The Role of the Stem-Loop A RNA Promoter in Flavivirus Replication
Abstract
1. Flavivirus
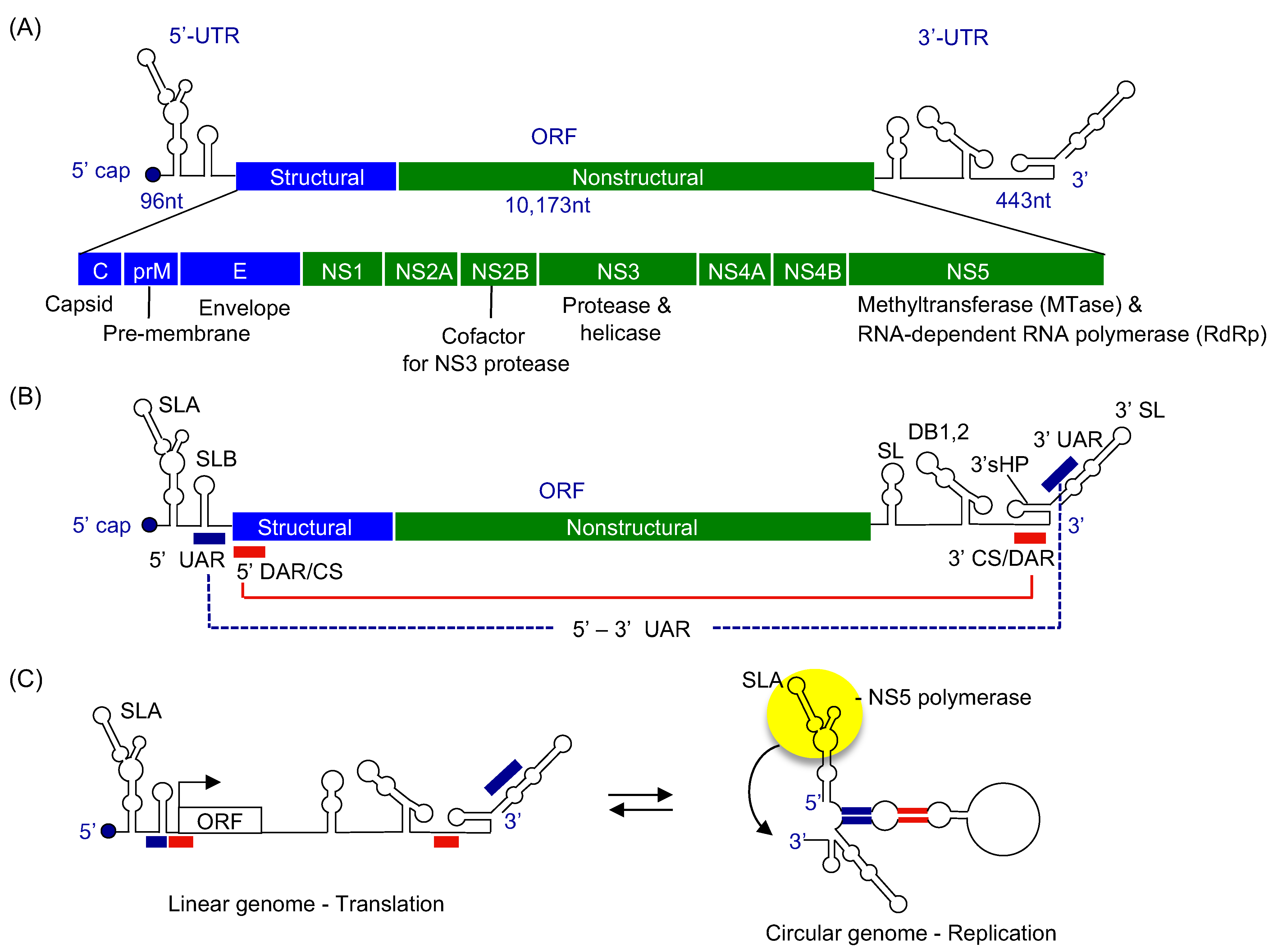
2. Flaviviral Genome Structure
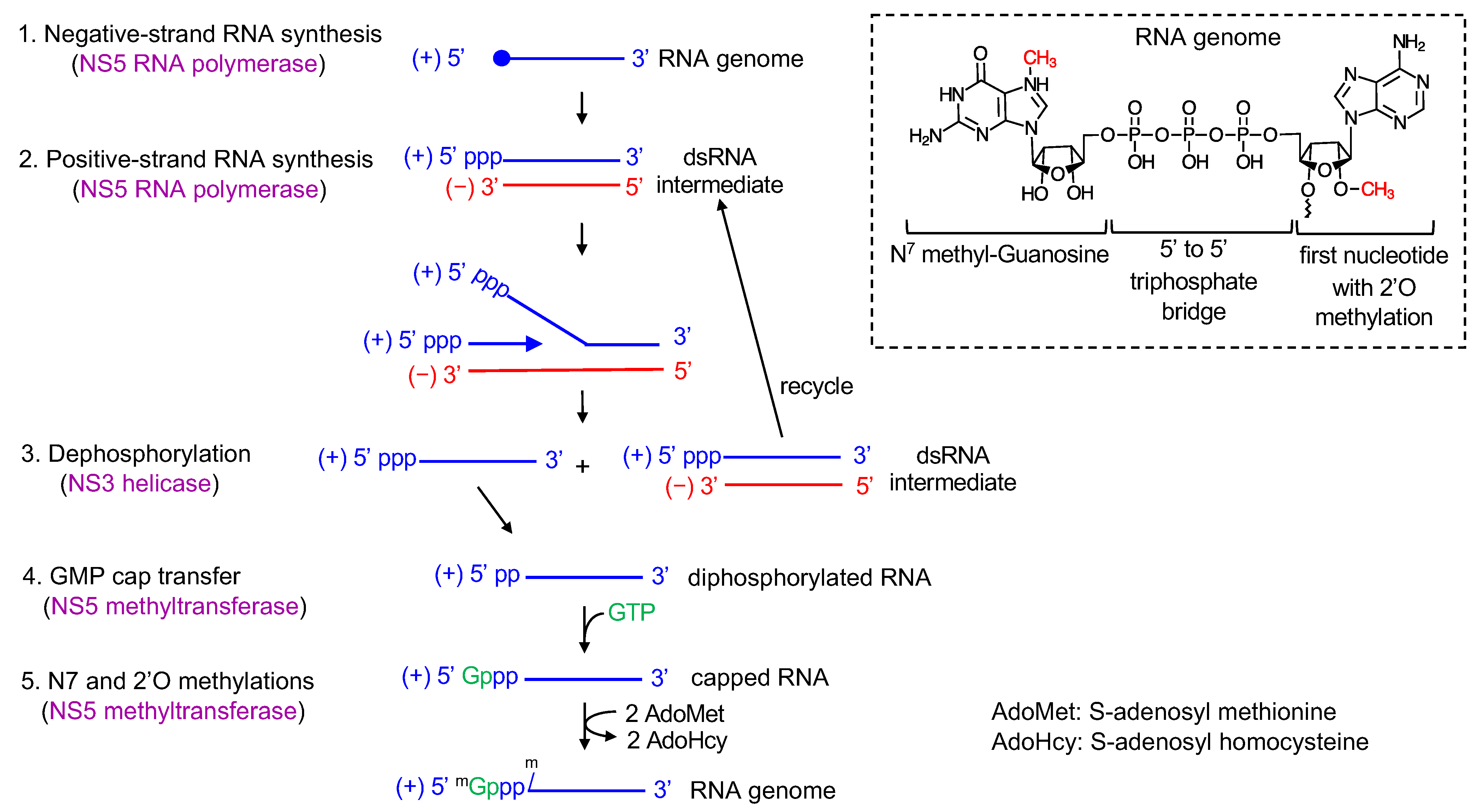
3. Flavivirus Genome Replication Steps
4. Viral RNA Elements Essential for Genome Replication
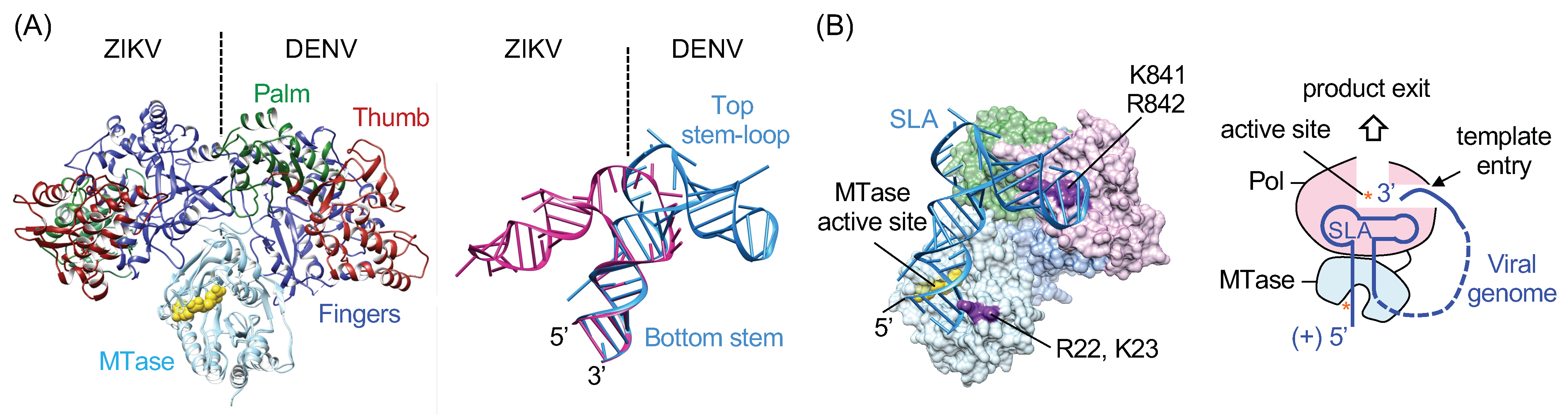
5. Structure of Viral RNA Promoter SLA

6. SLA Promoter-Mediated Negative-Strand RNA Synthesis
7. Positive-Strand RNA Synthesis
8. Conclusions and Perspectives
Funding
Conflicts of Interest
References
- Clyde, K.; Kyle, J.L.; Harris, E. Recent advances in deciphering viral and host determinants of dengue virus replication and pathogenesis. J. Virol. 2006, 80, 11418–11431. [Google Scholar] [CrossRef] [PubMed]
- Murray, N.E.; Quam, M.B.; Wilder-Smith, A. Epidemiology of dengue: Past, present and future prospects. Clin. Epidemiol. 2013, 5, 299–309. [Google Scholar] [PubMed]
- Miner, J.J.; Cao, B.; Govero, J.; Smith, A.M.; Fernandez, E.; Cabrera, O.H.; Garber, C.; Noll, M.; Klein, R.S.; Noguchi, K.K.; et al. Zika Virus Infection during Pregnancy in Mice Causes Placental Damage and Fetal Demise. Cell 2016, 165, 1081–1091. [Google Scholar] [CrossRef]
- Lindenbach, B.D.; Thiel, H.J.; Rice, C.M. Flaviviridae: The Viruses and Their Replication; Lippincott-Raven Publishers: Philadelphia, PA, USA, 2007; Volume 1, pp. 1101–1152. [Google Scholar]
- Ng, W.C.; Soto-Acosta, R.; Bradrick, S.S.; Garcia-Blanco, M.A.; Ooi, E.E. The 5′ and 3′ Untranslated Regions of the Flaviviral Genome. Viruses 2017, 9, 137. [Google Scholar] [CrossRef] [PubMed]
- Filomatori, C.V.; Lodeiro, M.F.; Alvarez, D.E.; Samsa, M.M.; Pietrasanta, L.; Gamarnik, A.V. A 5′ RNA element promotes dengue virus RNA synthesis on a circular genome. Genes Dev. 2006, 20, 2238–2249. [Google Scholar] [CrossRef]
- Gebhard, L.G.; Filomatori, C.V.; Gamarnik, A.V. Functional RNA elements in the dengue virus genome. Viruses 2011, 3, 1739–1756. [Google Scholar] [CrossRef] [PubMed]
- Lodeiro, M.F.; Filomatori, C.V.; Gamarnik, A.V. Structural and functional studies of the promoter element for dengue virus RNA replication. J. Virol. 2009, 83, 993–1008. [Google Scholar] [CrossRef]
- Pijlman, G.P.; Funk, A.; Kondratieva, N.; Leung, J.; Torres, S.; van der Aa, L.; Liu, W.J.; Palmenberg, A.C.; Shi, P.Y.; Hall, R.A.; et al. A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe 2008, 4, 579–591. [Google Scholar] [CrossRef] [PubMed]
- Slonchak, A.; Khromykh, A.A. Subgenomic flaviviral RNAs: What do we know after the first decade of research. Antivir. Res. 2018, 159, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Chapman, E.G.; Costantino, D.A.; Rabe, J.L.; Moon, S.L.; Wilusz, J.; Nix, J.C.; Kieft, J.S. The structural basis of pathogenic subgenomic flavivirus RNA (sfRNA) production. Science 2014, 344, 307–310. [Google Scholar] [CrossRef]
- Klema, V.J.; Padmanabhan, R.; Choi, K.H. Flaviviral Replication Complex: Coordination between RNA Synthesis and 5′-RNA Capping. Viruses 2015, 7, 4640–4656. [Google Scholar] [CrossRef]
- Welsch, S.; Miller, S.; Romero-Brey, I.; Merz, A.; Bleck, C.K.; Walther, P.; Fuller, S.D.; Antony, C.; Krijnse-Locker, J.; Bartenschlager, R. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 2009, 5, 365–375. [Google Scholar] [CrossRef] [PubMed]
- Chambers, T.J.; Grakoui, A.; Rice, C.M. Processing of the yellow fever virus nonstructural polyprotein: A catalytically active NS3 proteinase domain and NS2B are required for cleavages at dibasic sites. J. Virol. 1991, 65, 6042–6050. [Google Scholar] [CrossRef]
- Falgout, B.; Pethel, M.; Zhang, Y.M.; Lai, C.J. Both nonstructural proteins NS2B and NS3 are required for the proteolytic processing of dengue virus nonstructural proteins. J. Virol. 1991, 65, 2467–2475. [Google Scholar] [CrossRef]
- Li, H.; Clum, S.; You, S.; Ebner, K.E.; Padmanabhan, R. The serine protease and RNA-stimulated nucleoside triphosphatase and RNA helicase functional domains of dengue virus type 2 NS3 converge within a region of 20 amino acids. J. Virol. 1999, 73, 3108–3116. [Google Scholar] [CrossRef] [PubMed]
- Benarroch, D.; Selisko, B.; Locatelli, G.A.; Maga, G.; Romette, J.L.; Canard, B. The RNA helicase, nucleotide 5′-triphosphatase, and RNA 5′-triphosphatase activities of Dengue virus protein NS3 are Mg2+-dependent and require a functional Walker B motif in the helicase catalytic core. Virology 2004, 328, 208–218. [Google Scholar] [CrossRef]
- Issur, M.; Geiss, B.J.; Bougie, I.; Picard-Jean, F.; Despins, S.; Mayette, J.; Hobdey, S.E.; Bisaillon, M. The flavivirus NS5 protein is a true RNA guanylyltransferase that catalyzes a two-step reaction to form the RNA cap structure. RNA 2009, 15, 2340–2350. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, M.; Padmanabhan, R. De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymerase exhibits temperature dependence at the initiation but not elongation phase. J. Biol. Chem. 2001, 276, 39926–39937. [Google Scholar] [CrossRef] [PubMed]
- You, S.; Padmanabhan, R. A novel in vitro replication system for Dengue virus. Initiation of RNA synthesis at the 3′-end of exogenous viral RNA templates requires 5′- and 3′-terminal complementary sequence motifs of the viral RNA. J. Biol. Chem. 1999, 274, 33714–33722. [Google Scholar] [CrossRef] [PubMed]
- Padmanabhan, R.; Takhampunya, R.; Teramoto, T.; Choi, K.H. Flavivirus RNA synthesis in vitro. Methods 2015, 91, 20–34. [Google Scholar] [CrossRef] [PubMed]
- Wengler, G.; Wengler, G.; Gross, H.J. Studies on virus-specific nucleic acids synthesized in vertebrate and mosquito cells infected with flaviviruses. Virology 1978, 89, 423–437. [Google Scholar] [CrossRef]
- Gullberg, R.C.; Jordan Steel, J.; Moon, S.L.; Soltani, E.; Geiss, B.J. Oxidative stress influences positive strand RNA virus genome synthesis and capping. Virology 2015, 475, 219–229. [Google Scholar] [CrossRef]
- Dong, H.; Zhang, B.; Shi, P.Y. Flavivirus methyltransferase: A novel antiviral target. Antivir. Res 2008, 80, 1–10. [Google Scholar] [CrossRef]
- Egloff, M.P.; Benarroch, D.; Selisko, B.; Romette, J.L.; Canard, B. An RNA cap (nucleoside-2′-O-)-methyltransferase in the flavivirus RNA polymerase NS5: Crystal structure and functional characterization. EMBO J. 2002, 21, 2757–2768. [Google Scholar] [CrossRef]
- Ray, D.; Shah, A.; Tilgner, M.; Guo, Y.; Zhao, Y.; Dong, H.; Deas, T.S.; Zhou, Y.; Li, H.; Shi, P.Y. West Nile virus 5′-cap structure is formed by sequential guanine N-7 and ribose 2′-O methylations by nonstructural protein 5. J. Virol. 2006, 80, 8362–8370. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Ray, D.; Ren, S.; Zhang, B.; Puig-Basagoiti, F.; Takagi, Y.; Ho, C.K.; Li, H.; Shi, P.Y. Distinct RNA elements confer specificity to flavivirus RNA cap methylation events. J. Virol. 2007, 81, 4412–4421. [Google Scholar] [CrossRef]
- Bujalowski, P.J.; Bujalowski, W.; Choi, K.H. Interactions between the Dengue Virus Polymerase NS5 and Stem-Loop A. J. Virol. 2017, 91, e00047-17. [Google Scholar] [CrossRef] [PubMed]
- Khromykh, A.A.; Meka, H.; Guyatt, K.J.; Westaway, E.G. Essential role of cyclization sequences in flavivirus RNA replication. J. Virol. 2001, 75, 6719–6728. [Google Scholar] [CrossRef]
- Villordo, S.M.; Gamarnik, A.V. Genome cyclization as strategy for flavivirus RNA replication. Virus Res. 2009, 139, 230–239. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Nomaguchi, M.; Padmanabhan, R.; Markoff, L. Specific requirements for elements of the 5′ and 3′ terminal regions in flavivirus RNA synthesis and viral replication. Virology 2008, 374, 170–185. [Google Scholar] [CrossRef] [PubMed]
- Friebe, P.; Harris, E. Interplay of RNA elements in the dengue virus 5′ and 3′ ends required for viral RNA replication. J. Virol. 2010, 84, 6103–6118. [Google Scholar] [CrossRef] [PubMed]
- Filomatori, C.V.; Iglesias, N.G.; Villordo, S.M.; Alvarez, D.E.; Gamarnik, A.V. RNA sequences and structures required for the recruitment and activity of the dengue virus polymerase. J. Biol. Chem. 2011, 286, 6929–6939. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.; Bujalowski, P.J.; Teramoto, T.; Gottipati, K.; Scott, S.D.; Padmanabhan, R.; Choi, K.H. Structures of flavivirus RNA promoters suggest two binding modes with NS5 polymerase. Nat. Commun. 2021, 12, 2530. [Google Scholar] [CrossRef] [PubMed]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]
- Zuker, M.; Stiegler, P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981, 9, 133–148. [Google Scholar] [CrossRef]
- Li, X.F.; Jiang, T.; Yu, X.D.; Deng, Y.Q.; Zhao, H.; Zhu, Q.Y.; Qin, E.D.; Qin, C.F. RNA elements within the 5′ untranslated region of the West Nile virus genome are critical for RNA synthesis and virus replication. J. Gen. Virol 2010, 91, 1218–1223. [Google Scholar] [CrossRef]
- Bujalowski, P.J.; Bujalowski, W.; Choi, K.H. Identification of the viral RNA promoter stem loop A (SLA)-binding site on Zika virus polymerase NS5. Sci. Rep. 2020, 10, 13306. [Google Scholar] [CrossRef]
- Hodge, K.; Tunghirun, C.; Kamkaew, M.; Limjindaporn, T.; Yenchitsomanus, P.T.; Chimnaronk, S. Identification of a Conserved RNA-dependent RNA Polymerase (RdRp)-RNA Interface Required for Flaviviral Replication. J. Biol. Chem. 2016, 291, 17437–17449. [Google Scholar] [CrossRef] [PubMed]
- Zhong, W.; Ingravallo, P.; Wright-Minogue, J.; Skelton, A.; Uss, A.S.; Chase, R.; Yao, N.; Lau, J.Y.; Hong, Z. Nucleoside triphosphatase and RNA helicase activities associated with GB virus B nonstructural protein 3. Virology 1999, 261, 216–226. [Google Scholar] [CrossRef]
- Huang, Z.S.; Wang, C.C.; Wu, H.N. HCV NS3 protein helicase domain assists RNA structure conversion. FEBS Lett. 2010, 584, 2356–2362. [Google Scholar] [CrossRef]
- Ertel, K.J.; Brunner, J.E.; Semler, B.L. Mechanistic consequences of hnRNP C binding to both RNA termini of poliovirus negative-strand RNA intermediates. J. Virol. 2010, 84, 4229–4242. [Google Scholar] [CrossRef] [PubMed]
- Nomaguchi, M.; Teramoto, T.; Yu, L.; Markoff, L.; Padmanabhan, R. Requirements for West Nile virus (−)- and (+)-strand subgenomic RNA synthesis in vitro by the viral RNA-dependent RNA polymerase expressed in Escherichia coli. J. Biol. Chem. 2004, 279, 12141–12151. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, K.H. The Role of the Stem-Loop A RNA Promoter in Flavivirus Replication. Viruses 2021, 13, 1107. https://doi.org/10.3390/v13061107
Choi KH. The Role of the Stem-Loop A RNA Promoter in Flavivirus Replication. Viruses. 2021; 13(6):1107. https://doi.org/10.3390/v13061107
Chicago/Turabian StyleChoi, Kyung H. 2021. "The Role of the Stem-Loop A RNA Promoter in Flavivirus Replication" Viruses 13, no. 6: 1107. https://doi.org/10.3390/v13061107
APA StyleChoi, K. H. (2021). The Role of the Stem-Loop A RNA Promoter in Flavivirus Replication. Viruses, 13(6), 1107. https://doi.org/10.3390/v13061107





