Genetic Diversity and Possible Origins of the Hepatitis B Virus in Siberian Natives
Abstract
1. Introduction
2. Materials and Methods
2.1. HBV Strains
2.2. Phylogenetic Analysis
2.3. Determination of HBsAg Subtypes
2.4. Statistical Data Processing
| Group and Region | No. of Strains | Year(s) of Collection | GenBank Accession Numbers | Citation |
|---|---|---|---|---|
| Siberian Natives, n = 381 | ||||
| Altaians: Altai Republic, Ust-Kansky District (southwestern Siberia) | 21 | 1997 | JX090656-JX090674, JX125378, JX125379 | [25] |
| Altaian Kazakhs: Altai Republic, Kosh-Agachsky District (southwestern Siberia) | 7 | 1999 | JX090719-JX090724, JX125377 | [25] |
| Tuvans: Tyva (or Tuva) Republic (southern Siberia) | 18 | 2004–2008 | Author’s numbers (tuvXX, see Figure S1 in Supplementary Materials) | [35] |
| Teleutes: Kemerovo Region, Belovsky District (southwestern Siberia) | 5 | 2003 | JX090633-JX090637 | [25] |
| YNAR: combined group of Khants, Nenets, Komi, Kets in the Yamal-Nenets Autonomous Region (YNAR; northwestern Siberia) | 35 | 1992–2006 | JX090647, JX090675-JX090679, JX090688-JX090702, JX090711-JX090718, JX125366-JX125368, JX125382-JX125384 | [36] |
| Taimyr: combined group of Dolgans and Nganasans in the Dudinsky District (northernmost Siberia, Taimyr Peninsula) | 32 | 2000 | JX090626-JX090632, JX090638-JX090646, JX090703-JX090710, JX125370-JX125375, JX125381, JX125386 | [25] |
| Buryats: Irkutsk Region, Nukutsky and Alarsky Districts (southeastern Siberia) | 35 | 2005, 2006 | JX090605-JX090625, JX090680- JX090687, JX125364, JX125365, JX125369, JX125376, JX125380, JX125385 | [36] |
| Yakuts: Sakha Republic (or Yakutia) (eastern Siberia) | 17 | 2004–2006 | Author’s numbers (yakXX, see Figure S1 in Supplementary Materials) | [35] |
| 15 | 1997 | AY653781, AY653782, AY653787-AY653789, AY653796, AY653799, AY653828, EU594390-EU594395, EU594433 | [37,38] | |
| 35 | 2014 | KM212957, KP143742-KP143745, KP165597-KP165605, KP184495-KP184499, KP202936-KP202945, KP230541, KT962021-KT962025 | [39] | |
| 38 | 2008–2019 | OK143470, OK143474, OK143477, OK143478, OK143480, OK143482, OK143485, OK143489 OK143490, OK143494-OK143496, OK143498, OM025238-OM025253, author’s numbers (88–732, see Figure S1) | [40] | |
| Chukchi: Chukotka Autonomous Region (northeastern-most Siberia) | 123 | 1997–2008 | Author’s numbers (chXX, schXX, see Figure S1 in Supplementary Materials) | [35] |
| Urban populations of Russia and western republics of the former USSR (listed from west to east), n = 179 | ||||
| Estonia | 21 | 2003 | EU594383-EU594386, EU594400-EU594414, EU594434, EU594435 | [37,38] |
| Latvia | 33 | 2003–2005 | AY603447-AY603466, AY653829, AY653858, AY653885, AY653886, EU594387, EU594388, JX096952-JX096958 | [37,41]; Silamikelis & Legzdina, 2012 |
| Belarus | 31 | 2005 | EU414067, EU414075, EU414077, EU414078, EU414080-EU414085, EU414087, EU414090, EU414091, EU414093, EU414095-EU414097, EU414107-EU414110, EU414114-EU414122, EU414124-EU414129, EU414132-EU414143 | [42] |
| Russia (region not specified) | 2 | 2004 | AB205127, AB205128 | Nakajima et al., 2005 |
| Saint Petersburg | 2 | 2003, 2014 | AY509974, KT963508 | Morozov, 2004; Kalinina et al., 2016 |
| Moscow | 5 | 2003, 2004 | AB126580, AB126581, AY653786, AY653801, AY653847 | Abe & Tran, 2003; [38] |
| Volgograd city (southern European Russia) | 15 | 2003, 2004 | AY653776, AY653780, AY653791, AY653798, AY653800, AY653805, AY653808, AY653809, AY653825, AY653826, AY653830, AY653839, AY653843, EU594382, EU594389 | [37,38] |
| Tyumen city (eastern Ural) | 2 | 2004 | AY653871, AY653894 | [38] |
| Kemerovo city (western Siberia) | 8 | 2003, 2004 | AY653821, AY653859, AY653862, AY653866, AY653873, EU594415-EU594417 | [37,38] |
| Krasnoyarsk (southern Siberia) | 11 | 2003, 2004 | AY653810, AY653823, AY653824, AY653831, AY653870, AY653892, EU594429, EU594430-EU594432, EU594436 | [37,38] |
| Irkutsk Region (southeastern Siberia) | 8 | 2005 | JX090648-JX090655 | [36] |
| Chita city (eastern Siberia) | 7 | 2004 | AY653806, AY653811, AY653834, AY653849, AY653856, EU594398, EU594399 | [38] |
| Khabarovsk city (far eastern Russia) | 34 | 2003, 2004, 2015 | AY653822, AY653833, AY653850, AY653855, AY653876 AY653878, EU594418-EU594427, KX925286-KX925303 | [37,38]; Kotova et al., 2016 |
| Central Asian Republics of the Former USSR and Mongolia, n = 50 | ||||
| Uzbekistan | 19 | 2003, 2007 | AB222707-AB222715, AY653777, AY653814-AY653817, AY653819, AY653827, AY653832, AY653838, EU594397 | [37,43] |
| Kazakhstan | 2 | 2003 | EU594396, EU594428 | [38] |
| Tajikistan | 1 | 2003 | AY738889 | [17] |
| Mongolia | 28 | 2005–2006 | AB263404-AB263412, AB270534-AB270550, DQ111986, DQ111987 | [44,45] |
| Other Strains | ||||
| Prototype HBV strains with known (sub)genotypes from other parts of the world | 319 | 1984–2015 | See Figure S1 in Supplementary Materials for the accession numbers, locations, and years of collection | - |
| HBV strains from ancient tombs located in different parts of the world (including modern Russian and Central Asian territories) | 124 | up to 10,000 years ago | Author’s names; see the Figure S1 in Supplementary Materials for the names, locations and years of burials | [31] |
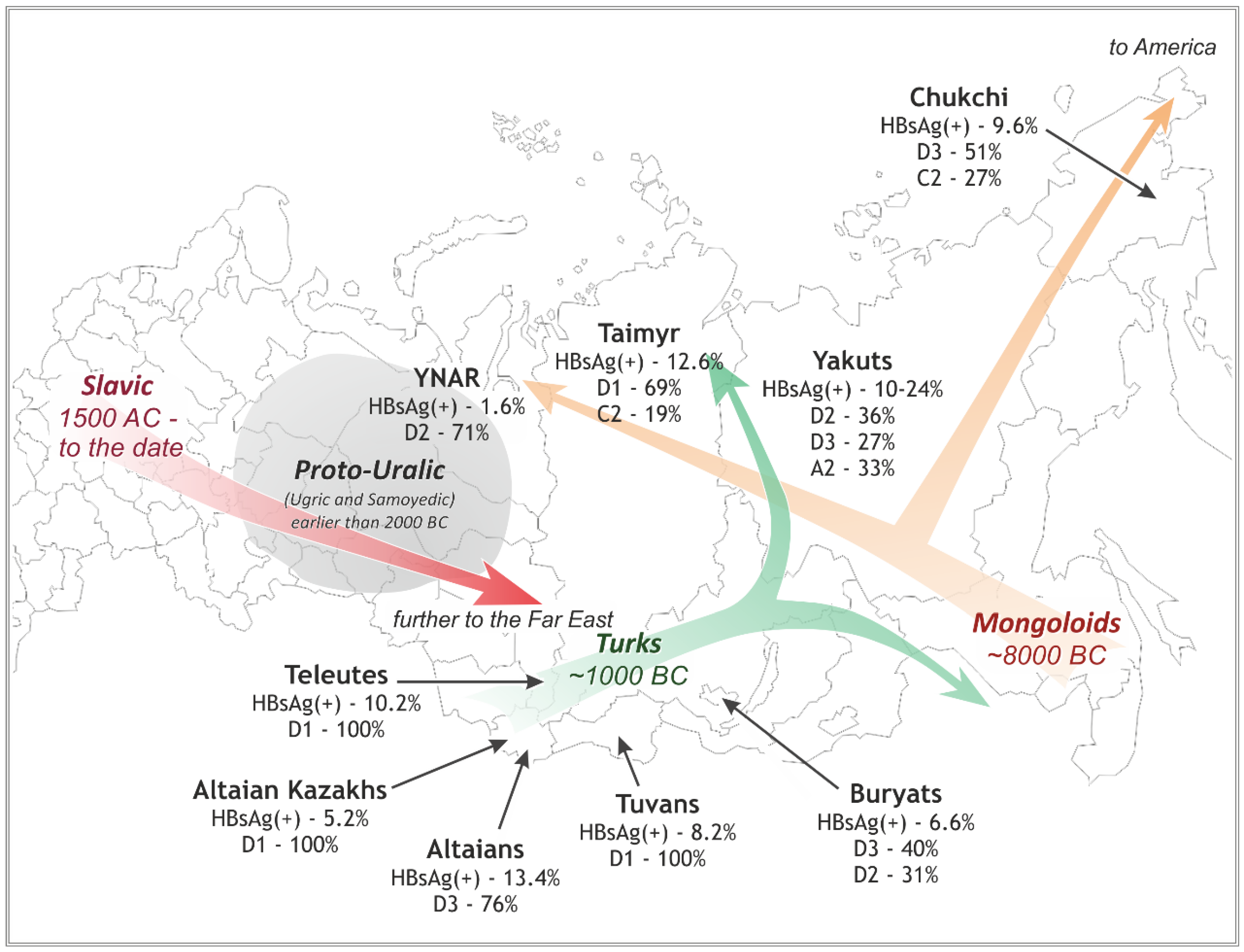
3. Results
3.1. HBV Viral Populations and Study Design
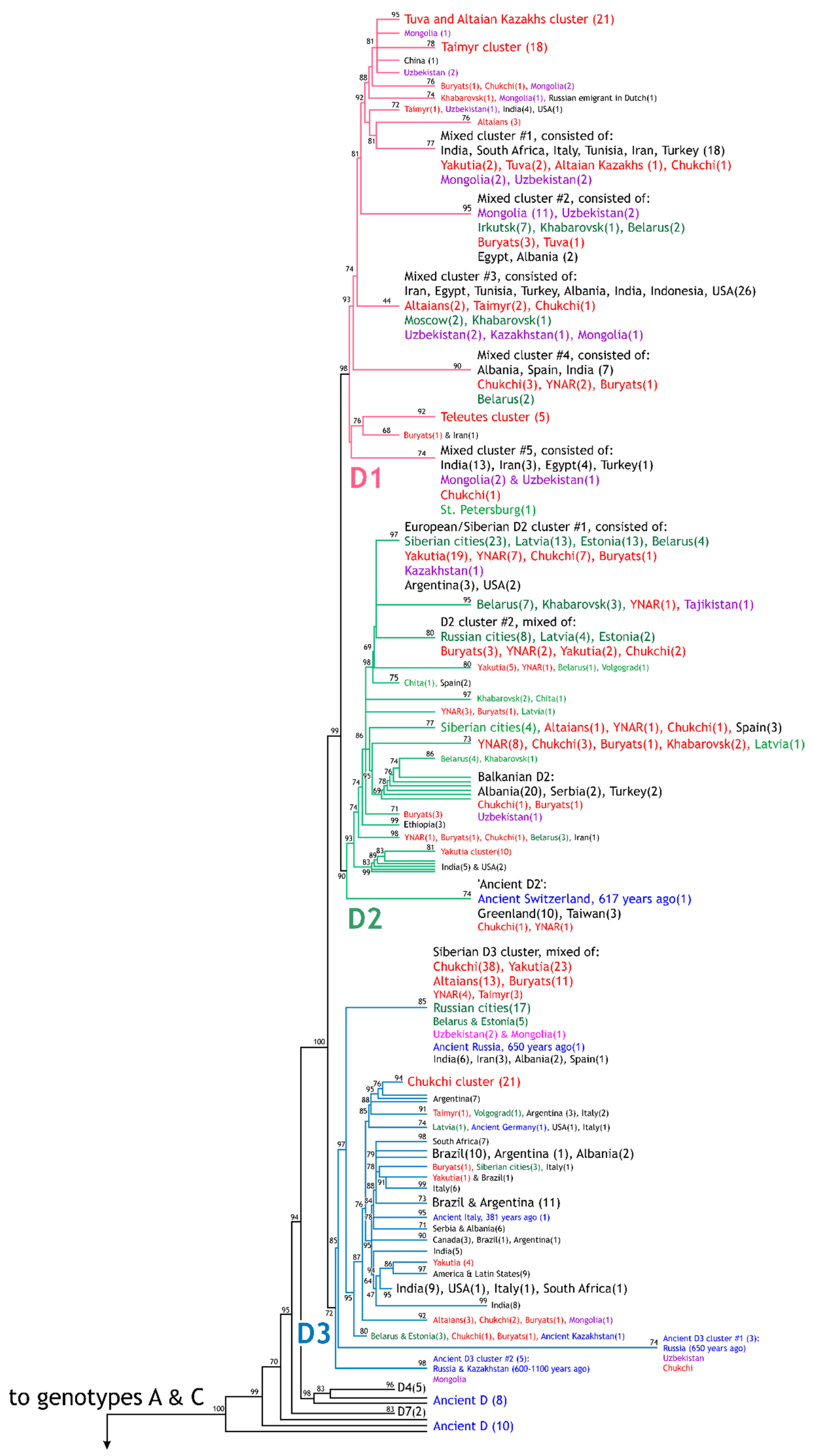

3.2. Differences in the HBV Subgenotypes in Siberian Natives and Urban Populations of the Former USSR
| Group | HBsAg (+) Rate in the Group 1 | N | HBV Subgenotypes | HBsAg Subtypes | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D1 | D2 | D3 | A2 | C2 | ayw2 | ayw3 | ayw4 | adw2 | adrq+ | N/D | |||
| Altaians | 1.4% | 21 | 4 19% | 1 5% | 16 76% | 0 | 0 | 20 95% | 1 5% | 0 | 0 | 0 | 0 |
| Altaian Kazakhs | 5.2% | 7 | 7 100% | 0 | 0 | 0 | 0 | 7 100% | 0 | 0 | 0 | 0 | 0 |
| Tuvans | 8.2% | 18 | 18 100% | 0 | 0 | 0 | 0 | 18 100% | 0 | 0 | 0 | 0 | 0 |
| Teleutes | 10.2% | 5 | 5 100% | 0 | 0 | 0 | 0 | 4 80% | 0 | 1 20% | 0 | 0 | 0 |
| YNAR | 0–1.6% | 35 | 2 6% | 25 71% | 5 14% | 2 6% | 1 3% | 9 26% | 21 60% | 1 3% | 3 8% | 1 3% | 0 |
| Taimyr | 12.6% | 32 | 22 69% | 0 | 4 12% | 0 | 6 19% | 25 78% | 0 | 1 3% | 0 | 6 19% | 0 |
| Buryats | 6.6% | 35 | 6 17% | 11 31% | 14 40% | 1 3% | 3 9% | 17 48% | 13 37% | 1 3% | 2 6% | 2 6% | 0 |
| Yakuts | 10.4–23.8% | 105 | 2 2% | 38 36% | 28 27% | 35 33% | 2 2% | 26 25% | 39 37% | 2 2% | 35 33% | 2 2% | 1 1% |
| Chukchi | 9.6% | 123 | 7 6% | 17 14% | 63 51% | 3 2% | 33 27% | 70 57% | 16 13% | 0 | 3 2% | 33 27% | 1 1% |
| Total: Siberian native populations | 4.4% | 381 | 73 19% | 92 24% | 130 34% | 41 11% | 45 12% | 196 51% | 90 24% | 6 2% | 43 11% | 44 11% | 2 1% |
| Urban populations of the former western USSR | 2–4% (estimation) | 179 | 17 9% | 100 56% | 30 17% | 31 17% | 1 1% | 47 26% | 87 49% | 8 4% | 33 18% | 1 1% | 3 2% |
| Central Asian Republics of the former USSR and Mongolia | 4.1–6.2%; up to 16% (Mongolia) | 50 | 31 62% | 3 6% | 7 14% | 5 10% | 4 8% | 39 78% | 4 8% | 0 | 5 10% | 2 4% | 0 |
3.3. Diversity of the HBV Characteristics and Variants in Different Native Groups in Siberia
3.3.1. Southwestern Siberia: D3 (ayw2) in Altaians v. D1 (ayw2) in Kazakhs, Tuvans, and Teleuts
3.3.2. Northwestern and Northern Siberia: D2 (ayw3) in YNAR v. D1 (ayw2) in Taimyr
3.3.3. Eastern Siberia: D3 (ayw2) in Buryats, D2 (ayw2) and A2 (adw2) in Yakuts, and D3 (awy2) and C2 (adrq+) in Chukchi
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Le Bouvier, G.L. The heterogeneity of Australia antigen. J. Infect. Dis. 1971, 123, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Bancroft, W.H.; Mundon, F.K.; Russell, P.K. Detection of additional antigenic determinants of hepatitis B antigen. J. Immunol. 1972, 109, 842–848. [Google Scholar] [PubMed]
- Couroucé-Pauty, A.M.; Plançon, A.; Soulier, J.P. Distribution of HBsAg subtypes in the world. Vox Sang. 1983, 44, 197–211. [Google Scholar] [CrossRef]
- Okamoto, H.; Imai, M.; Tsuda, F.; Tanaka, T.; Miyakawa, Y.; Mayumi, M. Point mutation in the S gene of hepatitis B virus for d/y or w/r subtypic change in two blood donors carrying a surface antigen of compound subtype adyr or adwr. J. Virol. 1987, 61, 3030–3034. [Google Scholar] [CrossRef] [PubMed]
- Norder, H.; Hammas, B.; Losfdahl, S.; Courouce, A.M.; Magnius, L.O. Comparison of the amino acid sequences of nine different serotypes of hepatitis B surface antigen and genomic classification of the corresponding hepatitis B virus strains. J. Gen. Virol. 1992, 73, 1201–1208. [Google Scholar] [CrossRef]
- Okamoto, H.; Tsuda, F.; Sakugawa, H.; Sastrosoewignjo, R.I.; Imai, M.; Miyakawa, Y.; Mayumi, M. Typing hepatitis B virus by homology in nucleotide sequence: Comparison of surface antigen subtypes. J. Gen. Virol. 1988, 69, 2575–2583. [Google Scholar] [CrossRef] [PubMed]
- Norder, H.; Courouce, A.M.; Magnius, L.O. Molecular basis of hepatitis B virus serotype variations within the four major subtypes. J. Gen. Virol. 1992, 73, 3141–3145. [Google Scholar] [CrossRef]
- Naumann, H.; Schaefer, S.; Yoshida, C.F.; Gaspar, A.M.; Repp, R.; Gerlich, W.H. Identification of a new hepatitis B virus (HBV) genotype from Brazil that expresses HBV surface antigen subtype adw4. J. Gen. Virol. 1993, 74, 1627–1632. [Google Scholar] [CrossRef]
- Norder, H.; Hammas, B.; Lee, S.D.; Bile, K.; Courouce, A.M.; Mushahwar, I.K.; Magnius, L.O. Genetic relatedness of hepatitis B viral strains of diverse geographical origin and natural variations in the primary structure of the surface antigen. J. Gen. Virol. 1993, 74, 1341–1348. [Google Scholar] [CrossRef]
- Norder, H.; Courouce, A.M.; Magnius, L.O. Complete genomes, phylogenetic relatedness, and structural proteins of six strains of the hepatitis B virus, four of which represent two new genotypes. Virology 1994, 198, 489–503. [Google Scholar] [CrossRef]
- Stuyver, L.; De Gendt, S.; Van Geyt, C.; Zoulim, F.; Fried, M.; Schinazi, R.F.; Rossau, R. A new genotype of hepatitis B virus: Complete genome and phylogenetic relatedness. J. Gen. Virol. 2000, 81, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Arauz-Ruiz, P.; Norder, H.; Robertson, B.H.; Magnius, L.O. Genotype H: A new Amerindian genotype of hepatitis B virus revealed in Central America. J. Gen. Virol. 2002, 83, 2059–2073. [Google Scholar] [CrossRef]
- McNaughton, A.L.; D’Arienzo, V.; Ansari, M.A.; Lumley, S.F.; Littlejohn, M.; Revill, P.; McKeating, J.A.; Matthews, P.C. Insights from deep sequencing of the HBV genome-unique, tiny, and misunderstood. Gastroenterology 2019, 156, 384–399. [Google Scholar] [CrossRef] [PubMed]
- Campos-Valdez, M.; Monroy-Ramírez, H.C.; Armendáriz-Borunda, J.; Sánchez-Orozco, L.V. Molecular mechanisms during hepatitis B infection and the effects of the virus variability. Viruses 2021, 13, 1167. [Google Scholar] [CrossRef]
- Kramvis, A.; Weitzmann, L.; Owiredu, W.K.; Kew, M.C. Analysis of the complete genome of subgroup A’ hepatitis B virus isolates from South Africa. J. Gen. Virol. 2002, 83, 835–839. [Google Scholar] [CrossRef]
- Huy, T.T.; Ushijima, H.; Quang, V.X.; Win, K.M.; Luengrojanakul, P.; Kikuchi, K.; Sata, T.; Abe, K. Genotype C of hepatitis B virus can be classified into at least two subgroups. J. Gen. Virol. 2004, 85, 283–292. [Google Scholar] [CrossRef]
- Norder, H.; Couroucé, A.M.; Coursaget, P.; Echevarria, J.M.; Lee, S.D.; Mushahwar, I.K.; Robertson, B.H.; Locarnini, S.; Magnius, L.O. Genetic diversity of hepatitis B virus strains derived worldwide: Genotypes, subgenotypes, and HBsAg subtypes. Intervirology 2004, 47, 289–309. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.H.; Yuen, L.; Revill, P. Clarification required for the definition of hepatitis B Virus subgenotypes C1 and C2. Intervirology 2009, 52, 321–322. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, S.; Magnius, L.; Norder, H. Under construction: Classification of hepatitis B virus genotypes and subgenotypes. Intervirology 2009, 52, 323–325. [Google Scholar] [CrossRef]
- Kurbanov, F.; Tanaka, Y.; Mizokami, M. Geographical and genetic diversity of the human hepatitis B virus. Hepatol. Res. 2010, 40, 14–30. [Google Scholar] [CrossRef]
- Flodgren, E.; Bengtsson, S.; Knutsson, M.; Strebkova, E.A.; Kidd, A.H.; Alexeyev, O.A.; Kidd-Ljunggren, K. Recent high incidence of fulminant hepatitis in Samara, Russia: Molecular analysis of prevailing hepatitis B and D virus strains. J. Clin. Microbiol. 2000, 38, 3311–3316. [Google Scholar] [CrossRef] [PubMed]
- Abe, K.; Hayakawa, E.; Sminov, A.V.; Rossina, A.L.; Ding, X.; Huy, T.T.; Sata, T.; Uchaikin, V.F. Molecular epidemiology of hepatitis B, C, D and E viruses among children in Moscow, Russia. J. Clin. Virol. 2004, 30, 57–61. [Google Scholar] [CrossRef] [PubMed]
- Kochneva, G.V.; Grazhdantseva, A.A.; Sivolobova, G.F.; Shustov, A.V.; Gavrilova, I.V.; Chub, E.V.; Bayandin, R.B.; Ternovoi, V.A.; Chausov, E.V.; Akinfeeva, L.A.; et al. Etiology of acute hepatitis and genotypic diversity of hepatitis A, B, C and E viruses in three regions of Siberia. Infect. Dis. 2005, 3, 26–31. (In Russian) [Google Scholar]
- Bayandin, R.B.; Shustov, A.V.; Kochneva, G.V.; Sivolobova, G.F.; Grazhdantseva, A.A.; Loktev, V.B.; Granitov, V.M.; Netesov, S.V. Incidence of markers, virus genotypes and risk factors of hepatitis B among patients of infectious department of Barnaul city municipal hospital. Infect. Dis. 2007, 5, 5–10. (In Russian) [Google Scholar]
- Manuilov, V.A.; Osipova, L.P.; Netesova, I.G.; Chub, E.V.; Bezuglova, L.V.; Norder, H.; Magnius, L.O.; Netesov, S.V. The prevalence of HBsAg subtypes and HBV genotypes in native population groups of Siberia. Mol. Genet. Microbiol. Virol. 2015, 30, 30–38. [Google Scholar] [CrossRef]
- Tran, T.T.; Trinh, T.N.; Abe, K. New complex recombinant genotype of hepatitis B virus identified in Vietnam. J. Virol. 2008, 82, 5657–5663. [Google Scholar]
- Tatematsu, K.; Tanaka, Y.; Kurbanov, F.; Sugauchi, F.; Mano, S.; Maeshiro, T.; Nakayoshi, T.; Wakuta, M.; Miyakawa, Y.; Mizokami, M. A genetic variant of hepatitis B virus divergent from known human and ape genotypes isolated from a Japanese patient and provisionally assigned to new genotype J. J. Virol. 2009, 83, 10538–10547. [Google Scholar] [CrossRef]
- Phung, T.B.; Alestig, E.; Nguyen, T.L.; Hannoun, C.; Lindh, M. Genotype X/C recombinant (putative genotype I) of hepatitis B virus is rare in Hanoi, Vietnam: Genotypes B4 and C1 predominate. J. Med. Virol. 2010, 82, 1327–1333. [Google Scholar] [CrossRef]
- Hannoun, C.; Norder, H.; Lindh, M. An aberrant genotype revealed in recombinant hepatitis B virus strains from Vietnam. J. Gen. Virol. 2000, 81, 2267–2272. [Google Scholar] [CrossRef]
- Haldipur, B.P.; Walimbe, A.M.; Arankalle, V.A. Circulation of genotype-I hepatitis B virus in the primitive tribes of Arunachal Pradesh in early sixties and molecular evolution of genotype-I. Infect. Genet. Evol. 2014, 27, 366–374. [Google Scholar] [CrossRef]
- Kocher, A.; Papac, L.; Barquera, R.; Key, F.M.; Spyrou, M.A.; Hübler, R.; Rohrlach, A.B.; Aron, F.; Stahl, R.; Wissgott, A.; et al. Ten millennia of hepatitis B virus evolution. Science 2021, 374, 182–188. [Google Scholar] [CrossRef] [PubMed]
- Valenzuela, P.; Quiroga, M.; Zalvidar, J.; Gray, P.; Rutter, W.J. The nucleotide sequence of the hepatitis B viral genome and the identification of the major viral genes. In Animal Virus Genetics; Fields, B.N., Jaenisch, R., Fox, C.F., Eds.; Academic Press: New York, NY, USA, 1980; pp. 57–70. [Google Scholar] [CrossRef]
- Anisimova, M.; Gil, M.; Dufayard, J.-F.; Dessimoz, C.; Gascuel, O. Survey of branch support methods demonstrates accuracy, power, and robustness of fast likelihood-based approximation schemes. Syst. Biol. 2011, 60, 685–699. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Chulanov, V.P. Epidemiological and Clinical Significance of Genetic Heterogeneity of Hepatitis A and B Viruses. Ph.D. Thesis, Central Research Institute for Epidemiology of the Rospotrebnadzor, Moscow, Russia, 2013. (In Russian). [Google Scholar]
- Manuilov, V.A.; Osipova, L.P.; Netesova, I.G.; Chub, E.V.; Tsoy, L.V.; Dul’beev, R.V.; Alexeeva, L.R.; Norder, H.; Magnius, L.O.; Netesov, S.V. Incidence of genotype of hepatitis B subvirus and HBsAg subtypes in native people of northern and southeastern Siberia. Mol. Genet. Microbiol. Virol. 2010, 25, 172–177. [Google Scholar] [CrossRef]
- Tallo, T.; Norder, H.; Tefanova, V.; Krispin, T.; Priimägi, L.; Mukomolov, S.; Mikhailov, M.; Magnius, L.O. Hepatitis B virus genotype D strains from Estonia share sequence similarity with strains from Siberia and may specify ayw4. J. Med. Virol. 2004, 74, 221–227. [Google Scholar] [CrossRef]
- Tallo, T.; Tefanova, V.; Priimagi, L.; Schmidt, J.; Katargina, O.; Michailov, M.; Mukomolov, S.; Magnius, L.; Norder, H. D2: Major subgenotype of hepatitis B virus in Russia and the Baltic region. J. Gen. Virol. 2008, 89, 1829–1839. [Google Scholar] [CrossRef]
- Semenov, A.V.; Ostankova, Y.V.; Gerasimova, V.V.; Bichurina, M.A.; Mukomolov, S.L.; Kozlov, A.V.; Totolian, A.A. For the question about molecular epidemiology of hepatitis B virus infection in the Republic Sakha (Yakutia). J. Infect. 2016, 8, 57–65. (In Russian) [Google Scholar]
- Karlsen, A.A.; Kyuregyan, K.K.; Isaeva, O.V.; Kichatova, V.S.; Asadi Mobarkhan, F.A.; Bezuglova, L.V.; Netesova, I.G.; Manuylov, V.A.; Pochtovyi, A.A.; Gushchin, V.A.; et al. Different evolutionary dynamics of hepatitis B virus genotypes A and D, and hepatitis D virus genotypes 1 and 2 in an endemic area of Yakutia, Russia. BMC Infect. Dis. 2022, 22, 452. [Google Scholar] [CrossRef]
- Sominskaya, I.; Mihailova, M.; Jansons, J.; Emelyanova, V.; Folkmane, I.; Smagris, E.; Dumpis, U.; Rozentals, R.; Pumpens, P. Hepatitis B and C virus variants in long-term immunosuppressed renal transplant patients in Latvia. Intervirology 2005, 48, 192–200. [Google Scholar] [CrossRef]
- Olinger, C.M.; Lazouskaya, N.V.; Eremin, V.F.; Muller, C.P. Multiple genotypes and subtypes of hepatitis B and C viruses in Belarus: Similarities with Russia and western European influences. Clin. Microbiol. Infect. 2008, 14, 575–581. [Google Scholar] [CrossRef]
- Avazova, D.; Kurbanov, F.; Tanaka, Y.; Sugiyama, M.; Radchenko, I.; Ruziev, D.; Musabaev, E.; Mizokami, M. Hepatitis B virus transmission pattern and vaccination efficiency in Uzbekistan. J. Med. Virol. 2008, 80, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Davaalkham, D.; Ojima, T.; Uehara, R.; Watanabe, M.; Oki, I.; Endo, K.; Takahashi, M.; Okamoto, H.; Nakamura, Y. Analysis of hepatitis B surface antigen mutations in Mongolia: Molecular epidemiology and implications for mass vaccination. Arch. Virol. 2007, 152, 575–584. [Google Scholar] [CrossRef] [PubMed]
- Odgerel, Z.; Choi, I.K.; Byun, K.S.; Pak, C.Y.; Bennett, S.; Gu, S.H.; Park, K.; Kee, S.H.; Song, K.J.; Song, J.W. Complete genome sequence and phylogenetic analysis of hepatitis B virus (HBV) isolated from Mongolian patients with chronic HBV infection. Virus Genes 2006, 33, 345–349. [Google Scholar] [CrossRef] [PubMed]
- Netesova, I.G.; Swenson, P.D.; Osipova, L.P.; Kisilev, N.N.; Posukh, O.L.; Cherepanova, N.S.; Kazakovtseva, M.A.; Kashinskaya, Y.O.; Netesov, S.V. Markers of viral hepatitis B among South Altaians of the settlement Mendur-Sokkon (Atlai Republic). Subtyping of HBsAg of HBV isolates using a panel of monoclonal antibodies. J. Microbiol. Epidemiol. Immunol. 2001, 1, 29–33. (In Russian) [Google Scholar]
- Netesova, I.G.; Swenson, P.D.; Osipova, L.P.; Gubina, M.A.; Posukh, O.L.; Netesov, S.V. Determination of HBsAg subtypes in Western Siberian part of Russia. J. Med. Virol. 2003, 71, 183–187. [Google Scholar] [CrossRef]
- Klushkina, V.V.; Kyuregyan, K.K.; Kozhanova, T.V.; Popova, O.E.; Dubrovina, P.G.; Isaeva, O.V.; Gordeychuk, I.V.; Mikhailov, M.I. Impact of universal hepatitis B vaccination on prevalence, infection-associated morbidity and mortality, and circulation of immune escape variants in Russia. PLoS ONE 2016, 11, e0157161. [Google Scholar] [CrossRef]
- Netesova, I.G.; Swenson, P.D.; Osipova, L.P.; Kalashnikova, T.V.; Netesov, S.V.; Favorov, M.O. Subtypes of HBsAg of hepatitis B virus in the Western Siberia. Vopr. Virusol. 2004, 1, 17–20. (In Russian) [Google Scholar]
- Dobrodeeva, L.K.; Kornienko, E.B.; Petrenya, N.N.; Lutfalieva, G.T.; Schegoleva, L.S.; Demeneva, L.V.; Duberman, B.L.; Tkachev, A.V.; Chiba, H.; Senoo, H.; et al. An unique seroepidemiological pattern of HBV, HCV and HTLV-I in Nenets and Komi in northwestern Russia. Asian Pac. J. Cancer Prev. 2005, 6, 342–345. [Google Scholar]
- Manuilov, V.A.; Netesova, I.G.; Osipova, L.P.; Shustov, A.V.; Bayandin, R.B.; Kochneva, G.V.; Netesov, S.V. Genetic variability of the hepatitis B virus isolates among the population of Shuryshkarsky district in Yamalo-Nenets autonomous region. Mol. Genet. Microbiol. Virol. 2005, 20, 30–34. (In Russian) [Google Scholar]
- Kuzin, S.N.; Pavlov, N.N.; Semenov, S.I.; Krivoshapkin, V.G.; Indeeva, L.D.; Savvin, R.G.; Nikolaev, A.V.; Chemezova, R.I.; Kozhevnikova, L.K.; Lavrov, V.F.; et al. The breadth of the spread of viral hepatitis among the various population groups in the Republic of Sakha (Yakutia). J. Microbiol. Epidemiol. Immunol. 2004, 1, 18–22. (In Russian) [Google Scholar]
- Zotova, A.V. Parenteral Viral Hepatitis in Southern Yakutia. Ph.D. Thesis, Central Research Institute for Epidemiology of the Rospotrebnadzor, Moscow, Russia, 2010. (In Russian). [Google Scholar]
- Vyazov, S.O.; Kompaniets, A.A.; Ananiev, V.A.; Listovskaya, E.K.; Dardik, F.G.; Pirozhkova, Z.P.; Leubenzon, A.S.; Mtchedlischvili, I.M.; Sakvaredidze, L.O.; Vorobey, V.S.; et al. Identification of HBsAg and anti-HBs in healthy population of different cities of the USSR. Vopr. Virusol. 1985, 30, 231–233. (In Russian) [Google Scholar]
- Somova, A.V.; Golosova, T.V.; Margolina, A.N.; Bagryantseva, S.Y. Serodiagnostics of viral hepatitis C and B in different groups of population. Vopr. Virusol. 1992, 37, 231–233. (In Russian) [Google Scholar]
- Shakhgil’dyan, I.V.; Mikhailov, M.I.; Onishchenko, G.G. Parenteral Viral Hepatitis: Epidemiology, Diagnosis, Prevention; GOU VUNMC MZ RF: Moscow, Russia, 2003; 384p. (In Russian) [Google Scholar]
- Davlidova, S.; Haley-Johnson, Z.; Nyhan, K.; Farooq, A.; Vermund, S.H.; Ali, S. Prevalence of HIV, HCV and HBV in Central Asia and the Caucasus: A systematic review. Int. J. Infect. Dis. 2021, 104, 510–525. [Google Scholar] [CrossRef] [PubMed]
- Dambadarjaa, D.; Mukhtar, Y.; Tsogzolbaatar, E.O.; Khuyag, S.O.; Dayan, A.; Oyunbileg, N.E.; Shagdarsuren, O.E.; Nyam, G.; Nakamura, Y.; Takahashi, M.; et al. Hepatitis B, C, and D virus infections and AFP tumor marker prevalence among the elderly population in Mongolia: A nationwide survey. J. Prev. Med. Public Health 2022, 55, 263–272. [Google Scholar] [CrossRef]
- Margolis, H.S.; Alter, M.J.; Hadler, S.C. Hepatitis B: Evolving epidemiology and implications for control. Semin. Liver Dis. 1991, 11, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Gordeichuk, I.V. Occult Infection Caused by Hepatitis B Virus: Virologic and Clinical-Morphological Characterization. Ph.D. Thesis, Institute for Poliomyelitis and Viral Encephalitis, Moscow, Russia, 2010. (In Russian). [Google Scholar]
- Gerasimova, V.V. Molecular and Epidemiological Characterization of Hepatitis B and C Viruses in Sakha Republic (Yakutia). Ph.D. Thesis, Pasteur Institute of the Rospotrebnadzor, St. Petersburg, Russia, 2016. (In Russian). [Google Scholar]
- Report of the Russian Census of 2010. Available online: http://www.gks.ru/free_doc/new_site/perepis2010/croc/perepis_itogi1612.htm# (accessed on 2 November 2022).
- Kao, J.H.; Chen, P.J.; Chen, D.S. Recent advances in the research of hepatitis B virus-related hepatocellular carcinoma: Epidemiologic and molecular biological aspects. Adv. Cancer Res. 2010, 108, 21–72. [Google Scholar] [CrossRef] [PubMed]
- Levin, M.G.; Potapov, L.P. (Eds.) Historical and Ethnographic Atlas of Siberia; Academy of Sciences of the USSR: Moscow, Russia, 1961; 498p. (In Russian) [Google Scholar]
- Trovão, N.S.; Thijssen, M.; Vrancken, B.; Pineda-Peña, A.C.; Mina, T.; Amini-Bavil-Olyaee, S.; Lemey, P.; Baele, G.; Pourkarim, M.R. Reconstruction of the origin and dispersal of the worldwide dominant hepatitis B virus subgenotype D1. Virus Evol. 2022, 8, veac028. [Google Scholar] [CrossRef]
- Amini-Bavil-Olyaee, S.; Alavian, S.M.; Adeli, A.; Sarrami-Forooshani, R.; Sabahi, F.; Sabouri, E.; Tavangar, H.R.; Azizi, M.; Mahboudi, F. Hepatitis B virus genotyping, core promoter, and precore/core mutations among Afghan patients infected with hepatitis B: A preliminary report. J. Med. Virol. 2006, 78, 358–364. [Google Scholar] [CrossRef]
- Pourkarim, M.R.; Vergote, V.; Amini-Bavil-Olyaee, S.; Sharifi, Z.; Sijmons, S.; Lemey, P.; Maes, P.; Alavian, S.M.; Van Ranst, M. Molecular characterization of hepatitis B virus (HBV) strains circulating in the northern coast of the Persian Gulf and its comparison with worldwide distribution of HBV subgenotype D1. J. Med. Virol. 2014, 86, 745–757. [Google Scholar] [CrossRef]
- Pineda-Peña, A.C.; Faria, N.R.; Mina, T.; Amini-Bavil-Olyaee, S.; Alavian, S.M.; Lemey, P.; Maes, P.; Van Ranst, M.; Pourkarim, M.R. Epidemiological history and genomic characterization of non-D1 HBV strains identified in Iran. J. Clin. Virol. 2015, 63, 38–41. [Google Scholar] [CrossRef]
- Sayan, M.; Dogan, C. Genotype/subgenotype distribution of hepatitis B virus among hemodialysis patients with chronical hepatitis B. Ann. Hepatol. 2012, 11, 849–854. [Google Scholar] [CrossRef]
- Sunbul, M.; Sugiyama, M.; Kurbanov, F.; Leblebicioglu, H.; Khan, A.; Elkady, A.; Tanaka, Y.; Mizokami, M. Specific mutations of basal core promoter are associated with chronic liver disease in hepatitis B virus subgenotype D1 prevalent in Turkey. Microbiol. Immunol. 2013, 57, 122–129. [Google Scholar] [CrossRef] [PubMed]
- Ciccozzi, M.; Ciccaglione, A.R.; Lo Presti, A.; Equestre, M.; Cella, E.; Ebranati, E.; Gabanelli, E.; Villano, U.; Bruni, R.; Yalcinkaya, T.; et al. Evolutionary dynamics of HBV-D1 genotype epidemic in Turkey. J. Med. Virol. 2014, 86, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Mojsiejczuk, L.N.; Torres, C.; Sevic, I.; Badano, I.; Malan, R.; Flichman, D.M.; Liotta, D.J.; Campos, R.H. Molecular epidemiology of hepatitis B virus in Misiones, Argentina. Infect. Genet. Evol. 2016, 44, 34–42. [Google Scholar] [CrossRef]
- Chachá, S.G.F.; Gomes-Gouvêa, M.S.; Malta, F.M.; Ferreira, S.D.C.; Villanova, M.G.; Souza, F.F.; Teixeira, A.C.; Passos, A.D.D.C.; Pinho, J.R.R.; Martinelli, A.L.C. Distribution of HBV subgenotypes in Ribeirão Preto, Southeastern Brazil: A region with history of intense Italian immigration. Braz. J. Infect. Dis. 2017, 21, 424–432. [Google Scholar] [CrossRef]
- Chandra, P.K.; Biswas, A.; Datta, S.; Banerjee, A.; Panigrahi, R.; Chakrabarti, S.; De, B.K.; Chakravarty, R. Subgenotypes of hepatitis B virus genotype D (D1, D2, D3 and D5) in India: Differential pattern of mutations, liver injury and occult HBV infection. J. Viral Hepatol. 2009, 16, 749–756. [Google Scholar] [CrossRef]
- Biswas, A.; Panigrahi, R.; Chandra, P.K.; Banerjee, A.; Datta, S.; Pal, M.; Chakraborty, S.; Bhattacharya, P.; Chakrabarti, S.; Chakravarty, R. Characterization of the occult hepatitis B virus variants circulating among the blood donors from eastern India. Sci. World J. 2013, 4, 212704. [Google Scholar] [CrossRef]
- Abu Zeid, W.M.; Ramadan, D.I.; Shemis, M.A. Prevalence of mutations within major hydrophilic region of hepatitis B virus and their correlation with genotypes among chronically infected patients in Egypt. Arab. J. Gastroenterol. 2016, 17, 34–40. [Google Scholar] [CrossRef]
- Kharkov, V.N.; Stepanov, V.A.; Medvedeva, O.F.; Spiridonova, M.F.; Voevoda, M.I.; Romaschenko, A.G.; Tadinova, V.N.; Puzyrev, V.P. Differences in structures of genetic funds of the Northern and Southern Altaians for haplogroups of Y-chromosome. Genetica 2007, 43, 1–13. (In Russian) [Google Scholar]
- Kharkov, V.N.; Medvedeva, O.F.; Luzina, F.A.; Kolbasko, A.V.; Gafarov, N.I.; Puzyrev, V.P.; Stepanov, V.A. Comparative characteristic of genetic funds of Teleuts by the Y-chromosome markers data. Genetica 2009, 43, 1–10. (In Russian) [Google Scholar]
- Kharkov, V.N.; Khamina, K.V.; Medvedeva, O.F.; Simonova, K.V.; Khitrinskaya, I.Y.; Stepanov, V.A. Structure of the genetic fund of Tuvans in the markers of Y-chromosome. Genetica 2013, 49, 1416–1425. (In Russian) [Google Scholar]
- Kharkov, V.N.; Khamina, K.V.; Medvedeva, O.F.; Simonova, K.V.; Eremina, E.R.; Stepanov, V.A. The genetic fund of Buryats: Clinal variability and territorial isolation of the markers of Y-chromosome. Genetica 2014, 50, 203–213. (In Russian) [Google Scholar]
- Stepanov, V.A. Evolution of genetic diversity and human diseases. Genetica 2016, 52, 852–864. (In Russian) [Google Scholar] [CrossRef]
- Glottolog. Comprehensive Reference Information for the World’s Languages, Especially the Lesser Known Languages. Available online: https://glottolog.org/glottolog/family (accessed on 2 November 2022).
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manuylov, V.; Chulanov, V.; Bezuglova, L.; Chub, E.; Karlsen, A.; Kyuregyan, K.; Ostankova, Y.; Semenov, A.; Osipova, L.; Tallo, T.; et al. Genetic Diversity and Possible Origins of the Hepatitis B Virus in Siberian Natives. Viruses 2022, 14, 2465. https://doi.org/10.3390/v14112465
Manuylov V, Chulanov V, Bezuglova L, Chub E, Karlsen A, Kyuregyan K, Ostankova Y, Semenov A, Osipova L, Tallo T, et al. Genetic Diversity and Possible Origins of the Hepatitis B Virus in Siberian Natives. Viruses. 2022; 14(11):2465. https://doi.org/10.3390/v14112465
Chicago/Turabian StyleManuylov, Victor, Vladimir Chulanov, Ludmila Bezuglova, Elena Chub, Anastasia Karlsen, Karen Kyuregyan, Yulia Ostankova, Alexander Semenov, Ludmila Osipova, Tatjana Tallo, and et al. 2022. "Genetic Diversity and Possible Origins of the Hepatitis B Virus in Siberian Natives" Viruses 14, no. 11: 2465. https://doi.org/10.3390/v14112465
APA StyleManuylov, V., Chulanov, V., Bezuglova, L., Chub, E., Karlsen, A., Kyuregyan, K., Ostankova, Y., Semenov, A., Osipova, L., Tallo, T., Netesova, I., Tkachuk, A., Gushchin, V., Netesov, S., Magnius, L. O., & Norder, H. (2022). Genetic Diversity and Possible Origins of the Hepatitis B Virus in Siberian Natives. Viruses, 14(11), 2465. https://doi.org/10.3390/v14112465







