.; Lesker, T.R.; Götting, J.; Ganzenmueller, T.; Sczyrba, A.; Dilthey, A.; Klawonn, F.; McHardy, A.C. Haploflow: Strain-Resolved de Novo Assembly of Viral Genomes. Genome Biol. 2021, 22, 212. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- UniProt. Available online: https://www.uniprot.org (accessed on 16 May 2025).
- ORFfinder Home-NCBI. Available online: https://www.ncbi.nlm.nih.gov/orffinder/ (accessed on 16 May 2025).
- National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov/ (accessed on 16 May 2025).
- Urban, M.; Cuzick, A.; Seager, J.; Nonavinakere, N.; Sahoo, J.; Sahu, P.; Iyer, V.L.; Khamari, L.; Martinez, M.C.; Hammond-Kosack, K.E. PHI-Base–the Multi-Species Pathogen–Host Interaction Database in 2025. Nucleic Acids Res. 2025, 53, D826–D838. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Matsuura, Y.; Ishiguro-Watanabe, M. KEGG: Biological Systems Database as a Model of the Real World. Nucleic Acids Res. 2025, 53, D672–D677. [Google Scholar] [CrossRef]
- Liu, B.; Zheng, D.; Jin, Q.; Chen, L.; Yang, J. VFDB 2019: A Comparative Pathogenomic Platform with an Interactive Web Interface. Nucleic Acids Res. 2019, 47, D687–D692. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Thorvaldsdóttir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-Performance Genomics Data Visualization and Exploration. Brief. Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef]
- Waskom, M.L. Seaborn: Statistical Data Visualization. J. Open Source Softw. 2021, 6, 3021. [Google Scholar] [CrossRef]
- Hunter, J.D. Matplotlib: A 2D Graphics Environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An Information Aesthetic for Comparative Genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Wang, J.; Yu, W.; Yang, Y.; Li, X.; Chen, T.; Liu, T.; Ma, N.; Yang, X.; Liu, R.; Zhang, B. Genome-Wide Analysis of Tomato Long Non-Coding RNAs and Identification as Endogenous Target Mimic for microRNA in Response to TYLCV Infection. Sci. Rep. 2015, 5, 16946. [Google Scholar] [CrossRef] [PubMed]
- Niu, X.; Sun, Y.; Chen, Z.; Li, R.; Padmanabhan, C.; Ruan, J.; Kreuze, J.F.; Ling, K.; Fei, Z.; Gao, S. Using Small RNA-Seq Data to Detect siRNA Duplexes Induced by Plant Viruses. Genes 2017, 8, 163. [Google Scholar] [CrossRef] [PubMed]
- Ramesh, S.V.; Williams, S.; Kappagantu, M.; Mitter, N.; Pappu, H.R. Transcriptome-Wide Identification of Host Genes Targeted by Tomato Spotted Wilt Virus-Derived Small Interfering RNAs. Virus Res. 2017, 238, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Samarskaya, V.O.; Spechenkova, N.; Ilina, I.; Suprunova, T.P.; Kalinina, N.O.; Love, A.J.; Taliansky, M.E. A Non-Canonical Pathway Induced by Externally Applied Virus-Specific dsRNA in Potato Plants. Int. J. Mol. Sci. 2023, 24, 15769. [Google Scholar] [CrossRef]
- Wang, J.; Zou, A.; Xiang, S.; Liu, C.; Peng, H.; Wen, Y.; Ma, X.; Chen, H.; Ran, M.; Sun, X. Transcriptome Analysis Reveals the Mechanism of Zinc Ion-Mediated Plant Resistance to TMV in Nicotiana benthamiana. Pestic. Biochem. Physiol. 2022, 184, 105100. [Google Scholar] [CrossRef]
- Samarskaya, V.; Kuznetsova, M.; Gryzunov, N.; Spechenkova, N.; Bagdasarova, P.; Ryabov, E.; Taliansky, M.; Kalinina, N.O. Identification of Two Novel Recombinant Types of Potato Virus Y from Solanum tuberosum Plants in Southern Region of Russia. Plant Dis. 2024, 109, 998–1003. [Google Scholar] [CrossRef]
- Gryzunov, N.; Morozov, S.Y.; Suprunova, T.; Samarskaya, V.; Spechenkova, N.; Yakunina, S.; Kalinina, N.O.; Taliansky, M. Genomes of Alphanucleorhabdovirus Physostegiae Isolates from Two Different Cultivar Groups of Solanum melongena. Viruses 2024, 16, 1538. [Google Scholar] [CrossRef]
- Jia, A.; Yan, C.; Yin, H.; Sun, R.; Xia, F.; Gao, L.; Zhang, Y.; Li, Y. Small RNA and Transcriptome Sequencing of a Symptomatic Peony Plant Reveals Mixed Infections with Novel Viruses. Plant Dis. 2021, 105, 3816–3828. [Google Scholar] [CrossRef]
- Sang, T.; Zhang, Z.; Liu, G.; Wang, P. Navigating the Landscape of Plant Proteomics. J. Integr. Plant Biol. 2025, 67, 740–761. [Google Scholar] [CrossRef]
- Chen, X.; Sun, Y.; Zhang, T.; Shu, L.; Roepstorff, P.; Yang, F. Quantitative Proteomics Using Isobaric Labeling: A Practical Guide. Genom. Proteom. Bioinform. 2021, 19, 689–706. [Google Scholar] [CrossRef]
- Ma, W.; Zhang, C.; Zhang, W.; Sheng, P.; Xu, M.; Ni, Y.; Chen, M.; Cheng, B.; Zhang, X. TMT-Based Comparative Peptidomics Analysis of Rice Seedlings under Salt Stress: An Accessible Method to Explore Plant Stress-Tolerance Processing. J. Proteom. Res. 2022, 21, 2905–2919. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Bi, X.; An, M.; Xia, Z.; Wu, Y. iTRAQ-Based Proteomic Analysis of Watermelon Fruits in Response to Cucumber Green Mottle Mosaic Virus Infection. Int. J. Mol. Sci. 2020, 21, 2541. [Google Scholar] [CrossRef] [PubMed]
- Lewandowska, D.; ten Have, S.; Hodge, K.; Tillemans, V.; Lamond, A.I.; Brown, J.W.S. Plant SILAC: Stable-Isotope Labelling with Amino Acids of Arabidopsis Seedlings for Quantitative Proteomics. PLoS ONE 2013, 8, e72207. [Google Scholar] [CrossRef]
- Zuniga, N.R.; Frost, D.C.; Kuhn, K.; Shin, M.; Whitehouse, R.L.; Wei, T.-Y.; He, Y.; Dawson, S.L.; Pike, I.; Bomgarden, R.D.; et al. Achieving a 35-Plex Tandem Mass Tag Reagent Set through Deuterium Incorporation. J. Proteom. Res. 2024, 23, 5153–5165. [Google Scholar] [CrossRef]
- Chen, C.; Hou, J.; Tanner, J.J.; Cheng, J. Bioinformatics Methods for Mass Spectrometry-Based Proteomics Data Analysis. Int. J. Mol. Sci. 2020, 21, 2873. [Google Scholar] [CrossRef]
- Perkins, D.N.; Pappin, D.J.; Creasy, D.M.; Cottrell, J.S. Probability-Based Protein Identification by Searching Sequence Databases Using Mass Spectrometry Data. Electrophoresis 1999, 20, 3551–3567. [Google Scholar] [CrossRef]
- Brodbelt, J.S.; Russell, D.H. Focus on the 20-Year Anniversary of SEQUEST. J. Am. Soc. Mass Spectrom. 2015, 26, 1797–1798. [Google Scholar] [CrossRef]
- Li, Y.F.; Arnold, R.J.; Li, Y.; Radivojac, P.; Sheng, Q.; Tang, H. A Bayesian Approach to Protein Inference Problem in Shotgun Proteomics. J. Comput. Biol. 2009, 16, 1183–1193. [Google Scholar] [CrossRef]
- Xu, Y.; Shi, Z.; Bao, L. An Expanding Repertoire of Protein Acylations. Mol. Cell. Proteom. 2022, 21, 100193. [Google Scholar] [CrossRef]
- Sang, T.; Xu, Y.; Qin, G.; Zhao, S.; Hsu, C.-C.; Wang, P. Highly Sensitive Site-Specific SUMOylation Proteomics in Arabidopsis. Nat. Plants 2024, 10, 1330–1342. [Google Scholar] [CrossRef]
- Yan, S.; Bhawal, R.; Yin, Z.; Thannhauser, T.W.; Zhang, S. Recent Advances in Proteomics and Metabolomics in Plants. Mol. Hortic. 2022, 2, 17. [Google Scholar] [CrossRef] [PubMed]
- Alexander, M.M.; Cilia, M. A Molecular Tug-of-War: Global Plant Proteome Changes during Viral Infection. Curr. Plant Biol. 2016, 5, 13–24. [Google Scholar] [CrossRef]
- Kamran, A.; Hussain, M.D.; Farooq, T.; Li, F.; Khan, M.; Li, X.; Yang, S.; Xie, X. Deciphering Intricate Plant-Virus Interactions: Potyvirids Orchestrate Protein Posttranslational Modifications to Regulate Pathogenicity. Microbiol. Res. 2025, 290, 127940. [Google Scholar] [CrossRef]
- Obrępalska-Stęplowska, A.; Renaut, J.; Planchon, S.; Przybylska, A.; Wieczorek, P.; Barylski, J.; Palukaitis, P. Effect of Temperature on the Pathogenesis, Accumulation of Viral and Satellite RNAs and on Plant Proteome in Peanut Stunt Virus and Satellite RNA-Infected Plants. Front. Plant Sci. 2015, 6, 903. [Google Scholar] [CrossRef]
- Rajamäki, M.-L.; Sikorskaite-Gudziuniene, S.; Sarmah, N.; Varjosalo, M.; Valkonen, J.P.T. Nuclear Proteome of Virus-Infected and Healthy Potato Leaves. BMC Plant Biol. 2020, 20, 355. [Google Scholar] [CrossRef]
- Anfoka, G.; Moshe, A.; Fridman, L.; Amrani, L.; Rotem, O.; Kolot, M.; Zeidan, M.; Czosnek, H.; Gorovits, R. Tomato Yellow Leaf Curl Virus Infection Mitigates the Heat Stress Response of Plants Grown at High Temperatures. Sci. Rep. 2016, 6, 19715. [Google Scholar] [CrossRef]
- Prasch, C.M.; Sonnewald, U. Simultaneous Application of Heat, Drought, and Virus to Arabidopsis Plants Reveals Significant Shifts in Signaling Networks. Plant Physiol. 2013, 162, 1849–1866. [Google Scholar] [CrossRef]
- Pang, Z.; Lu, Y.; Zhou, G.; Hui, F.; Xu, L.; Viau, C.; Spigelman, A.F.; MacDonald, P.E.; Wishart, D.S.; Li, S.; et al. MetaboAnalyst 6.0: Towards a Unified Platform for Metabolomics Data Processing, Analysis and Interpretation. Nucleic Acids Res. 2024, 52, W398–W406. [Google Scholar] [CrossRef]
- Cambiaghi, A.; Ferrario, M.; Masseroli, M. Analysis of Metabolomic Data: Tools, Current Strategies and Future Challenges for Omics Data Integration. Brief. Bioinform. 2017, 18, 498–510. [Google Scholar] [CrossRef]
- Sade, D.; Shriki, O.; Cuadros-Inostroza, A.; Tohge, T.; Semel, Y.; Haviv, Y.; Willmitzer, L.; Fernie, A.R.; Czosnek, H.; Brotman, Y. Comparative Metabolomics and Transcriptomics of Plant Response to Tomato Yellow Leaf Curl Virus Infection in Resistant and Susceptible Tomato Cultivars. Metabolomics 2015, 11, 81–97. [Google Scholar] [CrossRef]
- Liu, Y.; Li, Y.; Chen, E.; Xu, J.; Zhang, W.; Zeng, X.; Luo, X. Repeat and Haplotype Aware Error Correction in Nanopore Sequencing Reads with DeChat. Commun. Biol. 2024, 7, 1678. [Google Scholar] [CrossRef] [PubMed]
- Dillies, M.-A.; Rau, A.; Aubert, J.; Hennequet-Antier, C.; Jeanmougin, M.; Servant, N.; Keime, C.; Marot, G.; Castel, D.; Estelle, J.; et al. A Comprehensive Evaluation of Normalization Methods for Illumina High-Throughput RNA Sequencing Data Analysis. Brief. Bioinform. 2013, 14, 671–683. [Google Scholar] [CrossRef] [PubMed]
- Wagner, G.P.; Kin, K.; Lynch, V.J. Measurement of mRNA Abundance Using RNA-Seq Data: RPKM Measure Is Inconsistent among Samples. Theory. Biosci. 2012, 131, 281–285. [Google Scholar] [CrossRef]
- Leek, J.T.; Scharpf, R.B.; Bravo, H.C.; Simcha, D.; Langmead, B.; Johnson, W.E.; Geman, D.; Baggerly, K.; Irizarry, R.A. Tackling the Widespread and Critical Impact of Batch Effects in High-Throughput Data. Nat. Rev. Genet. 2010, 11, 733–739. [Google Scholar] [CrossRef]
- Risso, D.; Ngai, J.; Speed, T.P.; Dudoit, S. Normalization of RNA-Seq Data Using Factor Analysis of Control Genes or Samples. Nat. Biotechnol. 2014, 32, 896–902. [Google Scholar] [CrossRef]
- Yang, Y.; Saand, M.A.; Huang, L.; Abdelaal, W.B.; Zhang, J.; Wu, Y.; Li, J.; Sirohi, M.H.; Wang, F. Applications of Multi-Omics Technologies for Crop Improvement. Front. Plant Sci. 2021, 12, 563953. [Google Scholar] [CrossRef]
- Mostovenko, E.; Lichti, C.F.; Wang, Q. Similarity Measures between Proteomic and Transcriptomic Data as a Tool to Highlight Phenotypical Differences in 33 Glioma Stem Cell Lines. MOJ Proteom. Bioinform. 2015, 2, 186–191. [Google Scholar] [CrossRef]
- Dupont, C.; Armant, D.R.; Brenner, C.A. Epigenetics: Definition, Mechanisms and Clinical Perspective. Semin. Reprod. Med. 2009, 27, 351–357. [Google Scholar] [CrossRef]
- Mansoor, S.; Hamid, S.; Tuan, T.T.; Park, J.-E.; Chung, Y.S. Advance Computational Tools for Multiomics Data Learning. Biotechnol. Adv. 2024, 77, 108447. [Google Scholar] [CrossRef]
- Robles-Zazueta, C.A.; Pinto, F.; Molero, G.; Foulkes, M.J.; Reynolds, M.P.; Murchie, E.H. Prediction of Photosynthetic, Biophysical, and Biochemical Traits in Wheat Canopies to Reduce the Phenotyping Bottleneck. Front. Plant Sci. 2022, 13, 828451. [Google Scholar] [CrossRef]
- Awada, L.; Phillips, P.W.B.; Bodan, A.M. The Evolution of Plant Phenomics: Global Insights, Trends, and Collaborations (2000–2021). Front. Plant Sci. 2024, 15, 1410738. [Google Scholar] [CrossRef] [PubMed]
- Kaya, C. Optimizing Crop Production with Plant Phenomics Through High-Throughput Phenotyping and AI in Controlled Environments. Food Energy Secur. 2025, 14, e70050. [Google Scholar] [CrossRef]
- Jiang, T.; Du, K.; Xie, J.; Sun, G.; Wang, P.; Chen, X.; Cao, Z.; Wang, B.; Chao, Q.; Li, X.; et al. Activated Malate Circulation Contributes to the Manifestation of Light-Dependent Mosaic Symptoms. Cell Rep. 2023, 42, 112333. [Google Scholar] [CrossRef]
- Abdelkhalek, A.; Király, L.; Al-Mansori, A.-N.A.; Younes, H.A.; Zeid, A.; Elsharkawy, M.M.; Behiry, S.I. Defense Responses and Metabolic Changes Involving Phenylpropanoid Pathway and PR Genes in Squash (Cucurbita pepo L.) Following Cucumber Mosaic Virus Infection. Plants 2022, 11, 1908. [Google Scholar] [CrossRef]
- Rabie, M.; Aseel, D.G.; Younes, H.A.; Behiry, S.I.; Abdelkhalek, A. Transcriptional Responses and Secondary Metabolites Variation of Tomato Plant in Response to Tobacco Mosaic Virus Infestation. Sci. Rep. 2024, 14, 19565. [Google Scholar] [CrossRef]
- Yu, X.; Liu, Z.; Sun, X. Single-Cell and Spatial Multi-Omics in the Plant Sciences: Technical Advances, Applications, and Perspectives. Plant Commun. 2023, 4, 100508. [Google Scholar] [CrossRef]
- Maule, A.J.; Escaler, M.; Aranda, M.A. Programmed Responses to Virus Replication in Plants. Mol. Plant Pathol. 2000, 1, 9–15. [Google Scholar] [CrossRef]
- Huber, A.E.; Bauerle, T.L. Long-Distance Plant Signaling Pathways in Response to Multiple Stressors: The Gap in Knowledge. J. Exp. Bot. 2016, 67, 2063–2079. [Google Scholar] [CrossRef]
- Murmu, S.; Sinha, D.; Chaurasia, H.; Sharma, S.; Das, R.; Jha, G.K.; Archak, S. A Review of Artificial Intelligence-Assisted Omics Techniques in Plant Defense: Current Trends and Future Directions. Front. Plant Sci. 2024, 15, 1292054. [Google Scholar] [CrossRef]
- Rodriguez-Peña, R.; Mounadi, K.E.; Garcia-Ruiz, H. Changes in Subcellular Localization of Host Proteins Induced by Plant Viruses. Viruses 2021, 13, 677. [Google Scholar] [CrossRef]
- Shaw, J.; Yu, C.; Makhotenko, A.V.; Makarova, S.S.; Love, A.J.; Kalinina, N.O.; MacFarlane, S.; Chen, J.; Taliansky, M.E. Interaction of a Plant Virus Protein with the Signature Cajal Body Protein Coilin Facilitates Salicylic Acid-Mediated Plant Defence Responses. New Phytol. 2019, 224, 439–453. [Google Scholar] [CrossRef] [PubMed]
- Morriss, G.R.; Cooper, T.A. Protein Sequestration as a Normal Function of Long Noncoding RNAs and a Pathogenic Mechanism of RNAs Containing Nucleotide Repeat Expansions. Hum. Genet. 2017, 136, 1247–1263. [Google Scholar] [CrossRef] [PubMed]
- Kopp, F.; Mendell, J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Horlacher, M.; Cheng, L.; Winther, O. RNA Trafficking and Subcellular Localization-a Review of Mechanisms, Experimental and Predictive Methodologies. Brief. Bioinform. 2023, 24, bbad249. [Google Scholar] [CrossRef]
- Mäkinen, K.; Lõhmus, A.; Pollari, M. Plant RNA Regulatory Network and RNA Granules in Virus Infection. Front. Plant Sci. 2017, 8, 2093. [Google Scholar] [CrossRef]
- Núñez-Muñoz, L.A.; Calderón-Pérez, B.; Ruiz-Medrano, R.; Xoconostle-Cázares, B.; de la Torre-Almaraz, R. Viral Tropism in Plants, Reproductive Tissues, and Seeds. Arch. Microbiol. 2025, 207, 152. [Google Scholar] [CrossRef]
- Lucas, W.J.; Groover, A.; Lichtenberger, R.; Furuta, K.; Yadav, S.-R.; Helariutta, Y.; He, S.-Q.; Fukuda, H.; Kang, J.; Brady, S.M.; et al. The Plant Vascular System: Evolution, Development and Functions. J. Integr. Plant Biol. 2013, 55, 294–388. [Google Scholar] [CrossRef]
- Schmid, M.; Speiseder, T.; Dobner, T.; Gonzalez, R.A. DNA Virus Replication Compartments. J. Virol. 2014, 88, 1404–1420. [Google Scholar] [CrossRef]
- Xu, Y.; Ghanim, M.; Liu, Y. Editorial: Mixed Infections of Plant Viruses in Nature and the Impact on Agriculture. Front. Microbiol. 2022, 13, 922607. [Google Scholar] [CrossRef]

| Virus/Genus (RNA or DNA) | Plant | Examples of Modulated Pathways | References |
|---|---|---|---|
| Transcriptomics | |||
| TYLCV/Begomovirus (DNA) | Tomato | Gibberellin-mediated antiviral defence | [28] |
| RBSDV/Reovirus (RNA) | Rice | m6A methylation | [29] |
| RSV/Tennuivirus (RNA) | Rice | m6A methylation | [29] |
| CGMMV/Tobamovirus (RNA) | Watermelon | m6A methylation | [30] |
| PVY/Potyvirus (RNA) | Potato | Photosynthesis, carbohydrate synthesis | [31] |
| TYLCV/Begomovirus (DNA) | Tomato | Defence response, ubiquitination | [32] |
| PVY/Potyvirus (RNA) | Potato | Resistance, susceptibility, photosynthesis | [33] |
| RSV/Tennuivirus (RNA) | Rice | Photosynthesis, flowering, stress/defence responses | [34] |
| CaLCuV/Begomovirus (DNA) | Arabidopsis thaliana | Translation machinery | [35] |
| TuMV/Potyvirus (RNA) | |||
| CMV/Cucumovirus (RNA) | Arabidopsis halleri | Plant–pathogen interaction | [36] |
| BrYV/Polerovirus (RNA) | |||
| SVBV/Caulimovirus (DNA) | Strawberry | Pigment metabolism, plant–pathogen interactions | [37] |
| TMV/Tobamovirus (RNA) | Tobacco | Reprogramming auxin-regulated gene expression | [38] |
| CMV/Cucumovirus (RNA) | Tobacco | Photosynthesis and chlorophyll metabolism | [39] |
| CCYV/Crinivirus (RNA) | Cucumber | Phenylpropanoid synthesis, phenylalanine metabolism | [40] |
| MIMV/Nucleorhabdovirus (RNA) | Maize | Immune receptor signalling, RNA silencing, developmental processes | [41] |
| CBSV/Ipomovirus (RNA) | Cassava | [42] | |
| TYLCV/Begomovirus (DNA) | Tomato | Long non-coding and circular RNAs | [43] |
| CMV/Cucumovirus (RNA) | Capsicum annuum | Response to stress, defence response | [44] |
| BCTV/Geminivirus (DNA) | Sugar beet | Primary metabolic processes, volatile compounds | [45] |
| SLCCNV/Begomovirus (DNA) | Zucchini | Photosynthesis, plant–pathogen interactions | [46] |
| PVY/Potyvirus (RNA) | Potato | Poly(ADP)-ribosylation, methionine cycle | [47] |
| Proteomics | |||
| CWMV/Furovirus (RNA) | Nicotiana benthamiana | Abscisic acid-mediated antiviral defence | [48] |
| ORMV/Tobamovirus (RNA) | N. benthamiana | Jasmonic and abscisic acid signalling, intracellular | [49] |
| MNSV/Gammacarmovirus (RNA) | Melon | Processes controlling redox balance and cell death | [50] |
| RSV/Tennuivirus (RNA) | Rice | Chlorophyll biosynthesis and cell death processes | [51] |
| TMV/Tobamovirus (RNA) | Tobacco | Photosynthesis | [52] |
| TYLCCNV/Begomovirus (DNA) | Tobacco | Stress defence, energy production, photosynthesis | [53] |
| CSMV/Comovirus (RNA) | Vigna unguiculata | Redox homeostasis, protein synthesis, defence, stress | [54] |
| PSbMV/Potyvirus (RNA) | Pisum sativum | Plant–pathogen response, lipid metabolism | [55] |
| SRBSDV/Fijivirus (RNA) | Rice | Defence superoxide dismutase and catalase activities | [56] |
| CPSMV/Comovirus (RNA) | V. unguiculata | Photosynthesis, stress response, and oxidative burst | [57] |
| TSWV/Orthotospovirus (RNA) | Tobacco | Cell death, host defence, metabolism | [58] |
| PVY/Potyvirus (RNA) | Potato | Methionine cycle | [59] |
| Metabolomics | |||
| TRV/Tobravirus (RNA) | A. thaliana | Lipid and fatty acid metabolism | [60] |
| RBSDV/Reovirus (RNA) | Maize | Lipid and fatty acid metabolism | [61] |
| CCYV/Crinivirus (RNA) | Cucumber | Lipid metabolism | [62] |
| SCMV/Potyvirus (RNA) | Maize | Phenylpropanoid pathway | [63] |
| GLRaV-3/Ampelovirus (RNA) | Grapevine | Phenylalanine metabolism, salicylic acid pathway | [64] |
| HpMV/Carlavirus (RNA) | |||
| ApMV/Ilarvirus (RNA) | Hop | Accumulation of monoterpenes hydrocarbons | [65] |
| HLVd (viroid) | |||
| CMV/Cucumovirus (RNA) | Passion fruit | Levels of secondary metabolites and antioxidants | [66] |
| SaLV/Potyvirus (RNA) | Saffron | Composition of picrocrocin, safranal, and kaempferols | [67] |
| HgYMV/Begomovirus (DNA) | Horsegram | Accumulation of sugars, alkanes, and carboxylic acids | [68] |
| Transcriptomics and Proteomics | |||
| MYMIV/Begomovirus (DNA) | Soybean | Cell cycle, cell-wall biogenesis, hormone assimilation | [69] |
| PVY/Potyvirus (RNA) | Potato | Plant immunity regulation | [70,71] |
| SCMV/Potyvirus (RNA) | Sugarcane | Photosynthesis | [72] |
| SCMV/Potyvirus (RNA) | Sugarcane | Sugar metabolism | [73] |
| TuMV/Potyvirus (RNA) | Cabbage | Calcium signalling pathways, heat shock responses | [74] |
| GFLV/Nepovirus (RNA) | N. benthamiana | Chitinase activity and hypersensitive response | [75] |
| Transcriptomics and Metabolomics | |||
| PVY/Potyvirus (RNA) | Potato | Phenylpropanoids and antioxidant pathways | [76] |
| GRBaV/Grablovirus (DNA) | Grapevine | Abscisic acid, ethylene, and auxin pathways | [77] |
| GRBaV/Grablovirus (DNA) | Grapevine | Auxin-mediated pathways and photosynthesis | [78] |
| TuMV/Potyvirus (RNA) | Brassica rapa | Volatile organic compounds | [79] |
| TSWV/Orthotospovirus (RNA) | Tomato | Plant hormone signalling and flavonoid pathway | [80] |
| Proteomics and Metabolomics | |||
| SLCMV/Begomovirus (DNA) | Cassava | Plant–pathogen interaction, hormone signalling | [81] |
| Interactomics | |||
| TSWV/Orthotospovirus (RNA) | N. benthamiana | TSWV NSs-interacting proteins | [82] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Samarskaya, V.; Spechenkova, N.; Kalinina, N.O.; Love, A.J.; Taliansky, M. The Emerging Role of Omics-Based Approaches in Plant Virology. Viruses 2025, 17, 986. https://doi.org/10.3390/v17070986
Samarskaya V, Spechenkova N, Kalinina NO, Love AJ, Taliansky M. The Emerging Role of Omics-Based Approaches in Plant Virology. Viruses. 2025; 17(7):986. https://doi.org/10.3390/v17070986
Chicago/Turabian StyleSamarskaya, Viktoriya, Nadezhda Spechenkova, Natalia O. Kalinina, Andrew J. Love, and Michael Taliansky. 2025. "The Emerging Role of Omics-Based Approaches in Plant Virology" Viruses 17, no. 7: 986. https://doi.org/10.3390/v17070986
APA StyleSamarskaya, V., Spechenkova, N., Kalinina, N. O., Love, A. J., & Taliansky, M. (2025). The Emerging Role of Omics-Based Approaches in Plant Virology. Viruses, 17(7), 986. https://doi.org/10.3390/v17070986







