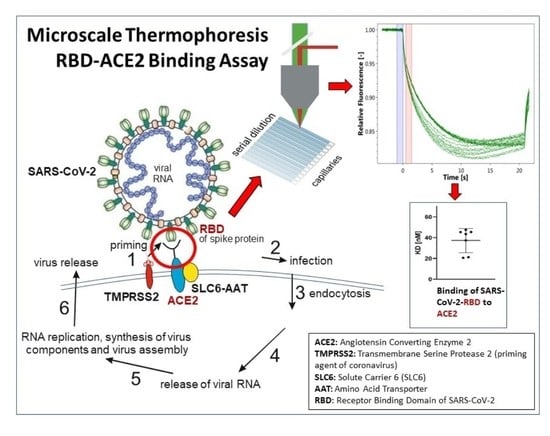
Applications of the Microscale Thermophoresis Binding Assay in COVID-19 Research
Abstract
1. Introduction
2. Materials and Methods
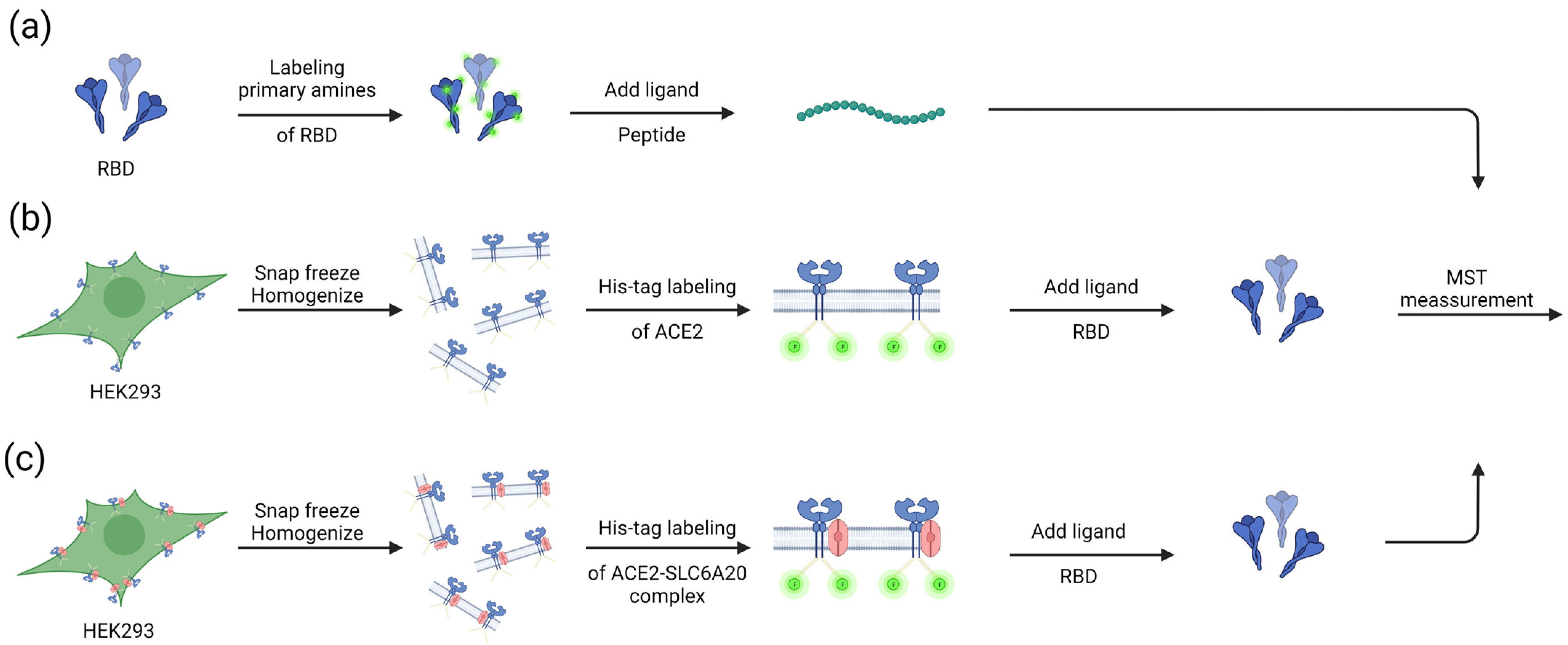
2.1. Labeling of Purified Proteins
2.2. Cell Culture and Lysate Preparation
2.3. His-Tag Labeling of Cell Lysate
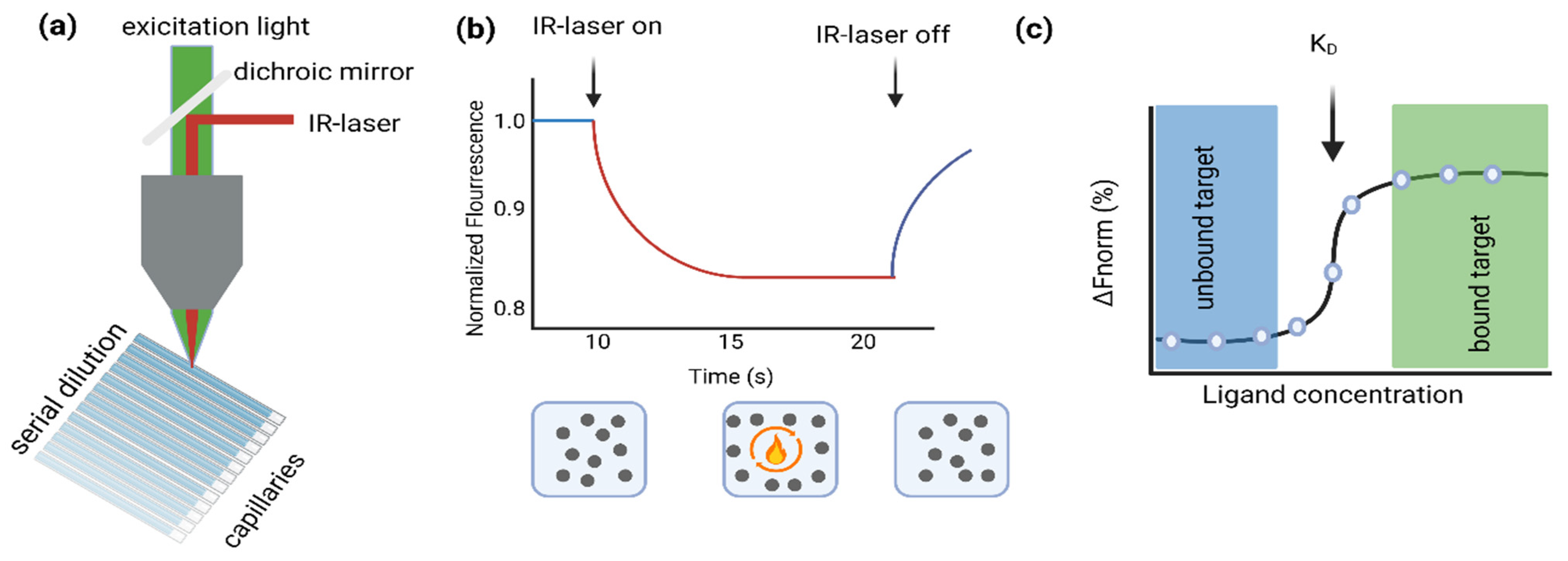
2.4. MST Measurements
2.5. Generation of the SLC6A20 Variant I529V (Ile529Val; rs61731475)
2.6. Amino Acid Sequences of the RBD-Binding Peptides That Were Tested with MST
3. Results
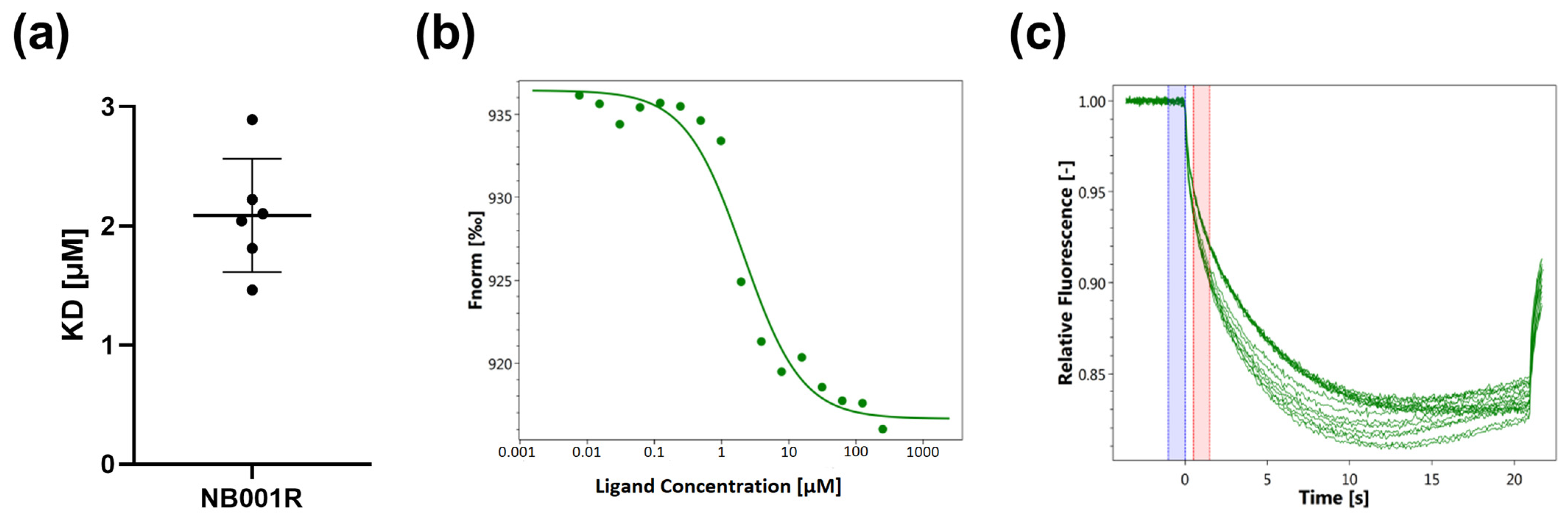
3.1. Determination of the Binding of SARS-CoV-2-S1-RBD to Antiviral Agents
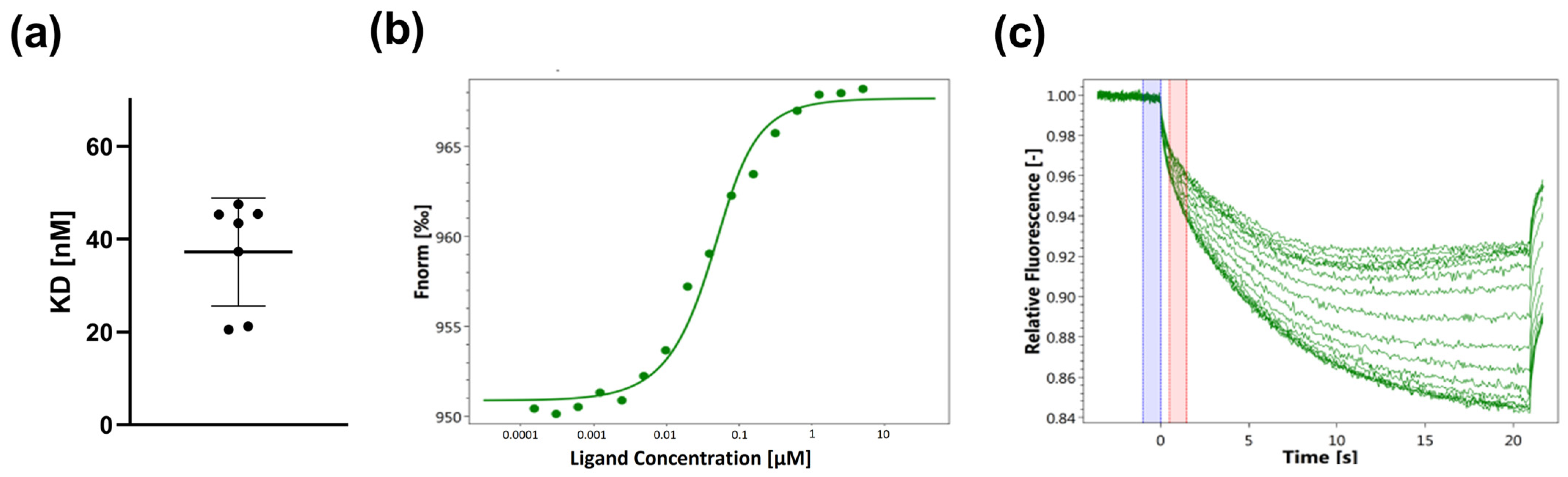
3.2. Determination of the Binding of SARS-CoV-2-S1-RBD to His-Tag-Labeled ACE2
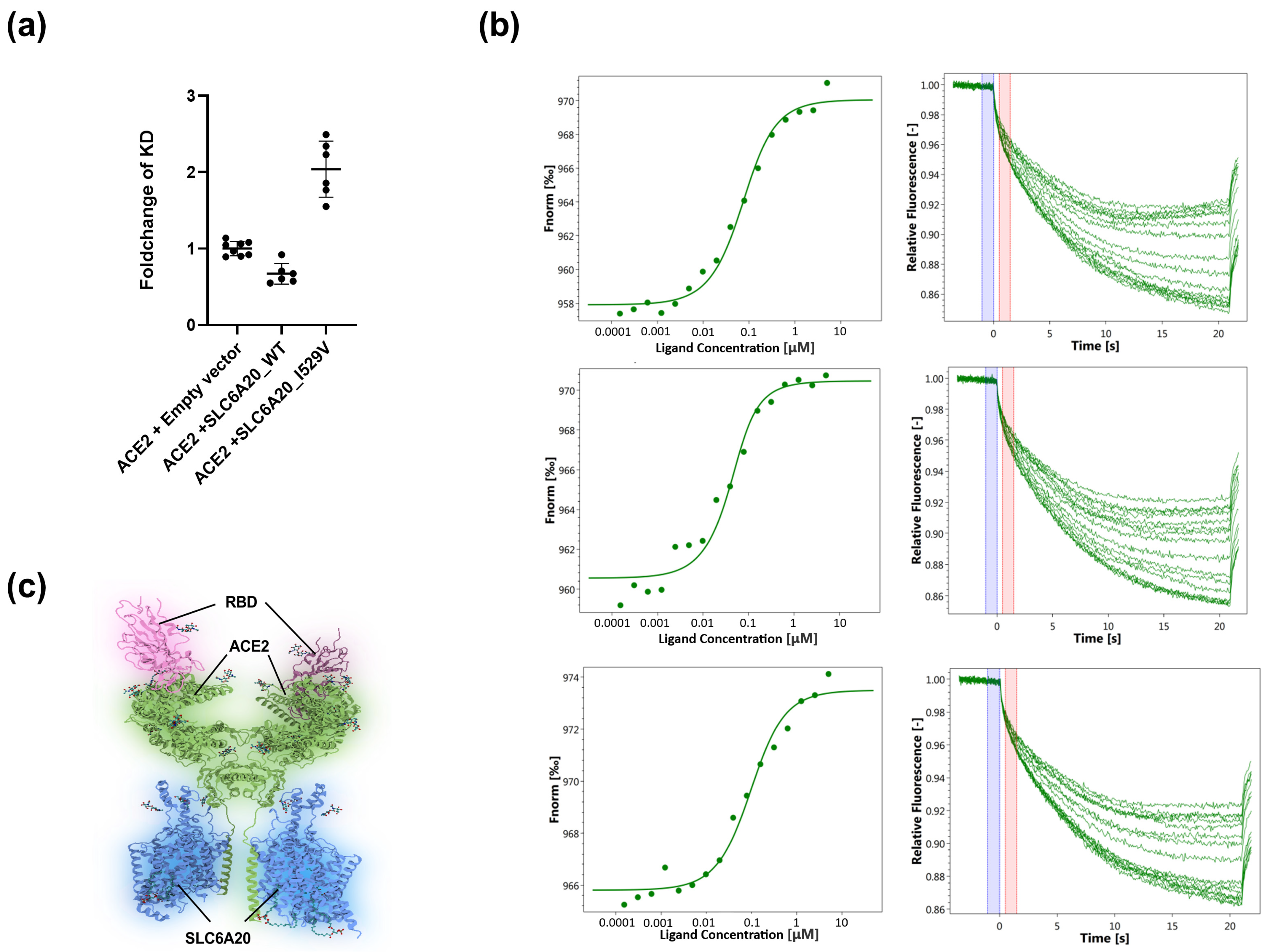
3.3. Determination of the Binding of SARS-CoV-2-S1-RBD to the ACE2-SLC6A20 Heterodimeric Complex in the Native Lipid Bilayer Environment
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, Y.; Grunewald, M.; Perlman, S. Coronaviruses: An Updated Overview of Their Replication and Pathogenesis. In Methods in Molecular Biology; Humana: New York, NY, USA, 2020; Volume 2203, pp. 1–29. [Google Scholar] [CrossRef]
- Maier, H.J.; Bickerton, E. Coronaviruses Methods and Protocols. In Methods in Molecular Biology; Humana: New York, NY, USA, 2020; Volume 2203, ISBN 9781071608999. [Google Scholar]
- Chilamakuri, R.; Agarwal, S. COVID-19: Characteristics and therapeutics. Cells 2021, 10, 206. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Farouk, I.A.; Lal, S.K. COVID-19: A Review on the Novel Coronavirus Disease. Viruses 2021, 13, 202. [Google Scholar] [CrossRef]
- Khan, M.; Adil, S.F.; Alkhathlan, H.Z.; Tahir, M.N.; Saif, S.; Khan, M.; Khan, S.T. COVID-19: A Global Challenge with Old History, Epidemiology and Progress So Far. Molecules 2021, 26, 39. [Google Scholar] [CrossRef]
- Chen, Y.; Klein, S.L.; Garibaldi, B.T.; Li, H.; Wu, C.; Osevala, N.M.; Li, T.; Margolick, J.B.; Pawelec, G.; Leng, S.X. Aging in COVID-19: Vulnerability, immunity and intervention. Ageing Res. Rev. 2021, 65, 101205. [Google Scholar] [CrossRef] [PubMed]
- WHO Coronavirus (COVID-19) Dashboard. WHO Coronavirus (COVID-19) Dashboard With Vaccination Data. Available online: https://covid19.who.int/ (accessed on 7 April 2023).
- Seidel, S.A.I.; Dijkman, P.M.; Lea, W.A.; Van Den Bogaart, G.; Jerabek-willemsen, M.; Lazic, A.; Joseph, J.S.; Srinivasan, P.; Baaske, P.; Simeonov, A.; et al. Microscale thermophoresis quantifies biomolecular interactions under previously challenging conditions. Methods 2023, 59, 301–315. [Google Scholar] [CrossRef] [PubMed]
- El Deeb, S.; Al-Harrasi, A.; Khan, A.; Al-Broumi, M.; Al-Thani, G.; Alomairi, M.; Elumalai, P.; Sayed, R.A.; Ibrahim, A.E. Microscale thermophoresis as a powerful growing analytical technique for the investigation of biomolecular interaction and the determination of binding parameters. Methods Appl. Fluoresc. 2022, 10, 042001. [Google Scholar] [CrossRef]
- Magnez, R.; Bailly, C.; Thuru, X. Microscale Thermophoresis as a Tool to Study Protein Interactions and Their Implication in Human Diseases. Int. J. Mol. Sci. 2022, 23, 7672. [Google Scholar] [CrossRef]
- Jerabek-Willemsen, M.; Wienken, C.J.; Braun, D.; Baaske, P.; Duhr, S. Molecular interaction studies using microscale thermophoresis. Assay Drug Dev. Technol. 2011, 9, 342–353. [Google Scholar] [CrossRef]
- Asmari, M.; Ratih, R.; Alhazmi, H.A.; El Deeb, S. Thermophoresis for characterizing biomolecular interaction. Methods 2018, 146, 107–119. [Google Scholar] [CrossRef]
- Jerabek-Willemsen, M.; André, T.; Wanner, R.; Roth, H.M.; Duhr, S.; Baaske, P.; Breitsprecher, D. MicroScale Thermophoresis: Interaction analysis and beyond. J. Mol. Struct. 2014, 1077, 101–113. [Google Scholar] [CrossRef]
- Chatzikyriakidou, Y.; Ahn, D.H.; Nji, E.; Drew, D. The GFP thermal shift assay for screening ligand and lipid interactions to solute carrier transporters. Nat. Protoc. 2021, 16, 5357–5376. [Google Scholar] [CrossRef] [PubMed]
- Homola, J. Present and future of surface plasmon resonance biosensors. Anal. Bioanal. Chem. 2003, 377, 528–539. [Google Scholar] [CrossRef] [PubMed]
- Ghai, R.; Falconer, R.J.; Collins, B.M. Applications of isothermal titration calorimetry in pure and applied research-survey of the literature from 2010. J. Mol. Recognit. 2012, 25, 32–52. [Google Scholar] [CrossRef]
- Zhang, J.; Jones, C.P.; Amaré, A.R.F. Biochimica et Biophysica Acta Global analysis of riboswitches by small-angle X-ray scattering and calorimetry. BBA-Gene Regul. Mech. 2014, 1839, 1020–1029. [Google Scholar] [CrossRef]
- Becker, W.; Bhattiprolu, K.C.; Zangger, K. Investigating Protein—Ligand Interactions by Solution Nuclear Magnetic Resonance Spectroscopy. ChemPhysChem 2018, 19, 895–906. [Google Scholar] [CrossRef] [PubMed]
- Clémençon, B.; Lüscher, B.P.; Hediger, M.A. Establishment of a novel microscale thermophoresis ligand-binding assay for characterization of SLC solute carriers using oligopeptide transporter PepT1 (SLC15 family) as a model system. J. Pharmacol. Toxicol. Methods 2018, 92, 67–76. [Google Scholar] [CrossRef]
- Romain, M.; Thiroux, B.; Tardy, M.; Quesnel, B.; Thuru, X. Measurement of Protein-Protein Interactions through Microscale Thermophoresis (MST). Bio-Protocol 2020, 10, e3574. [Google Scholar] [CrossRef]
- Dijkman, P.M.; Lea, W.A.; Gmbh, N.T.; Lazic, A.; Gmbh, N.T. Microscale Thermophoresis Quantifies Biomolecular Interactions under Previously Challenging Conditions. Methods 2014, 59, 301–315. [Google Scholar]
- Li, J.; Li, C.; Xiao, W.; Yuan, D.; Wan, G.; Ma, L. Site-directed mutagenesis by combination of homologous recombination and DpnI digestion of the plasmid template in Escherichia coli. Anal. Biochem. 2008, 373, 389–391. [Google Scholar] [CrossRef]
- Yan, R.; Zhang, Y.; Li, Y.; Xia, L.; Guo, Y.; Zhou, Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020, 367, 1444–1448. [Google Scholar] [CrossRef]
- Shang, J.; Ye, G.; Shi, K.; Wan, Y.; Luo, C.; Aihara, H.; Geng, Q.; Auerbach, A.; Li, F. Structural basis of receptor recognition by SARS-CoV-2. Nature 2020, 581, 221–224. [Google Scholar] [CrossRef]
- Thompson, J.L.; Mignen, O.; Shuttleworth, T.J. The ARC Channel—An Endogenous Store-Independent Orai Channel. Curr. Top. Membr. 2013, 71, 125–148. [Google Scholar] [CrossRef] [PubMed]
- Rutz, S.; Deneka, D.; Dittmann, A.; Sawicka, M.; Dutzler, R. Structure of a volume-regulated heteromeric LRRC8A/C channel. Nat. Struct. Mol. Biol. 2022, 30, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Borroto-Escuela, D.O.; Fuxe, K. Oligomeric Receptor Complexes and Their Allosteric Receptor-Receptor Interactions in the Plasma Membrane Represent a New Biological Principle for Integration of Signals in the CNS. Front. Mol. Neurosci. 2019, 12, 230. [Google Scholar] [CrossRef] [PubMed]
- Semiz, S. SIT1 transporter as a potential novel target in treatment of COVID-19. Biomol. Concepts 2021, 12, 156–163. [Google Scholar] [CrossRef]
- Kasela, S.; Daniloski, Z.; Bollepalli, S.; Jordan, T.X.; tenOever, B.R.; Sanjana, N.E.; Lappalainen, T. Integrative approach identifies SLC6A20 and CXCR6 as putative causal genes for the COVID-19 GWAS signal in the 3p21.31 locus. Genome Biol. 2021, 22, 1–10. [Google Scholar] [CrossRef]
- Ling, Y.; Van Herpt, T.T.W.; van Hoek, M.; Dehghan, A.; Hofman, A.; Uitterlinden, A.G.; Jiang, S.; Lieverse, A.G.; Bravenboer, B.; Lu, D.; et al. A genetic variant in SLC6A20 is associated with Type 2 diabetes in white-European and Chinese populations. Diabet. Med. 2014, 31, 1350–1356. [Google Scholar] [CrossRef]
- Severe Covid-19 GWAS Group. Genomewide Association Study of Severe COVID-19 with Respiratory Failure. N. Engl. J. Med. 2020, 383, 1522–1534. [Google Scholar] [CrossRef] [PubMed]
- Azzarà, A.; Cassano, I.; Paccagnella, E.; Tirindelli, M.C.; Nobile, C.; Schittone, V.; Lintas, C.; Sacco, R.; Gurrieri, F. Genetic variants determine intrafamilial variability of SARS-CoV-2 clinical outcomes in 19 Italian families. PLoS ONE 2022, 17, e0275988. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Wang, J.; Li, Y.; Zhang, Y.; Tian, R.; Yan, R. Structures of ACE2–SIT1 recognized by Omicron variants of SARS-CoV-2. Cell Discov. 2022, 8, 123. [Google Scholar] [CrossRef]
- Stevens, B.R.; Ellory, J.C.; Preston, R.L. B0AT1 Amino Acid Transporter Complexed With SARS-CoV-2 Receptor ACE2 Forms a Heterodimer Functional Unit: In Situ Conformation Using Radiation Inactivation Analysis. Function 2021, 2, zqab027. [Google Scholar] [CrossRef] [PubMed]
- Bröer, S. The role of the neutral amino acid transporter B0AT1 (SLC6A19) in Hartnup disorder and protein nutrition. IUBMB Life 2009, 61, 591–599. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Deng, F.; Shi, K.; Ye, G.; Wang, G.; Fang, L.; Xiao, S.; Fu, Z.; Peng, G. Dimerization of Coronavirus nsp9 with Diverse Modes Enhances Its Nucleic Acid Binding Affinity. J. Virol. 2018, 92, e00692-18. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, L.; Yan, L.; Ming, Z.; Jia, Z.; Lou, Z.; Rao, Z. Structural and Biochemical Characterization of Endoribonuclease Nsp15 Encoded by Middle East Respiratory Syndrome Coronavirus. J. Virol. 2018, 92, e00893-18. [Google Scholar] [CrossRef]
- Shahhamzehei, N.; Abdelfatah, S.; Efferth, T. In Silico and In Vitro Identification of Pan-Coronaviral Main Protease Inhibitors from a Large Natural Product Library. Pharmaceuticals 2022, 15, 308. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nydegger, D.T.; Pujol-Giménez, J.; Kandasamy, P.; Vogt, B.; Hediger, M.A. Applications of the Microscale Thermophoresis Binding Assay in COVID-19 Research. Viruses 2023, 15, 1432. https://doi.org/10.3390/v15071432
Nydegger DT, Pujol-Giménez J, Kandasamy P, Vogt B, Hediger MA. Applications of the Microscale Thermophoresis Binding Assay in COVID-19 Research. Viruses. 2023; 15(7):1432. https://doi.org/10.3390/v15071432
Chicago/Turabian StyleNydegger, Damian T., Jonai Pujol-Giménez, Palanivel Kandasamy, Bruno Vogt, and Matthias A. Hediger. 2023. "Applications of the Microscale Thermophoresis Binding Assay in COVID-19 Research" Viruses 15, no. 7: 1432. https://doi.org/10.3390/v15071432
APA StyleNydegger, D. T., Pujol-Giménez, J., Kandasamy, P., Vogt, B., & Hediger, M. A. (2023). Applications of the Microscale Thermophoresis Binding Assay in COVID-19 Research. Viruses, 15(7), 1432. https://doi.org/10.3390/v15071432