The Kaumoebavirus LCC10 Genome Reveals a Unique Gene Strand Bias among “Extended Asfarviridae”
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Cultures
2.2. Genome Sequencing and Analysis
3. Results and Discussion
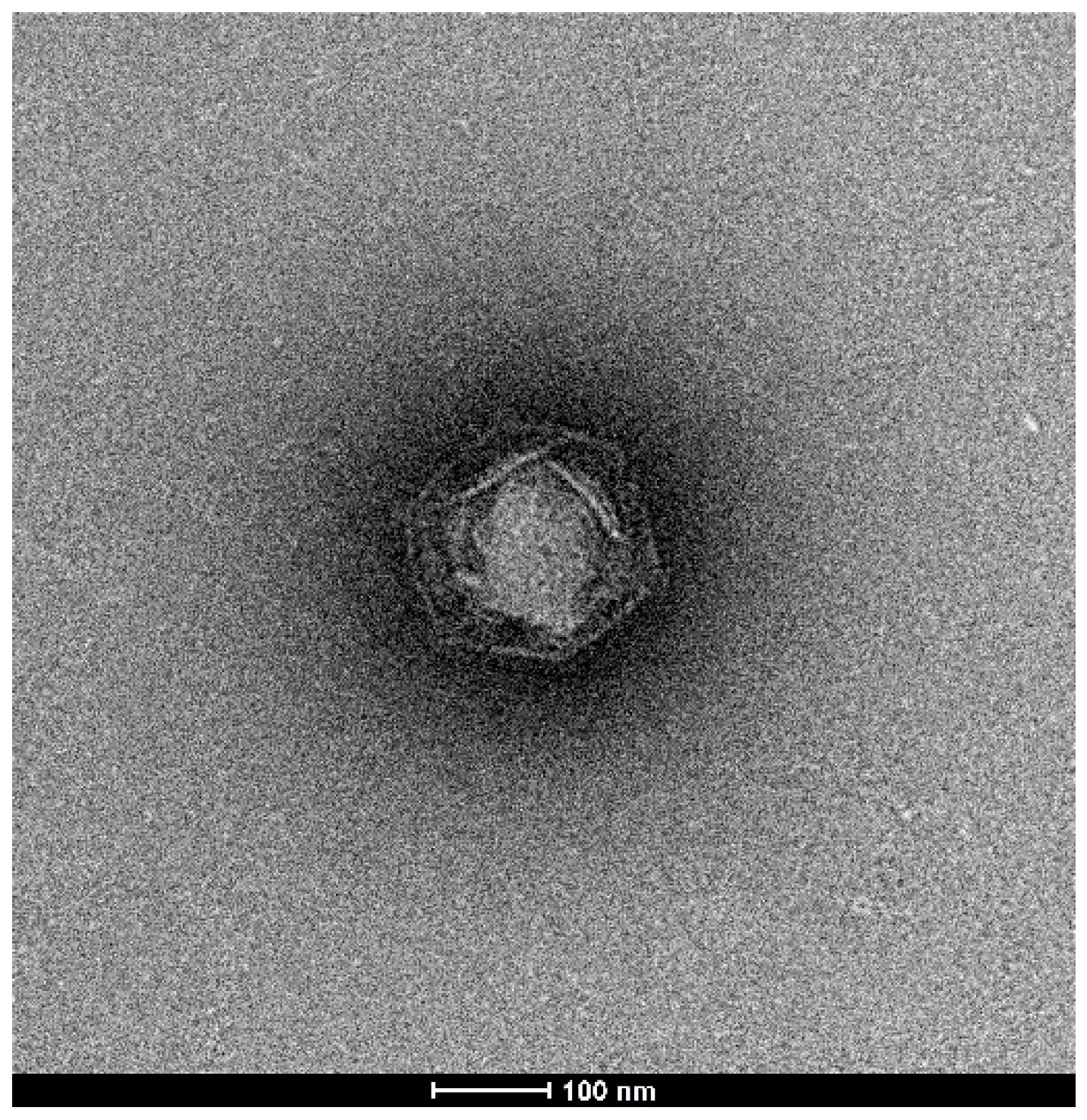
3.1. Isolation and Electron Microscopy
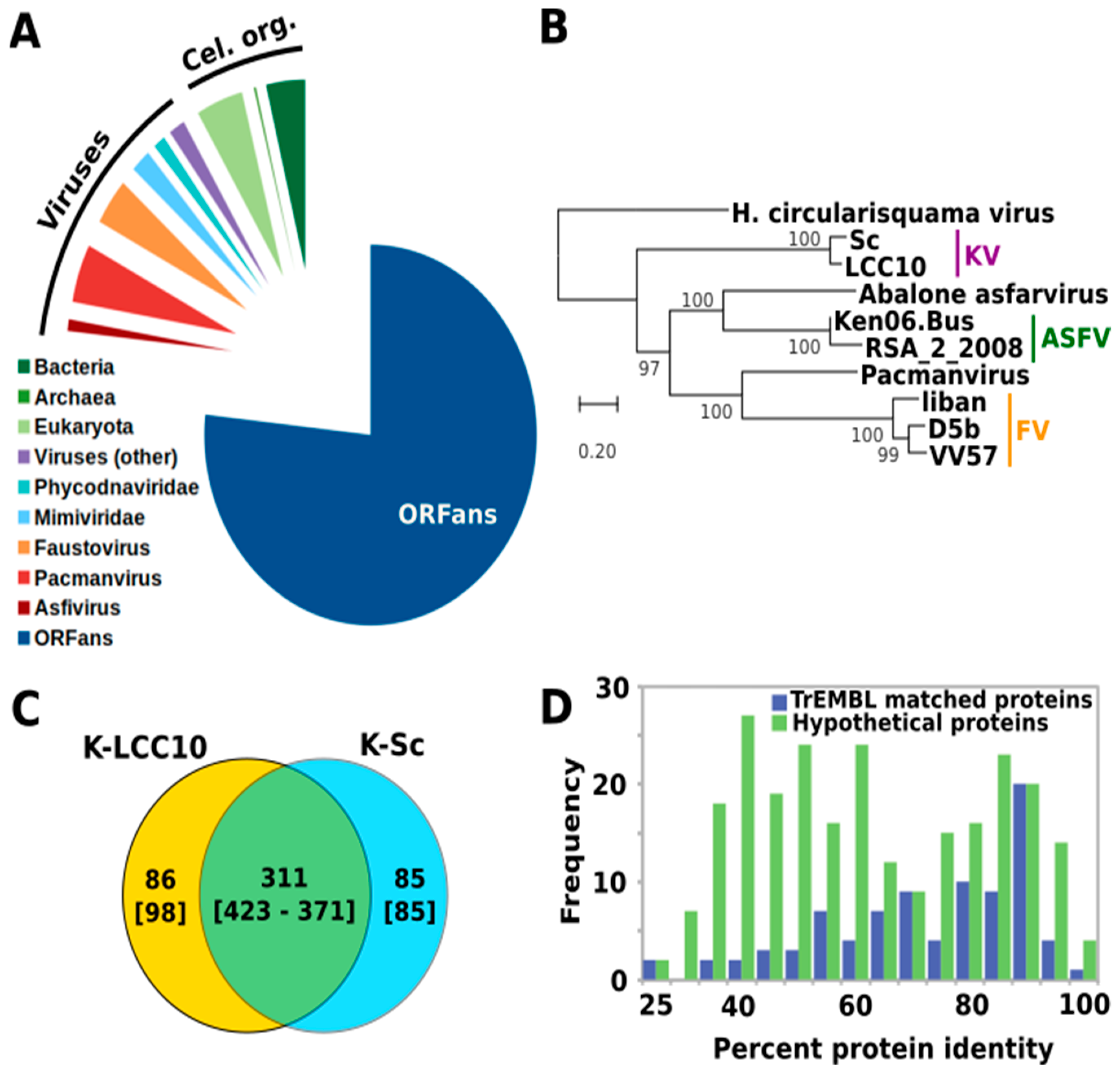
3.2. KV Genome Content
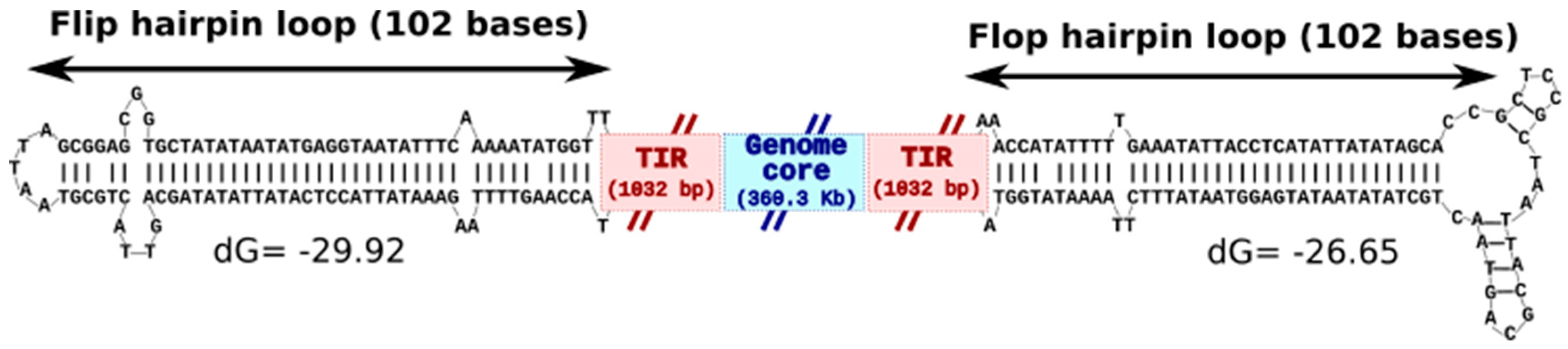
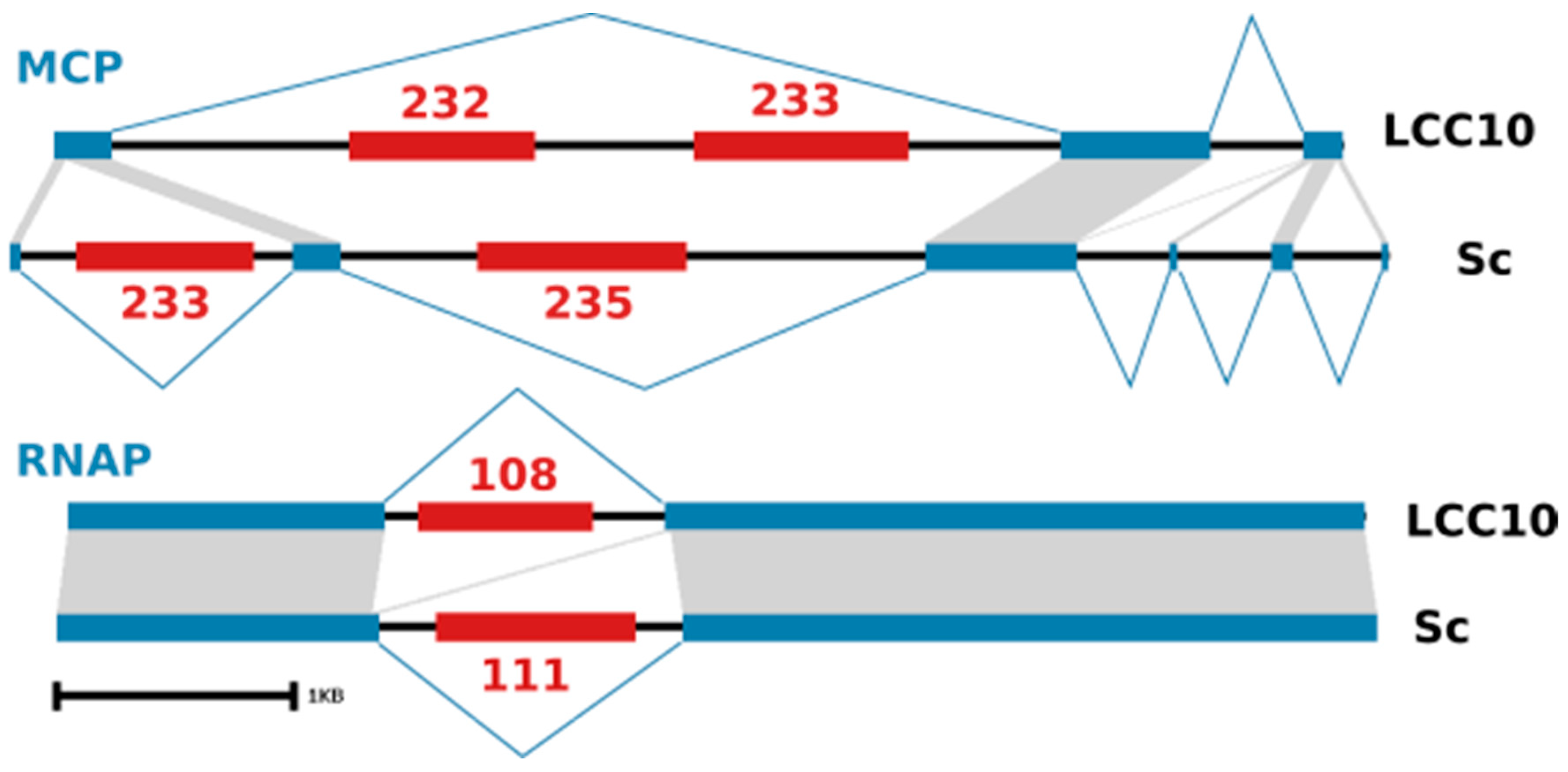
3.3. KV Genome Organization
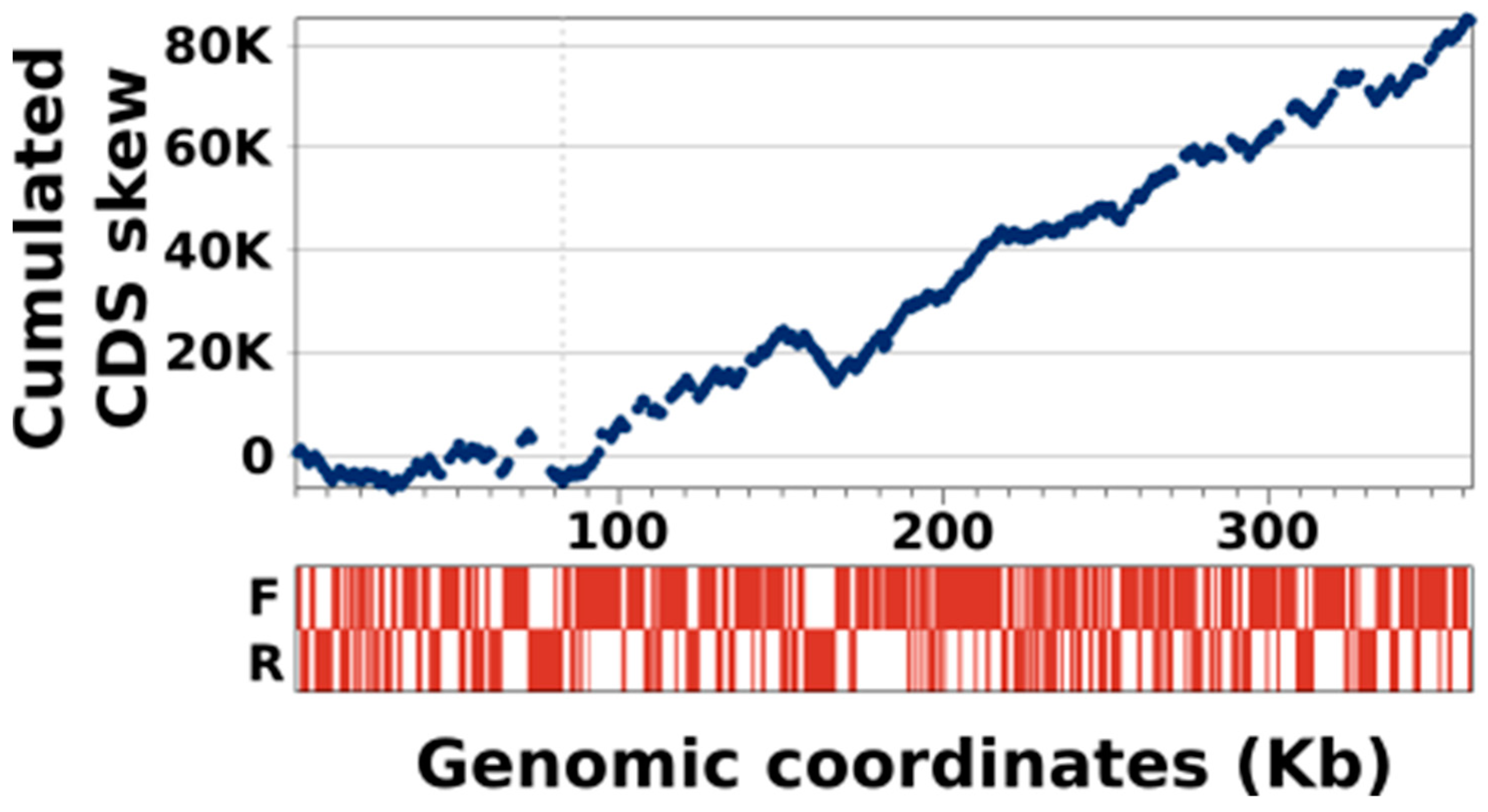
3.4. KVs Genome Has a Unique Gene Strand Bias
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- La Scola, B. A Giant Virus in Amoebae. Science 2003, 299, 2033. [Google Scholar] [CrossRef] [PubMed]
- Walker, P.J.; Siddell, S.G.; Lefkowitz, E.J.; Mushegian, A.R.; Dempsey, D.M.; Dutilh, B.E.; Harrach, B.; Harrison, R.L.; Hendrickson, R.C.; Junglen, S.; et al. Changes to virus taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2019). Arch. Virol. 2019, 164, 2417–2429. [Google Scholar] [CrossRef] [PubMed]
- Reteno, D.G.; Benamar, S.; Khalil, J.B.; Andreani, J.; Armstrong, N.; Klose, T.; Rossmann, M.; Colson, P.; Raoult, D.; La Scola, B. Faustovirus, an Asfarvirus-Related New Lineage of Giant Viruses Infecting Amoebae. J. Virol. 2015, 89, 6585–6594. [Google Scholar] [CrossRef] [PubMed]
- Benamar, S.; Reteno, D.G.I.; Bandaly, V.; Labas, N.; Raoult, D.; La Scola, B. Faustoviruses: Comparative Genomics of New Megavirales Family Members. Front. Microbiol. 2016, 7, 3. [Google Scholar] [CrossRef] [PubMed]
- Khalil, J.Y.B.; Andreani, J.; Raoult, D.; La Scola, B. A Rapid Strategy for the Isolation of New Faustoviruses from Environmental Samples Using Vermamoeba vermiformis. J. Vis. Exp. 2016, 54104. [Google Scholar] [CrossRef] [PubMed]
- Louazani, A.C.; Andreani, J.; Ouarhache, M.; Aherfi, S.; Baptiste, E.; Levasseur, A.; La Scola, B. Genome Sequences of New Faustovirus Strains ST1 and LC9, Isolated from the South of France. Genome Announc. 2017, 5. [Google Scholar] [CrossRef]
- Geballa-Koukoulas, K.; Boudjemaa, H.; Andreani, J.; La Scola, B.; Blanc, G. Comparative Genomics Unveils Regionalized Evolution of the Faustovirus Genomes. Viruses 2020, 12, 577. [Google Scholar] [CrossRef]
- Bajrai, L.H.; Benamar, S.; Azhar, E.I.; Robert, C.; Levasseur, A.; Raoult, D.; La Scola, B. Kaumoebavirus, a New Virus That Clusters with Faustoviruses and Asfarviridae. Viruses 2016, 8, 278. [Google Scholar] [CrossRef]
- Andreani, J.; Khalil, J.Y.B.; Baptiste, E.; Hasni, I.; Michelle, C.; Raoult, D.; Levasseur, A.; La Scola, B. Orpheovirus IHUMI-LCC2: A New Virus among the Giant Viruses. Front. Microbiol. 2018, 8, 2643. [Google Scholar] [CrossRef]
- Abrahão, J.; Silva, L.; Silva, L.S.; Khalil, J.Y.B.; Rodrigues, R.; Arantes, T.; Assis, F.; Boratto, P.; Andrade, M.; Kroon, E.G.; et al. Tailed giant Tupanvirus possesses the most complete translational apparatus of the known virosphere. Nat. Commun. 2018, 9, 1–12. [Google Scholar] [CrossRef]
- Bajrai, L.H.; Mougari, S.; Andreani, J.; Baptiste, E.; Delerce, J.; Raoult, D.; Azhar, E.I.; La Scola, B.; Levasseur, A. Isolation of Yasminevirus, the First Member of Klosneuvirinae Isolated in Coculture with Vermamoeba vermiformis, Demonstrates an Extended Arsenal of Translational Apparatus Components. J. Virol. 2019, 94. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Yutin, N. Evolution of the Large Nucleocytoplasmic DNA Viruses of Eukaryotes and Convergent Origins of Viral Gigantism. Adv. Virus Res. 2019, 103, 167–202. [Google Scholar] [CrossRef] [PubMed]
- Alonso, C.; Borca, M.V.; Dixon, L.; Revilla, Y.; Rodriguez, F.; Escribano, J.M. ICTV Report Consortium ICTV Virus Taxonomy Profile: Asfarviridae. J. Gen. Virol. 2018, 99, 613–614. [Google Scholar] [CrossRef] [PubMed]
- Dixon, L.K.; Nash, R.; Hawes, P.C.; Netherton, C.L. African Swine Fever Virus. In Reference Module in Life Sciences; Elsevier BV: Amsterdam, The Netherlands, 2019; p. 9780128096338209000. [Google Scholar]
- Klose, T.; Reteno, D.G.; Benamar, S.; Hollerbach, A.; Colson, P.; La Scola, B.; Rossmann, M.G. Structure of faustovirus, a large dsDNA virus. Proc. Natl. Acad. Sci. USA 2016, 113, 6206–6211. [Google Scholar] [CrossRef] [PubMed]
- Criscuolo, A.; Brisse, S. AlienTrimmer: A tool to quickly and accurately trim off multiple short contaminant sequences from high-throughput sequencing reads. Genomics 2013, 102, 500–506. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef]
- Baroudy, B.M.; Venkatesan, S.; Moss, B. Incompletely base-paired flip-flop terminal loops link the two DNA strands of the vaccinia virus genome into one uninterrupted polynucleotide chain. Cell 1982, 28, 315–324. [Google Scholar] [CrossRef]
- Besemer, J.; Lomsadze, A.; Borodovsky, M. GeneMarkS: A self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001, 29, 2607–2618. [Google Scholar] [CrossRef]
- Laslett, D.; Canback, B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004, 32, 11–16. [Google Scholar] [CrossRef]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef] [PubMed]
- Emms, D.M.; Kelly, S. OrthoFinder: Solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015, 16, 157. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Bateman, A.; Finn, R.D. Predicting active site residue annotations in the Pfam database. BMC Bioinform. 2007, 8, 298. [Google Scholar] [CrossRef] [PubMed]
- Zdobnov, E.M.; Apweiler, R. InterProScan—An integration platform for the signature-recognition methods in InterPro. Bioinformatics 2001, 17, 847–848. [Google Scholar] [CrossRef] [PubMed]
- Kuraku, S.; Zmasek, C.M.; Nishimura, O.; Katoh, K. aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Res. 2013, 41, W22–W28. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Zhang, Y.; Strasser, P.; Grabherr, R.; Van Etten, J.L. Hairpin Loop Structure at the Termini of the Chlorella Virus PBCV-1 Genome. Virology 1994, 202, 1079–1082. [Google Scholar] [CrossRef]
- González, A.; Talavera, A.; Almendral, J.M.; Viñuela, E. Hairpin loop structure of African swine fever virus DNA. Nucleic Acids Res. 1986, 14, 6835–6844. [Google Scholar] [CrossRef]
- Du, S.; Traktman, P. Vaccinia virus DNA replication: Two hundred base pairs of telomeric sequence confer optimal replication efficiency on minichromosome templates. Proc. Natl. Acad. Sci. USA 1996, 93, 9693–9698. [Google Scholar] [CrossRef]
- Stuart, D.; Graham, K.; Schreiber, M.; Macaulay, C.; McFadden, G. The target DNA sequence for resolution of poxvirus replicative intermediates is an active late promoter. J. Virol. 1991, 65, 61–70. [Google Scholar] [CrossRef]
- Fan, H.; Sakuraba, K.; Komuro, A.; Kato, S.; Harada, F.; Hirose, Y. PCIF1, a novel human WW domain-containing protein, interacts with the phosphorylated RNA polymerase II. Biochem. Biophys. Res. Commun. 2003, 301, 378–385. [Google Scholar] [CrossRef]
- De Carvalho, M.O.; Ferreira, H.B. Quantitative determination of gene strand bias in prokaryotic genomes. Genomics 2007, 90, 733–740. [Google Scholar] [CrossRef] [PubMed]
- Kano-Sueoka, T.; Lobry, J.R.; Sueoka, N. Intra-strand biases in bacteriophage T4 genome. Gene 1999, 238, 59–64. [Google Scholar] [CrossRef]
- Kreuzer, K.N.; Brister, J.R. Initiation of bacteriophage T4 DNA replication and replication fork dynamics: A review in the Virology Journal series on bacteriophage T4 and its relatives. Virol. J. 2010, 7, 358. [Google Scholar] [CrossRef] [PubMed]
- Baroudy, B.; Venkatesan, S.; Moss, B. Structure and Replication of Vaccinia Virus Telomeres. Cold Spring Harb. Symp. Quant. Biol. 1983, 47, 723–729. [Google Scholar] [CrossRef]
- Moyer, R.W.; Graves, R.L. The mechanism of cytoplasmic orthopoxvirus DNA replication. Cell 1981, 27, 391–401. [Google Scholar] [CrossRef]
- Senkevich, T.G.; Bruno, D.; Martens, C.; Porcella, S.F.; Wolf, Y.I.; Moss, B. Mapping vaccinia virus DNA replication origins at nucleotide level by deep sequencing. Proc. Natl. Acad. Sci. USA 2015, 112, 10908–10913. [Google Scholar] [CrossRef]
- Moss, B. Poxvirus DNA Replication. Cold Spring Harb. Perspect. Biol. 2013, 5, a010199. [Google Scholar] [CrossRef]

| Strain | LCC10 | Sc |
|---|---|---|
| Genome length | 362,586 | 350,731 |
| GC% | 43.1 | 43.7 |
| TIRs (bp) | 1032 | 1407 |
| Predicted Genes (Gene families) | 507 | 456 |
| Hypothetical proteins | 385 (76%) | 304 (67%) |
| Paralogs (Gene families) | 183 (44) | 116 (35) |
| Strain-specific genes | 98 | 82 |
| Species | Accession Number | Gene Number | Strand Excess (a) | p-Val (b) | CDS Cumul. Length | Genome CDS Skew | p-Val (c) |
|---|---|---|---|---|---|---|---|
| Kaumoebavirus LCC10 (KV-LCC10) | MT334784 | 507 | 11.3% | 2 × 10−7 | 342,156 | 26% | 9 × 10−5 |
| w/o hypothetical protein genes | 120 | 11.7% | 7 × 10−3 | 152,698 | 34% | 2 × 10−3 | |
| w/o tandemly duplicated genes | 463 | 10% | 9 × 10−6 | 316,152 | 24% | 3 × 10−4 | |
| Kaumoebavirus Sc (KV-Sc) | KX552040 | 429 | 11.5% | 1 × 10−6 | 280,782 | 29% | 2 × 10−5 |
| African swine fever virus (ASFV) | ASU18466 | 152 | −2% | 0.34 | 150,903 | −15% | 0.19 |
| Faustovirus (FV) | MN830295 | 503 | 4% | 0.03 | 439,093 | 9% | 0.19 |
| Pacmanvirus (PV) | LT706986 | 465 | 4% | 0.03 | 353,586 | 5% | 0.29 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Geballa-Koukoulas, K.; Andreani, J.; La Scola, B.; Blanc, G. The Kaumoebavirus LCC10 Genome Reveals a Unique Gene Strand Bias among “Extended Asfarviridae”. Viruses 2021, 13, 148. https://doi.org/10.3390/v13020148
Geballa-Koukoulas K, Andreani J, La Scola B, Blanc G. The Kaumoebavirus LCC10 Genome Reveals a Unique Gene Strand Bias among “Extended Asfarviridae”. Viruses. 2021; 13(2):148. https://doi.org/10.3390/v13020148
Chicago/Turabian StyleGeballa-Koukoulas, Khalil, Julien Andreani, Bernard La Scola, and Guillaume Blanc. 2021. "The Kaumoebavirus LCC10 Genome Reveals a Unique Gene Strand Bias among “Extended Asfarviridae”" Viruses 13, no. 2: 148. https://doi.org/10.3390/v13020148
APA StyleGeballa-Koukoulas, K., Andreani, J., La Scola, B., & Blanc, G. (2021). The Kaumoebavirus LCC10 Genome Reveals a Unique Gene Strand Bias among “Extended Asfarviridae”. Viruses, 13(2), 148. https://doi.org/10.3390/v13020148






