Novel Porcine Circoviruses in View of Lessons Learned from Porcine Circovirus Type 2-Epidemiology and Threat to Pigs and Other Species
Abstract
1. Introduction
2. Occurrence and Spread of New Porcine Circoviruses
3. Diversity and Evolutionary Rate of Porcine Circoviruses
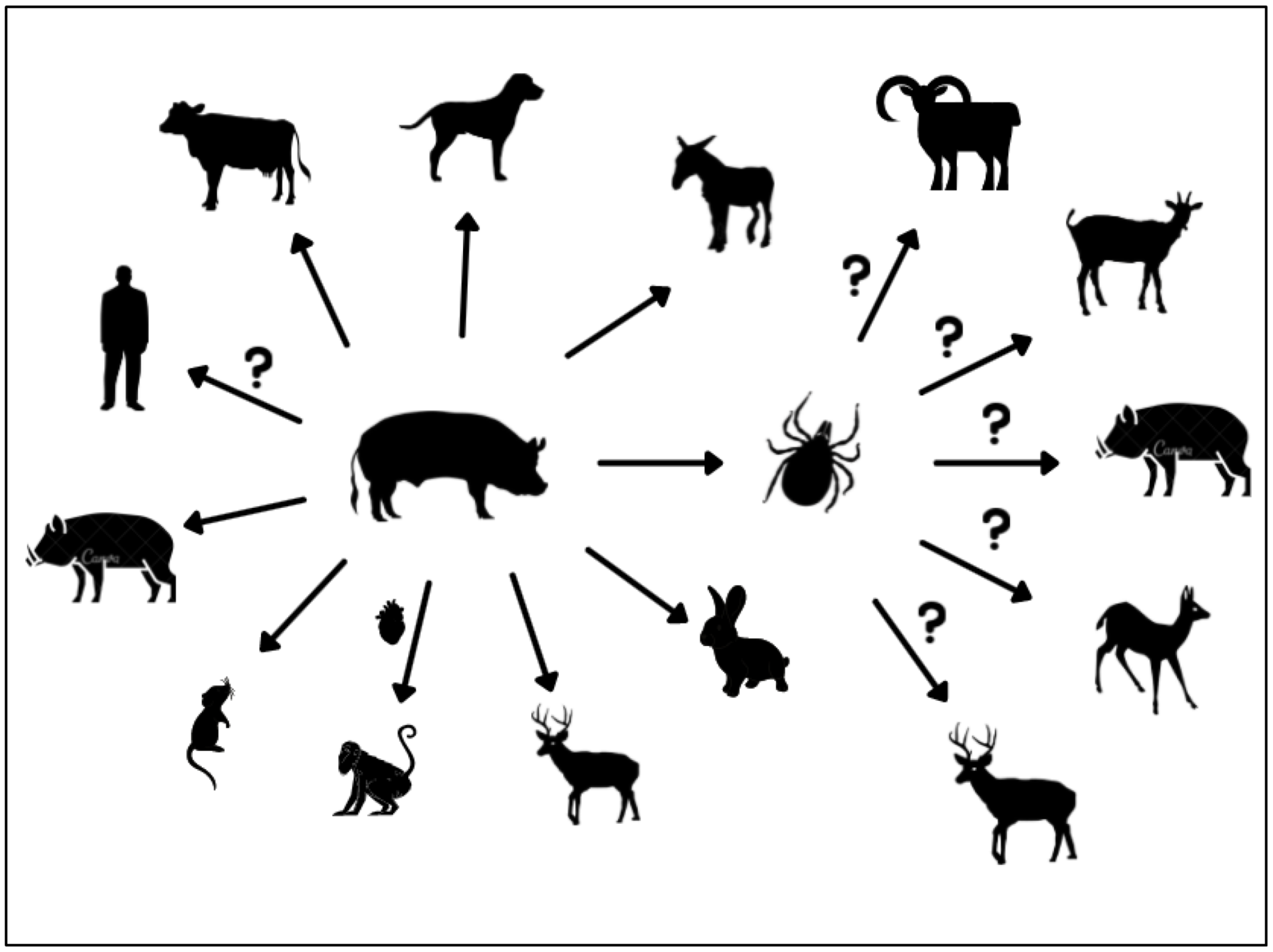
4. Porcine Circoviruses as a Potential Threat to Other Species
5. Zoonotic Potential of Porcine Circoviruses
6. Pathogenicity of New Porcine Circoviruses for Pigs
7. Development of Vaccine against New PCVs—Current Data
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Rosario, K.; Breitbart, M.; Harrach, B.; Segalés, J.; Delwart, E.; Biagini, P.; Varsani, A. Revisiting the Taxonomy of the Family Circoviridae: Establishment of the Genus Cyclovirus and Removal of the Genus Gyrovirus. Arch. Virol. 2017, 162, 1447–1463. [Google Scholar] [CrossRef] [PubMed]
- Mankertz, A.; Persson, F.; Mankertz, J.; Blaess, G.; Buhk, H.J. Mapping and Characterization of the Origin of DNA Replication of Porcine Circovirus. J. Virol. 1997, 71, 2562–2566. [Google Scholar] [CrossRef] [PubMed]
- Garigliany, M.-M.; Börstler, J.; Jöst, H.; Badusche, M.; Desmecht, D.; Schmidt-Chanasit, J.; Cadar, D. Characterization of a Novel Circo-like Virus in Aedes Vexans Mosquitoes from Germany: Evidence for a New Genus within the Family Circoviridae. J. Gen. Virol. 2015, 96, 915–920. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Kapoor, A.; Slikas, B.; Bamidele, O.S.; Wang, C.; Shaukat, S.; Masroor, M.A.; Wilson, M.L.; Ndjango, J.-B.N.; Peeters, M.; et al. Multiple Diverse Circoviruses Infect Farm Animals and Are Commonly Found in Human and Chimpanzee Feces. J. Virol. 2010, 84, 1674–1682. [Google Scholar] [CrossRef] [PubMed]
- Tischer, I.; Rasch, R.; Tochtermann, G. Characterization of Papovavirus-and Picornavirus-like Particles in Permanent Pig Kidney Cell Lines. Zenibl. Bukt. 1974, 226, 153–167. [Google Scholar]
- Ellis, J.; Hassard, L.; Clark, E.; Harding, J.; Allan, G.; Willson, P.; Strokappe, J.; Martin, K.; McNeilly, F.; Meehan, B.; et al. Isolation of Circovirus from Lesions of Pigs with Postweaning Multisystemic Wasting Syndrome. Can. Vet. J. Rev. Vet. Can. 1998, 39, 44–51. [Google Scholar]
- Palinski, R.; Piñeyro, P.; Shang, P.; Yuan, F.; Guo, R.; Fang, Y.; Byers, E.; Hause, B.M. A Novel Porcine Circovirus Distantly Related to Known Circoviruses Is Associated with Porcine Dermatitis and Nephropathy Syndrome and Reproductive Failure. J. Virol. 2017, 91, e01879-16. [Google Scholar] [CrossRef]
- Phan, T.G.; Giannitti, F.; Rossow, S.; Marthaler, D.; Knutson, T.P.; Li, L.; Deng, X.; Resende, T.; Vannucci, F.; Delwart, E. Detection of a Novel Circovirus PCV3 in Pigs with Cardiac and Multi-Systemic Inflammation. Virol. J. 2016, 13, 184. [Google Scholar] [CrossRef]
- Zhang, H.; Hu, W.; Li, J.; Liu, T.; Zhou, J.; Opriessnig, T.; Xiao, C. Novel Circovirus Species Identified in Farmed Pigs Designated as Porcine Circovirus 4, Hunan Province, China. Transbound. Emerg. Dis. 2020, 67, 1057–1061. [Google Scholar] [CrossRef]
- Opriessnig, T.; Karuppannan, A.K.; Castro, A.M.; Xiao, C.-T. Porcine Circoviruses: Current Status, Knowledge Gaps and Challenges. Virus Res. 2020, 286, 198044. [Google Scholar] [CrossRef]
- Finsterbusch, T.; Steinfeldt, T.; Caliskan, R.; Mankertz, A. Analysis of the Subcellular Localization of the Proteins Rep, Rep′ and Cap of Porcine Circovirus Type 1. Virology 2005, 343, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Tischer, I.; Mields, W.; Wolff, D.; Vagt, M.; Griem, W. Studies on Epidemiology and Pathogenicity of Porcine Circovirus. Arch. Virol. 1986, 91, 271–276. [Google Scholar] [CrossRef] [PubMed]
- Alarcon, P.; Rushton, J.; Wieland, B. Cost of Post-Weaning Multi-Systemic Wasting Syndrome and Porcine Circovirus Type-2 Subclinical Infection in England—An Economic Disease Model. Prev. Vet. Med. 2013, 110, 88–102. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Guo, J.; Zhang, G.; Jin, Q.; Liu, Y.; Jia, R.; Wang, A. Fine Mapping of Linear B Cell Epitopes on Capsid Protein of Porcine Circovirus 3. Appl. Microbiol. Biotechnol. 2020, 104, 6223–6234. [Google Scholar] [CrossRef]
- Segalés, J.; Kekarainen, T.; Cortey, M. The Natural History of Porcine Circovirus Type 2: From an Inoffensive Virus to a Devastating Swine Disease? Vet. Microbiol. 2013, 165, 13–20. [Google Scholar] [CrossRef]
- Ku, X.; Chen, F.; Li, P.; Wang, Y.; Yu, X.; Fan, S.; Qian, P.; Wu, M.; He, Q. Identification and Genetic Characterization of Porcine Circovirus Type 3 in China. Transbound. Emerg. Dis. 2017, 64, 703–708. [Google Scholar] [CrossRef]
- Stadejek, T.; Woźniak, A.; Miłek, D.; Biernacka, K. First Detection of Porcine Circovirus Type 3 on Commercial Pig Farms in Poland. Transbound. Emerg. Dis. 2017, 64, 1350–1353. [Google Scholar] [CrossRef] [PubMed]
- Tochetto, C.; Lima, D.A.; Varela, A.P.M.; Loiko, M.R.; Paim, W.P.; Scheffer, C.M.; Herpich, J.I.; Cerva, C.; Schmitd, C.; Cibulski, S.P.; et al. Full-Genome Sequence of Porcine Circovirus Type 3 Recovered from Serum of Sows with Stillbirths in Brazil. Transbound. Emerg. Dis. 2018, 65, 5–9. [Google Scholar] [CrossRef]
- Molini, U.; Marruchella, G.; Matheus, F.; Hemberger, Y.M.; Chiwome, B.; Khaiseb, S.; Cattoli, G.; Franzo, G. Molecular Investigation of Porcine Circovirus Type 3 Infection in Pigs in Namibia. Pathogens 2021, 10, 585. [Google Scholar] [CrossRef]
- Kwon, T.; Yoo, S.J.; Park, C.-K.; Lyoo, Y.S. Prevalence of Novel Porcine Circovirus 3 in Korean Pig Populations. Vet. Microbiol. 2017, 207, 178–180. [Google Scholar] [CrossRef]
- Bera, B.C.; Choudhary, M.; Anand, T.; Virmani, N.; Sundaram, K.; Choudhary, B.; Tripathi, B.N. Detection and Genetic Characterization of Porcine Circovirus 3 (PCV3) in Pigs in India. Transbound. Emerg. Dis. 2020, 67, 1062–1067. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Zhang, Y.-H.; Tian, R.-B.; Hou, C.-Y.; Li, X.-S.; Zheng, L.-L.; Wang, L.-Q.; Chen, H.-Y. Prevalence and Genetic Analysis of Porcine Circovirus Type 2 (PCV2) and Type 3 (PCV3) between 2018 and 2020 in Central China. Infect. Genet. Evol. 2021, 94, 105016. [Google Scholar] [CrossRef]
- Hayashi, S.; Ohshima, Y.; Furuya, Y.; Nagao, A.; Oroku, K.; Tsutsumi, N.; Sasakawa, C.; Sato, T. First Detection of Porcine Circovirus Type 3 in Japan. J. Vet. Med. Sci. 2018, 80, 1468–1472. [Google Scholar] [CrossRef] [PubMed]
- Kedkovid, R.; Woonwong, Y.; Arunorat, J.; Sirisereewan, C.; Sangpratum, N.; Lumyai, M.; Kesdangsakonwut, S.; Teankum, K.; Jittimanee, S.; Thanawongnuwech, R. Porcine Circovirus Type 3 (PCV3) Infection in Grower Pigs from a Thai Farm Suffering from Porcine Respiratory Disease Complex (PRDC). Vet. Microbiol. 2018, 215, 71–76. [Google Scholar] [CrossRef]
- Yuzhakov, A.G.; Raev, S.A.; Alekseev, K.P.; Grebennikova, T.V.; Verkhovsky, O.A.; Zaberezhny, A.D.; Aliper, T.I. First Detection and Full Genome Sequence of Porcine Circovirus Type 3 in Russia. Virus Genes 2018, 54, 608–611. [Google Scholar] [CrossRef]
- Chang, C.-C.; Wu, C.-W.; Chang, Y.-C.; Wu, C.-Y.; Chien, M.-S.; Huang, C. Detection and Phylogenetic Analysis of Porcine Circovirus Type 3 in Taiwan. Arch. Virol. 2021, 166, 259–263. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.Y.; Opaskornkul, K.; Thanawongnuwech, R.; Arshad, S.S.; Hassan, L.; Ooi, P.T. First Molecular Detection and Complete Sequence Analysis of Porcine Circovirus Type 3 (PCV3) in Peninsular Malaysia. PLoS ONE 2020, 15, e0235832. [Google Scholar] [CrossRef]
- Vargas-Bermudez, D.S.; Campos, F.S.; Bonil, L.; Mogollon, D.; Jaime, J. First Detection of Porcine Circovirus Type 3 in Colombia and the Complete Genome Sequence Demonstrates the Circulation of PCV 3a1 and PCV 3a2. Vet. Med. Sci. 2019, 5, 182–188. [Google Scholar] [CrossRef]
- Serena, M.S.; Cappuccio, J.A.; Barrales, H.; Metz, G.E.; Aspitia, C.G.; Lozada, I.; Perfumo, C.J.; Quiroga, M.A.; Piñeyro, P.; Echeverría, M.G. First Detection and Genetic Characterization of Porcine Circovirus Type 3 (PCV3) in Argentina and Its Association with Reproductive Failure. Transbound. Emerg. Dis. 2021, 68, 1761–1766. [Google Scholar] [CrossRef]
- Faccini, S.; Barbieri, I.; Gilioli, A.; Sala, G.; Gibelli, L.R.; Moreno, A.; Sacchi, C.; Rosignoli, C.; Franzini, G.; Nigrelli, A. Detection and Genetic Characterization of Porcine Circovirus Type 3 in Italy. Transbound. Emerg. Dis. 2017, 64, 1661–1664. [Google Scholar] [CrossRef]
- Franzo, G.; Legnardi, M.; Hjulsager, C.K.; Klaumann, F.; Larsen, L.E.; Segales, J.; Drigo, M. Full-Genome Sequencing of Porcine Circovirus 3 Field Strains from Denmark, Italy and Spain Demonstrates a High within-Europe Genetic Heterogeneity. Transbound. Emerg. Dis. 2018, 65, 602–606. [Google Scholar] [CrossRef] [PubMed]
- Collins, P.J.; McKillen, J.; Allan, G. Porcine Circovirus Type 3 in the UK. Vet. Rec. 2017, 181, 599. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Berg, M.; Fossum, C.; Wallgren, P.; Blomström, A.-L. Detection and Genetic Characterisation of Porcine Circovirus 3 from Pigs in Sweden. Virus Genes 2018, 54, 466–469. [Google Scholar] [CrossRef] [PubMed]
- Fux, R.; Söckler, C.; Link, E.K.; Renken, C.; Krejci, R.; Sutter, G.; Ritzmann, M.; Eddicks, M. Full Genome Characterization of Porcine Circovirus Type 3 Isolates Reveals the Existence of Two Distinct Groups of Virus Strains. Virol. J. 2018, 15, 25. [Google Scholar] [CrossRef]
- Deim, Z.; Dencső, L.; Erdélyi, I.; Valappil, S.K.; Varga, C.; Pósa, A.; Makrai, L.; Rákhely, G. Porcine Circovirus Type 3 Detection in a Hungarian Pig Farm Experiencing Reproductive Failures. Vet. Rec. 2019, 185, 84. [Google Scholar] [CrossRef]
- Savic, B.; Milicevic, V.; Radanovic, O.; Zdravkovic, N.; Stevancevic, O.; Kureljusic, B.; Nesic, K. Identification and Genetic Characterization of Porcine Circovirus 3 on Pig Farms in Serbia. Arch. Virol. 2020, 165, 193–199. [Google Scholar] [CrossRef]
- Rudova, N.; Lymanska, O.; Stegniy, B.; Bolotin, V.; Solodiankin, O.; Gerilovych, A. First Detection of Porcine Circovirus Type 3 in Ukraine. Agric. Sci. Pract. 2021, 8, 16–23. [Google Scholar] [CrossRef]
- Rodrigues, I.L.F.; Cruz, A.C.M.; Souza, A.E.; Knackfuss, F.B.; Costa, C.H.C.; Silveira, R.L.; Castro, T.X. Retrospective Study of Porcine Circovirus 3 (PCV3) in Swine Tissue from Brazil (1967–2018). Braz. J. Microbiol. 2020, 51, 1391–1397. [Google Scholar] [CrossRef]
- Sun, J.; Wei, L.; Lu, Z.; Mi, S.; Bao, F.; Guo, H.; Tu, C.; Zhu, Y.; Gong, W. Retrospective Study of Porcine Circovirus 3 Infection in China. Transbound. Emerg. Dis. 2018, 65, 607–613. [Google Scholar] [CrossRef]
- Franzo, G.; Tucciarone, C.M.; Drigo, M.; Cecchinato, M.; Martini, M.; Mondin, A.; Menandro, M.L. First Report of Wild Boar Susceptibility to Porcine Circovirus Type 3: High Prevalence in the Colli Euganei Regional Park (Italy) in the Absence of Clinical Signs. Transbound. Emerg. Dis. 2018, 65, 957–962. [Google Scholar] [CrossRef]
- Hou, C.; Zhang, L.; Zhang, Y.; Cui, J.; Zhao, L.; Zheng, L.; Chen, H. Phylogenetic Analysis of Porcine Circovirus 4 in Henan Province of China: A Retrospective Study from 2011 to 2021. Transbound. Emerg. Dis. 2021. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Du, Q.; Han, Z.; Bi, J.; Lan, T.; Wang, W.; Zheng, M. Detection and Genetic Characterization of Porcine Circovirus 4 (PCV4) in Guangxi, China. Gene 2021, 773, 145384. [Google Scholar] [CrossRef] [PubMed]
- Tian, R.; Zhao, Y.; Cui, J.; Zheng, H.; Xu, T.; Hou, C.; Wang, Z.; Li, X.; Zheng, L.; Chen, H. Molecular Detection and Phylogenetic Analysis of Porcine Circovirus 4 in Henan and Shanxi Provinces of China. Transbound. Emerg. Dis. 2021, 68, 276–282. [Google Scholar] [CrossRef] [PubMed]
- Ha, Z.; Yu, C.; Xie, C.; Wang, G.; Zhang, Y.; Hao, P.; Li, J.; Li, Z.; Li, Y.; Rong, F.; et al. Retrospective Surveillance of Porcine Circovirus 4 in Pigs in Inner Mongolia, China, from 2016 to 2018. Arch. Virol. 2021, 166, 1951–1959. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Xiao, Y.; Li, X.; Li, S.; Xie, N.; Yan, X.; Li, X.; Zhu, J. Development and Application of a Quadruplex Real-time PCR Assay for Differential Detection of Porcine Circoviruses (PCV1 to PCV4) in Jiangsu Province of China from 2016 to 2020. Transbound. Emerg. Dis. 2021, 68, 1615–1624. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.-G.; Do, H.-Q.; Huynh, T.-M.-L.; Park, Y.-H.; Park, B.-K.; Chung, H.-C. Molecular-Based Detection, Genetic Characterization and Phylogenetic Analysis of Porcine Circovirus 4 from Korean Domestic Swine Farms. Transbound. Emerg. Dis. 2021. [Google Scholar] [CrossRef]
- Franzo, G.; Ruiz, A.; Grassi, L.; Sibila, M.; Drigo, M.; Segalés, J. Lack of Porcine Circovirus 4 Genome Detection in Pig Samples from Italy and Spain. Pathogens 2020, 9, 433. [Google Scholar] [CrossRef]
- Olvera, A.; Cortey, M.; Segalés, J. Molecular Evolution of Porcine Circovirus Type 2 Genomes: Phylogeny and Clonality. Virology 2007, 357, 175–185. [Google Scholar] [CrossRef]
- Wang, F.; Guo, X.; Ge, X.; Wang, Z.; Chen, Y.; Cha, Z.; Yang, H. Genetic Variation Analysis of Chinese Strains of Porcine Circovirus Type 2. Virus Res. 2009, 145, 151–156. [Google Scholar] [CrossRef]
- Guo, L.J.; Lu, Y.H.; Wei, Y.W.; Huang, L.P.; Liu, C.M. Porcine Circovirus Type 2 (PCV2): Genetic Variation and Newly Emerging Genotypes in China. Virol. J. 2010, 7, 273. [Google Scholar] [CrossRef]
- Franzo, G.; Segalés, J. Porcine Circovirus 2 (PCV-2) Genotype Update and Proposal of a New Genotyping Methodology. PLoS ONE 2018, 13, e0208585. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Noll, L.; Lu, N.; Porter, E.; Stoy, C.; Zheng, W.; Liu, X.; Peddireddi, L.; Niederwerder, M.; Bai, J. Genetic Diversity and Prevalence of Porcine Circovirus Type 3 (PCV3) and Type 2 (PCV2) in the Midwest of the USA during 2016–2018. Transbound. Emerg. Dis. 2020, 67, 1284–1294. [Google Scholar] [CrossRef] [PubMed]
- Franzo, G.; Tucciarone, C.M.; Cecchinato, M.; Drigo, M. Porcine Circovirus Type 2 (PCV2) Evolution before and after the Vaccination Introduction: A Large Scale Epidemiological Study. Sci. Rep. 2016, 6, 39458. [Google Scholar] [CrossRef] [PubMed]
- Turlewicz-Podbielska, H.; Pomorska-Mól, M. Choroby Świń Wywoływane Przez Cirkowirusy Oraz Grypa Świń w Świetle Danych 11. Sympozjum Europejskiego Stowarzyszenia Zarządzania Zdrowiem Świń. Życie Weter. 2019, 94, 804–809. [Google Scholar]
- Wei, R.; Xie, J.; Theuns, S.; Nauwynck, H.J. Changes on the Viral Capsid Surface during the Evolution of Porcine Circovirus Type 2 (PCV2) from 2009 till 2018 May Lead to a Better Receptor Binding. Virus Evol. 2019, 5, vez026. [Google Scholar] [CrossRef] [PubMed]
- Firth, C.; Charleston, M.A.; Duffy, S.; Shapiro, B.; Holmes, E.C. Insights into the Evolutionary History of an Emerging Livestock Pathogen: Porcine Circovirus 2. J. Virol. 2009, 83, 12813–12821. [Google Scholar] [CrossRef]
- Franzo, G.; Cortey, M.; Segalés, J.; Hughes, J.; Drigo, M. Phylodynamic Analysis of Porcine Circovirus Type 2 Reveals Global Waves of Emerging Genotypes and the Circulation of Recombinant Forms. Mol. Phylogenet. Evol. 2016, 100, 269–280. [Google Scholar] [CrossRef]
- Rose, N.; Opriessnig, T.; Grasland, B.; Jestin, A. Epidemiology and Transmission of Porcine Circovirus Type 2 (PCV2). Virus Res. 2012, 164, 78–89. [Google Scholar] [CrossRef]
- Franzo, G.; Delwart, E.; Fux, R.; Hause, B.; Su, S.; Zhou, J.; Segalés, J. Genotyping Porcine Circovirus 3 (PCV-3) Nowadays: Does It Make Sense? Viruses 2020, 12, 265. [Google Scholar] [CrossRef]
- Li, G.; He, W.; Zhu, H.; Bi, Y.; Wang, R.; Xing, G.; Zhang, C.; Zhou, J.; Yuen, K.-Y.; Gao, G.F.; et al. Origin, Genetic Diversity, and Evolutionary Dynamics of Novel Porcine Circovirus 3. Adv. Sci. 2018, 5, 1800275. [Google Scholar] [CrossRef]
- Klaumann, F.; Correa-Fiz, F.; Franzo, G.; Sibila, M.; Núñez, J.I.; Segalés, J. Current Knowledge on Porcine Circovirus 3 (PCV-3): A Novel Virus With a Yet Unknown Impact on the Swine Industry. Front. Vet. Sci. 2018, 5, 315. [Google Scholar] [CrossRef] [PubMed]
- Pomorska-Mól, M.; Podgórska, K.; Urbaniak, K. Wybrane Dane Prezentowane Podczas 25. Kongresu IPVS. Część III. Cirkowirusy i Koronawirusy Świń. Życie Weter. 2019, 94, 348–352. [Google Scholar]
- Franzo, G.; He, W.; Correa-Fiz, F.; Li, G.; Legnardi, M.; Su, S.; Segalés, J. A Shift in Porcine Circovirus 3 (PCV-3) History Paradigm: Phylodynamic Analyses Reveal an Ancient Origin and Prolonged Undetected Circulation in the Worldwide Swine Population. Adv. Sci. 2019, 6, 1901004. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Sun, N.; Tian, F.; Wen, Y.; Xu, C.; Li, J.; Chen, Q.; Wang, J. Genetic Analysis of Porcine Circovirus Type 2 from Dead Minks. J. Gen. Virol. 2016, 97, 2316–2322. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Shan, T.; Soji, O.B.; Alam, M.M.; Kunz, T.H.; Zaidi, S.Z.; Delwart, E. Possible Cross-Species Transmission of Circoviruses and Cycloviruses among Farm Animals. J. Gen. Virol. 2011, 92, 768–772. [Google Scholar] [CrossRef] [PubMed]
- Kappe, E.C.; Halami, M.Y.; Schade, B.; Alex, M.; Hoffmann, D.; Gangl, A.; Meyer, K.; Dekant, W.; Schwarz, B.-A.; Johne, R.; et al. Bone Marrow Depletion with Haemorrhagic Diathesis in Calves in Germany: Characterization of the Disease and Preliminary Investigations on Its Aetiology. Berl. Munch. Tierarztl. Wochenschr. 2010, 123, 31–41. [Google Scholar]
- Lőrincz, M.; Cságola, A.; Biksi, I.; Szeredi, L.; Dán, Á.; Tuboly, T. Detection of Porcine Circovirus in Rodents—Short Communication. Acta Vet. Hung. 2010, 58, 265–268. [Google Scholar] [CrossRef]
- Halami, M.Y.; Müller, H.; Böttcher, J.; Vahlenkamp, T.W. Whole-Genome Sequences of Two Strains of Porcine Circovirus 2 Isolated from Calves in Germany. Genome Announc. 2014, 2, e01150-13. [Google Scholar] [CrossRef]
- Zhai, S.-L.; Lu, S.-S.; Wei, W.-K.; Lv, D.-H.; Wen, X.-H.; Zhai, Q.; Chen, Q.-L.; Sun, Y.-W.; Xi, Y. Reservoirs of Porcine Circoviruses: A Mini Review. Front. Vet. Sci. 2019, 6, 319. [Google Scholar] [CrossRef]
- Song, T.; Hao, J.; Zhang, R.; Tang, M.; Li, W.; Hui, W.; Fu, Q.; Wang, C.; Xin, S.; Zhang, S.; et al. First Detection and Phylogenetic Analysis of Porcine Circovirus Type 2 in Raccoon Dogs. BMC Vet. Res. 2019, 15, 107. [Google Scholar] [CrossRef]
- Song, T.; Zhang, S.; Hao, J.; Xin, S.; Hui, W.; Tang, M.; Li, W.; Tian, R.; Liu, X.; Rui, P.; et al. First Detection and Genetic Analysis of Fox-origin Porcine Circovirus Type 2. Transbound. Emerg. Dis. 2019, 66, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Franzo, G.; Grassi, L.; Tucciarone, C.M.; Drigo, M.; Martini, M.; Pasotto, D.; Mondin, A.; Menandro, M.L. A Wild Circulation: High Presence of Porcine Circovirus 3 in Different Mammalian Wild Hosts and Ticks. Transbound. Emerg. Dis. 2019, 66, 1548–1557. [Google Scholar] [CrossRef] [PubMed]
- Klaumann, F.; Dias-Alves, A.; Cabezón, O.; Mentaberre, G.; Castillo-Contreras, R.; López-Béjar, M.; Casas-Díaz, E.; Sibila, M.; Correa-Fiz, F.; Segalés, J. Porcine Circovirus 3 Is Highly Prevalent in Serum and Tissues and May Persistently Infect Wild Boar (Sus Scrofa Scrofa). Transbound. Emerg. Dis. 2019, 66, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Dei Giudici, S.; Franzoni, G.; Bonelli, P.; Angioi, P.P.; Zinellu, S.; Deriu, V.; Carta, T.; Sechi, A.M.; Salis, F.; Balzano, F.; et al. Genetic Characterization of Porcine Circovirus 3 Strains Circulating in Sardinian Pigs and Wild Boars. Pathogens 2020, 9, 344. [Google Scholar] [CrossRef]
- Prinz, C.; Stillfried, M.; Neubert, L.K.; Denner, J. Detection of PCV3 in German Wild Boars. Virol. J. 2019, 16, 25. [Google Scholar] [CrossRef] [PubMed]
- de Souza, T.C.; Gava, D.; Schaefer, R.; Leme, R.A.; da Silva Porto, G.; Alfieri, A.A. Porcine Circovirus 3a Field Strains in Free-Living Wild Boars in Paraná State, Brazil. Animals 2021, 11, 1634. [Google Scholar] [CrossRef]
- Dhandapani, G.; Yoon, S.; Noh, J.Y.; Jang, S.S.; Han, S.; Jeong, D.G.; Kim, H.K. Detection of Porcine Circovirus 3 from Captured Wild Boars in Korea. Vet. Med. Sci. 2021, 7, 1807–1814. [Google Scholar] [CrossRef]
- Song, S.; Park, K.-N.; Choe, S.; Cha, R.M.; Shin, J.; Hyun, B.-H.; An, D.-J. Complete Genome Sequences of Two Type 3 Porcine Circoviruses, WB17KW and WB20GG, Isolated from Korean Wild Boar. Microbiol. Resour. Announc. 2021, 10, e01386-20. [Google Scholar] [CrossRef]
- Czyżewska-Dors, E.; Núñez, J.I.; Saporiti, V.; Huerta, E.; Riutord, C.; Cabezón, O.; Segalés, J.; Sibila, M. Detection of Porcine Circovirus 3 in Wildlife Species in Spain. Pathogens 2020, 9, 341. [Google Scholar] [CrossRef]
- Wang, W.; Sun, W.; Cao, L.; Zheng, M.; Zhu, Y.; Li, W.; Liu, C.; Zhuang, X.; Xing, J.; Lu, H.; et al. An Epidemiological Investigation of Porcine Circovirus 3 Infection in Cattle in Shandong Province, China. BMC Vet. Res. 2019, 15, 60. [Google Scholar] [CrossRef]
- Wang, T.; Chai, W.; Wang, Y.; Liu, W.; Huang, Z.; Chen, L.; Guo, R.; Dong, Y.; Liu, M.; Zheng, Q.; et al. First Detection and Phylogenetic Analysis of Porcine Circovirus 3 in Female Donkeys with Reproductive Disorders. BMC Vet. Res. 2021, 17, 308. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, Z.; Zou, Y.; Zhang, N.; Wang, D.; Tu, D.; Yang, L.; Deng, Z.; Yang, Y.; Jiang, P.; et al. First Molecular Detection of Porcine Circovirus Type 3 in Dogs in China. Virus Genes 2018, 54, 140–144. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; McGraw, S.; Zhu, K.; Leutenegger, C.M.; Marks, S.L.; Kubiski, S.; Gaffney, P.; Dela Cruz, F.N., Jr.; Wang, C.; Delwart, E.; et al. Circovirus in Tissues of Dogs with Vasculitis and Hemorrhage. Emerg. Infect. Dis. 2013, 19, 534–541. [Google Scholar] [CrossRef] [PubMed]
- Decaro, N.; Martella, V.; Desario, C.; Lanave, G.; Circella, E.; Cavalli, A.; Elia, G.; Camero, M.; Buonavoglia, C. Genomic Characterization of a Circovirus Associated with Fatal Hemorrhagic Enteritis in Dog, Italy. PLoS ONE 2014, 9, e105909. [Google Scholar] [CrossRef]
- Jiang, S.; Zhou, N.; Li, Y.; An, J.; Chang, T. Detection and Sequencing of Porcine Circovirus 3 in Commercially Sourced Laboratory Mice. Vet. Med. Sci. 2019, 5, 176–181. [Google Scholar] [CrossRef]
- Franzo, G.; Segales, J.; Tucciarone, C.M.; Cecchinato, M.; Drigo, M. The Analysis of Genome Composition and Codon Bias Reveals Distinctive Patterns between Avian and Mammalian Circoviruses Which Suggest a Potential Recombinant Origin for Porcine Circovirus 3. PLoS ONE 2018, 13, e0199950. [Google Scholar] [CrossRef]
- Liu, X.; Ouyang, T.; Ouyang, H.; Liu, X.; Niu, G.; Huo, W.; Yin, W.; Pang, D.; Ren, L. Human Cells Are Permissive for the Productive Infection of Porcine Circovirus Type 2 in Vitro. Sci. Rep. 2019, 9, 5638. [Google Scholar] [CrossRef]
- Denner, J. Recent Progress in Xenotransplantation, with Emphasis on Virological Safety. Ann. Transplant. 2016, 21, 717–727. [Google Scholar] [CrossRef]
- Krüger, L.; Längin, M.; Reichart, B.; Fiebig, U.; Kristiansen, Y.; Prinz, C.; Kessler, B.; Egerer, S.; Wolf, E.; Abicht, J.-M.; et al. Transmission of Porcine Circovirus 3 (PCV3) by Xenotransplantation of Pig Hearts into Baboons. Viruses 2019, 11, 650. [Google Scholar] [CrossRef]
- Bernstein, C.N.; Nayar, G.; Hamel, A.; Blanchard, J.F. Study of Animal-Borne Infections in the Mucosas of Patients with Inflammatory Bowel Disease and Population-Based Controls. J. Clin. Microbiol. 2003, 41, 4986–4990. [Google Scholar] [CrossRef][Green Version]
- Esona, M.D.; Mijatovic-Rustempasic, S.; Yen, C.; Parashar, U.D.; Gentsch, J.R.; Bowen, M.D.; LaRussa, P. Detection of PCV-2 DNA in Stool Samples from Infants Vaccinated with RotaTeq®. Hum. Vaccines Immunother. 2014, 10, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Mijatovic-Rustempasic, S.; Immergluck, L.C.; Parker, T.C.; Laghaie, E.; Mohammed, A.; McFadden, T.; Parashar, U.D.; Bowen, M.D.; Cortese, M.M. Shedding of Porcine Circovirus Type 1 DNA and Rotavirus RNA by Infants Vaccinated with Rotarix®. Hum. Vaccines Immunother. 2017, 13, 928–935. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Borkenhagen, L.K.; Mallinson, K.A.; Tsao, R.W.; Ha, S.-J.; Lim, W.-H.; Toh, T.-H.; Anderson, B.D.; Fieldhouse, J.K.; Philo, S.E.; Chong, K.-S.; et al. Surveillance for Respiratory and Diarrheal Pathogens at the Human-Pig Interface in Sarawak, Malaysia. PLoS ONE 2018, 13, e0201295. [Google Scholar] [CrossRef] [PubMed]
- Rakibuzzaman, A.; Ramamoorthy, S. Comparative Immunopathogenesis and Biology of Recently Discovered Porcine Circoviruses. Transbound. Emerg. Dis. 2021, 68, 2957–2968. [Google Scholar] [CrossRef] [PubMed]
- Segalés, J. Porcine Circovirus Type 2 (PCV2) Infections: Clinical Signs, Pathology and Laboratory Diagnosis. Virus Res. 2012, 164, 10–19. [Google Scholar] [CrossRef]
- Temeeyasen, G.; Lierman, S.; Arruda, B.L.; Main, R.; Vannucci, F.; Gimenez-Lirola, L.G.; Piñeyro, P.E. Pathogenicity and Immune Response against Porcine Circovirus Type 3 Infection in Caesarean-Derived, Colostrum-Deprived Pigs. J. Gen. Virol. 2020, 102. [Google Scholar] [CrossRef]
- Dal Santo, A.C.; Cezario, K.C.; Bennemann, P.E.; Machado, S.A.; Martins, M. Full-Genome Sequences of Porcine Circovirus 3 (PCV3) and High Prevalence in Mummified Fetuses from Commercial Farms in Brazil. Microb. Pathog. 2020, 141, 104027. [Google Scholar] [CrossRef]
- Byrd, A.L.; Segre, J.A. Adapting Koch’s Postulates. Science 2016, 351, 224–226. [Google Scholar] [CrossRef]
- Jiang, H.; Wang, D.; Wang, J.; Zhu, S.; She, R.; Ren, X.; Tian, J.; Quan, R.; Hou, L.; Li, Z.; et al. Induction of Porcine Dermatitis and Nephropathy Syndrome in Piglets by Infection with Porcine Circovirus Type 3. J. Virol. 2019, 93, e02045-18. [Google Scholar] [CrossRef]
- Mora-Díaz, J.; Piñeyro, P.; Shen, H.; Schwartz, K.; Vannucci, F.; Li, G.; Arruda, B.; Giménez-Lirola, L. Isolation of PCV3 from Perinatal and Reproductive Cases of PCV3-Associated Disease and In Vivo Characterization of PCV3 Replication in CD/CD Growing Pigs. Viruses 2020, 12, 219. [Google Scholar] [CrossRef]
- Alomar, J.; Saporiti, V.; Pérez, M.; Gonçalvez, D.; Sibila, M.; Segalés, J. Multisystemic Lymphoplasmacytic Inflammation Associated with PCV-3 in Wasting Pigs. Transbound. Emerg. Dis. 2021, 68, 2969–2974. [Google Scholar] [CrossRef] [PubMed]
- Vargas-Bermúdez, D.S.; Vargas-Pinto, M.A.; Mogollón, J.D.; Jaime, J. Field Infection of a Gilt and Its Litter Demonstrates Vertical Transmission and Effect on Reproductive Failure Caused by Porcine Circovirus Type 3 (PCV3). BMC Vet. Res. 2021, 17, 150. [Google Scholar] [CrossRef] [PubMed]
- Mai, J.; Wang, D.; Zou, Y.; Zhang, S.; Meng, C.; Wang, A.; Wang, N. High Co-Infection Status of Novel Porcine Parvovirus 7 with Porcine Circovirus 3 in Sows That Experienced Reproductive Failure. Front. Vet. Sci. 2021, 8, 695553. [Google Scholar] [CrossRef]
- Saporiti, V.; Franzo, G.; Sibila, M.; Segalés, J. Porcine Circovirus 3 (PCV-3) as a Causal Agent of Disease in Swine and a Proposal of PCV-3 Associated Disease Case Definition. Transbound. Emerg. Dis. 2021, 68, 2936–2948. [Google Scholar] [CrossRef]
- Kedkovid, R.; Woonwong, Y.; Arunorat, J.; Sirisereewan, C.; Sangpratum, N.; Kesdangsakonwut, S.; Tummaruk, P.; Teankum, K.; Assavacheep, P.; Jittimanee, S.; et al. Porcine Circovirus Type 3 (PCV3) Shedding in Sow Colostrum. Vet. Microbiol. 2018, 220, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Zhang, N.; Zhang, J.; Zhang, S.; Jiang, Y.; Wang, D.; Tan, Q.; Yang, Y.; Wang, N. Molecular Detection and Sequence Analysis of Porcine Circovirus Type 3 in Sow Sera from Farms with Prolonged Histories of Reproductive Problems in Hunan, China. Arch. Virol. 2018, 163, 2841–2847. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Wu, X.; Zhang, L.; Xin, C.; Liu, Y.; Shi, J.; Peng, Z.; Xu, S.; Fu, F.; Yu, J.; et al. The Occurrence of Porcine Circovirus 3 without Clinical Infection Signs in Shandong Province. Transbound. Emerg. Dis. 2017, 64, 1337–1341. [Google Scholar] [CrossRef]
- Nielsen, E.O.; Enøe, C.; Jorsal, S.E.; Barfod, K.; Svensmark, B.; Bille-Hansen, V.; Vigre, H.; Bøtner, A.; Baekbo, P. Postweaning Multisystemic Wasting Syndrome in Danish Pig Herds: Productivity, Clinical Signs and Pathology. Vet. Rec. 2008, 162, 505–508. [Google Scholar] [CrossRef]
- Ilha, M.; Nara, P.; Ramamoorthy, S. Early Antibody Responses Map to Non-Protective, PCV2 Capsid Protein Epitopes. Virology 2020, 540, 23–29. [Google Scholar] [CrossRef]
- Sno, M.; Cox, E.; Holtslag, H.; Nell, T.; Pel, S.; Segers, R.; Fachinger, V.; Witvliet, M. Efficacy and Safety of a New Intradermal PCV2 Vaccine in Pigs. Trials Vaccinol. 2016, 5, 24–31. [Google Scholar] [CrossRef]
- Woźniak, A.; Miłek, D.; Matyba, P.; Stadejek, T. Real-Time PCR Detection Patterns of Porcine Circovirus Type 2 (PCV2) in Polish Farms with Different Statuses of Vaccination against PCV2. Viruses 2019, 11, 1135. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Sun, W.; Lu, H.; Tian, M.; Xie, C.; Zhao, G.; Han, J.; Wang, W.; Zheng, M.; Du, R.; et al. Genetic Variation Analysis of PCV1 Strains Isolated from Guangxi Province of China in 2015. BMC Vet. Res. 2018, 14, 43. [Google Scholar] [CrossRef] [PubMed]
- Saporiti, V.; Huerta, E.; Correa-Fiz, F.; Grosse Liesner, B.; Duran, O.; Segalés, J.; Sibila, M. Detection and Genotyping of Porcine Circovirus 2 (PCV-2) and Detection of Porcine Circovirus 3 (PCV-3) in Sera from Fattening Pigs of Different European Countries. Transbound. Emerg. Dis. 2020, 67, 2521–2531. [Google Scholar] [CrossRef] [PubMed]
- Puvanendiran, S.; Stone, S.; Yu, W.; Johnson, C.R.; Abrahante, J.; Jimenez, L.G.; Griggs, T.; Haley, C.; Wagner, B.; Murtaugh, M.P. Absence of Porcine Circovirus Type 1 (PCV1) and High Prevalence of PCV 2 Exposure and Infection in Swine Finisher Herds. Virus Res. 2011, 157, 92–98. [Google Scholar] [CrossRef]
- Patterson, A.R.; Opriessnig, T. Epidemiology and Horizontal Transmission of Porcine Circovirus Type 2 (PCV2). Anim. Health Res. Rev. 2010, 11, 217–234. [Google Scholar] [CrossRef]
- Cadar, D.; Cságola, A.; Spinu, M.; Dán, Á.; Ursu, K.; Lőrincz, M.; Tuboly, T. Prevalence of Porcine Circoviruses in Transylvanian Wild Boars, Detected by Real-Time PCR—Short Communication. Acta Vet. Hung. 2010, 58, 475–481. [Google Scholar] [CrossRef]
- Feng, H.; Blanco, G.; Segalés, J.; Sibila, M. Can Porcine Circovirus Type 2 (PCV2) Infection Be Eradicated by Mass Vaccination? Vet. Microbiol. 2014, 172, 92–99. [Google Scholar] [CrossRef]
- Deng, J.; Li, X.; Zheng, D.; Wang, Y.; Chen, L.; Song, H.; Wang, T.; Huang, Y.; Pang, W.; Tian, K. Establishment and Application of an Indirect ELISA for Porcine Circovirus 3. Arch. Virol. 2018, 163, 479–482. [Google Scholar] [CrossRef]
- Wang, D.; Mai, J.; Lei, B.; Zhang, Y.; Yang, Y.; Wang, N. Structure, Antigenic Properties, and Highly Efficient Assembly of PCV4 Capsid Protein. Front. Vet. Sci. 2021, 8, 695466. [Google Scholar] [CrossRef]
- Liu, B.Y.; Gao, B.; Liu, M.Z.; Zhang, T.T.; Liu, B.S.; Chen, Z.L. High Repetitive Arginine in the Anterior of PCV3 Capsid Protein Is a Severe Obstacle for Its Expression in E. Coli. AMB Express 2020, 10, 214. [Google Scholar] [CrossRef]
- Yan, D.; Wei, Y.-Q.; Guo, H.-C.; Sun, S.-Q. The Application of Virus-like Particles as Vaccines and Biological Vehicles. Appl. Microbiol. Biotechnol. 2015, 99, 10415–10432. [Google Scholar] [CrossRef] [PubMed]
- Weibel, H.; Sydler, T.; Brugnera, E.; Voets, H.; Grosse Liesner, B.; Sidler, X. Efficacy of Simultaneous Vaccination with Enterisol® Ileitis and Ingelvac® CircoFLEXTM in a Swiss Breeding Farm. Schweiz. Arch. Tierheilkd. 2012, 154, 445–450. [Google Scholar] [CrossRef] [PubMed]
- Koinig, H.C.; Talker, S.C.; Stadler, M.; Ladinig, A.; Graage, R.; Ritzmann, M.; Hennig-Pauka, I.; Gerner, W.; Saalmüller, A. PCV2 Vaccination Induces IFN-γ/TNF-α Co-Producing T Cells with a Potential Role in Protection. Vet. Res. 2015, 46, 20. [Google Scholar] [CrossRef] [PubMed]
- Balka, G.; Dreckmann, K.; Papp, G.; Kraft, C. Vaccination of Piglets at 2 and 3 Weeks of Age with Ingelvac PRRSFLEX® EU Provides Protection against Heterologous Field Challenge in the Face of Homologous Maternally Derived Antibodies. Porc. Health Manag. 2016, 2, 24. [Google Scholar] [CrossRef]
- Bi, M.; Li, X.; Zhai, W.; Yin, B.; Tian, K.; Mo, X. Structural Insight into the Type-Specific Epitope of Porcine Circovirus Type 3. Biosci. Rep. 2020, 40, BSR20201109. [Google Scholar] [CrossRef]
- Li, X.; Bai, Y.; Zhang, H.; Zheng, D.; Wang, T.; Wang, Y.; Deng, J.; Sun, Z.; Tian, K. Production of a Monoclonal Antibody against Porcine Circovirus Type 3 Cap Protein. J. Virol. Methods 2018, 261, 10–13. [Google Scholar] [CrossRef]
- Peswani, A.R.; Narkpuk, J.; Krueger, A.; Bracewell, D.G.; Lekcharoensuk, P.; Haslam, S.M.; Dell, A.; Jaru-Ampornpan, P.; Robinson, C. Novel Constructs and 1-Step Chromatography Protocols for the Production of Porcine Circovirus 2d (PCV2d) and Circovirus 3 (PCV3) Subunit Vaccine Candidates. Food Bioprod. Process. 2022, 131, 125–135. [Google Scholar] [CrossRef]
- Jiang, Z.; Wu, J.; Jiang, M.; Xie, Y.; Bu, W.; Liu, C.; Zhang, G.; Luo, M. A Novel Technique for Constructing Infectious Cloning of Type 3 Porcine Circovirus. Front. Microbiol. 2020, 11, 1067. [Google Scholar] [CrossRef]


| Species | PCV1 | PCV2 | PCV3 | PCV4 | References |
|---|---|---|---|---|---|
| Year of description | 1974 | 1998 | 2016 | 2019 | [5,6,7,8,9] |
| Mortality in pigs | Not reported | Weaners: 11.2% (1.4–50%) Finisher pigs: 5.2% | Up to 7th day of life: 40%, Later: 16% within 6 months | Unknown | [12,21,108] |
| Pathogenicity for pigs | no | Yes | yes | Unknown | [6,7,8,9,10,12,85] |
| Seroconversion | 7 dpi | 7 dpi | IgG 3 dpi IgM 7 dpi | Unknown | [12,96,100,109] |
| Humoral immunity duration | Up to 39–47 weeks pi | 23 weeks | IgM up to 28 dpi IgG up to 42 dpi | Unknown | [12,96,100,110] |
| Vaccine | No vaccine needed | Subunit; chimeric PCV1-2 vaccines | Not available (work in progress) | Not available | [12,111] |
| Occurrence | Worldwide | Worldwide | Worldwide | China, South Korea | [7,9,16,18,30,46,52,111,112,113] |
| World prevalence | Low | High | High | Unknown | [30,114,115] |
| Other species with detectable virus | Wild boars | Humans Wild boars Mice Cattle Racoon Dogs Foxes Minks Flies Mosquitoes | Wild boars Ticks Donkeys Cattle Dogs Mice Baboons Wild ruminants (e.g., red deer, fallow deer, mouflon) | Unknown | [40,64,65,66,67,68,69,70,71,72,79,81,82,85,89,116] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Turlewicz-Podbielska, H.; Augustyniak, A.; Pomorska-Mól, M. Novel Porcine Circoviruses in View of Lessons Learned from Porcine Circovirus Type 2-Epidemiology and Threat to Pigs and Other Species. Viruses 2022, 14, 261. https://doi.org/10.3390/v14020261
Turlewicz-Podbielska H, Augustyniak A, Pomorska-Mól M. Novel Porcine Circoviruses in View of Lessons Learned from Porcine Circovirus Type 2-Epidemiology and Threat to Pigs and Other Species. Viruses. 2022; 14(2):261. https://doi.org/10.3390/v14020261
Chicago/Turabian StyleTurlewicz-Podbielska, Hanna, Agata Augustyniak, and Małgorzata Pomorska-Mól. 2022. "Novel Porcine Circoviruses in View of Lessons Learned from Porcine Circovirus Type 2-Epidemiology and Threat to Pigs and Other Species" Viruses 14, no. 2: 261. https://doi.org/10.3390/v14020261
APA StyleTurlewicz-Podbielska, H., Augustyniak, A., & Pomorska-Mól, M. (2022). Novel Porcine Circoviruses in View of Lessons Learned from Porcine Circovirus Type 2-Epidemiology and Threat to Pigs and Other Species. Viruses, 14(2), 261. https://doi.org/10.3390/v14020261







