Abstract
The overarching structure of the type I interferon (IFN) system is conserved across vertebrates. However, the variable numbers of whole genome duplication events during fish evolution offer opportunities for the expansion, diversification, and new functionalization of the genes that are involved in antiviral immunity. In this review, we examine how fish models provide new insights about the implication of virus-driven inflammation in immunity and hematopoiesis. Mechanisms that have been discovered in fish, such as the strong adjuvant effect of type I IFN that is used with DNA vaccination, constitute good models to understand how virus-induced inflammatory mechanisms can interfere with adaptive responses. We also comment on new discoveries regarding the role of pathogen-induced inflammation in the development and guidance of hematopoietic stem cells in zebrafish. These findings raise issues about the potential interferences of viral infections with the establishment of the immune system. Finally, the recent development of genome editing provides new opportunities to dissect the roles of the key players involved in the antiviral response in fish, hence enhancing the power of comparative approaches.
Keywords:
fish; type I interferon; vaccine; adjuvant; hematopoiesis; genome editing; Crispr; innate immunity 1. Introduction
Most of the basic features of the immunity processes are conserved across vertebrates, but the anatomical and physiological fundamental differences between fish and tetrapods make comparative analysis interesting, as it reveals many specific adaptations and original mechanisms.
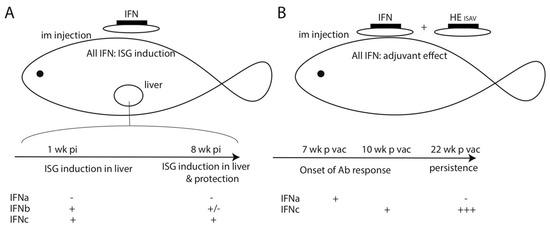
Type I and type II interferons (IFN) have been described in fish [1,2,3]. While structural studies have definitely demonstrated that fish type I IFN encoded by intron-containing genes are the true counterparts of mammalian type I IFN [4], these cytokines have diversified independently during fish and tetrapod evolution [5]. Type I IFN is highly variable across fish species, with zebrafish laboratory strains having only four genes coding type I IFN (named IFNφ) and salmonids having over 30 type I IFN genes [6]. These IFN are classified into two groups, depending on the number of cysteine bridges present in the protein, and further divided into subtypes based on sequence similarity [7]. For example, in Atlantic salmon, six subtypes have been distinguished (a–e): IFNa, d, and e have two cysteines and belong to group 1, while IFNb, c, and f have four cysteines and belong to group 2 [6]. Even in fish species in which type I IFN are relatively well described such as Atlantic salmon, rainbow trout, and zebrafish, the specific functions of genes or subtypes remain elusive. In Atlantic salmon, Robertsen et al. have shown that IFNa1 (group 1) and IFNc (group 2) had strong antiviral activity against IPNV in vitro, while IFNb (group 2) was less potent, and IFNd (group 1) was not active at all [8]. While type I IFN receptors have been identified in zebrafish and salmonids, the detailed structure of signaling pathways within the fish IFN remain poorly characterized, and is mainly inferred from what is known in mammals.
Most functional studies of IFN antiviral activity have been performed in vitro using fish cell lines, and only a few studies have compared different type I IFN in vivo. In one example of such studies performed in zebrafish, Lopez-Munoz reported that while IFNs from both group 1 (IFNφ1) and group 2 (IFNφ2 and φ3) have antiviral activity in vivo, only IFNφ1 provides efficient protection against the fish bacterial pathogen Streptococcus iniae. This study also suggested different kinetics of induction of effector IFN-stimulated genes (ISG) by group 1 and 2 IFNs [9]. The in vivo characterization of type I interferon activity constitutes a major challenge to better understanding fish/virus interactions, identifying IFN producing cell subtypes, and obtaining an integrated view of the fish IFN contribution to antiviral defenses. In particular, the work performed on cell lines has completely overlooked the role of IFN in the interaction between innate and adaptive antiviral responses, as well as all its potential functions in development, cell differentiation, and regulation of the cell cycle. A lot of in-depth research is still required to fully understand the regulation mechanisms and the functional properties of these type I IFNs across the great diversity of teleost fish.
The antiviral functions of fish type I IFNs and their mechanisms have been recently reviewed [5,10,11,12,13]. In this review, we focus on the new developments of three different topics that will likely be important in the fields of fish interferons in the coming years. We first discuss the role of type I IFN activity in the development of protection against viral diseases provided by DNA vaccination in fish. In a second section, we examine the role of sterile and pathogen-induced inflammation on the regulation of hematopoietic stem cell differentiation, and the potential impact of viruses on the maturation of immunity. We discuss the available data in zebrafish and the perspectives offered by this model. Finally, we present the recent development in genome editing, especially those based on CRISPR/cas9 technologies and their contribution to the understanding of antiviral mechanisms in fish.
3. Inflammation and Hematopoietic Stem Cell Differentiation
Besides its antiviral and antiproliferative properties, type I IFN is also an important factor for immune cell differentiation [55]. IFN-dependent mechanisms of leukocyte differentiation may be activated by the inflammatory context of innate responses to infections. However, the maintenance of inflammation leads to tissue damage, and sometimes evolves to chronic inflammatory diseases, which also affect immune cell dynamics [56]. IFN impact on cell differentiation has also been described in sterile conditions, as part of the physiological differentiation program. The description of mechanisms sustaining hematopoiesis regulation by pro-inflammatory signaling is an active field of research, but many open questions remain, especially regarding the resilience mechanisms after infections. In this regard, the zebrafish embryo constitutes an interesting vertebrate model, which is based on the high similarity between the zebrafish and human antiviral innate immune response [57] and conserved hematopoiesis processes [58]. As in mammals, blood development occurs through primitive and definitive waves in zebrafish [59,60], which relies on conserved genetic pathways, even though hematopoietic organs are partly different from those of mammals [61,62]. Embryonic primitive hematopoiesis gives rise to primitive erythrocytes and primitive myeloid cells (neutrophils and macrophages) in the lateral mesoderm, which is the equivalent of the mammalian yolk sac before 24 hours post-fertilization (hpf). From 26 hpf, hematopoietic stem cells (HSC) emerge from the transition of dorsal aorta endothelium to hematopoietic cells during the definitive wave [63,64]. HSC then migrate to colonize caudal hematopoietic tissue, which is the equivalent of the mouse fetal liver and placenta, to give rise to erythrocytes, thrombocytes, and myeloid cells. Starting at 48 hpf, HSC colonize the pronephric marrow, which is the equivalent of mammalian bone marrow, and differentiate in all of the blood cell lineages: erythrocytes, thrombocytes myeloid cells, and lymphoid cells. From 54 hpf, lymphoid progenitors reach the thymus to differentiate in lymphocyte T cells, which colonize the pronephros and the rest of the body [65].
Advantages of the zebrafish model (optical transparency, genome editing tools, and the transgenic animals library) have been largely exploited to describe in real time the molecular and cellular processes occurring during hematopoiesis, combining microscopy on transgenic lines expressing fluorescent reporters specifically in HSC (runx1, cmyb), vascular endothelium (kdrl), and immune cell populations (mpeg, mpx) [66,67]. Gain and loss of function experiments based on transgenesis, mRNA, or morpholinos injection [68] have been instrumental to identifying the genetic factors involved in the emergence of HSC from the hemogenic endothelium in the dorsal aorta, as well as HSC specification, proliferation, and/or differentiation. Regulatory mechanisms sustaining these processes have been studied in comparative gene expression analyses between fish and mammals HSC [69], using zebrafish to trigger the functional analysis of mammalian HSC regulator candidates [68,70,71] or even for the screening of biological compounds enhancing HSC production [72,73]. Importantly, these studies in zebrafish contributed to demonstrating the important role of pro-inflammatory signaling in hematopoiesis during normal embryogenesis or in the context of pathogen infections [56,74,75]. In this context, the zebrafish embryo is a valuable model to investigate the impact of infections or IFN on hematopoiesis.
3.1. Inflammation, IFNs, and Zebrafish HSC Differentiation in Aseptic Conditions
In homeostatic conditions, the emergence of HSC from the hemogenic endothelium depends on the exposure to inflammatory cytokines and the modulation of conserved transcription factors (scl, lmo2, gata1, ikaros, and spi1).
In zebrafish, primitive myeloid cells (neutrophils and macrophages) have been identified as major sources of inflammatory cytokines such as TNFα. Primitive myeloid cells are believed to play a major role by producing inflammatory cytokines, thus activating signaling cascade in both endothelial and hemogenic endothelial cells [74] or through direct interactions, as demonstrated by the involvement of macrophages in breaking down the extracellular matrix, allowing the migration of HSC migration and the colonization of hematopoietic organs [76]. TNFα induces jag1a-Notch and NFκB signaling pathways, which are involved in the emergence of HSC [77,78]. Similar to TNFα, type I [78] d type II IFNs [78,79], as well as interleukin IL1β [78] signaling, trigger the emergence of HSC. IFN-γ signaling also acts on the transition from endothelial to HSC, which is a signal transducer and activator of transcription (stat3)-dependent process involving Notch signaling and blood flow [79]. Beside cytokines, miRNA have also been identified as regulators of hematopoiesis. Among the 639 mature miRNA described in zebrafish in the miRBase, miR-126, miR-144, miR-451, and miR-142-3p were respectively involved in erythroid/megakaryocytic lineage, erythropoiesis, and the specification and differentiation of HSC ([80]). The impact of miRNAs on type I or type II IFN responses is still an expanding field.
3.2. Impact of Infections and Innate Responses on HSC Differentiation
During infections, pathogen detection rapidly triggers inflammatory responses and hematopoiesis activation, through several non exclusive mechanisms: (1) PAMP sensing by immune cells in hematopoietic sites (2) PAMP sensing in periphery leading proinflammatory cytokines, (3) direct infection of hematopoietic cells and (4) activation through hematopoietic microenvironment [56,74].
These mechanisms were first characterized in the context of bacterial infections in mammalian models. Recruited neutrophils ingest and kill invading bacteria, promoting inflammatory mediator synthesis and secondary recruitment macrophages and professional Antigen Presenting cells (APCs) at the site of infection. The hematopoietic system responds to the increased demand of neutrophils and myeloid cells by switching from steady-state to emergency granulopoiesis [81]. Under Pathogen Associated Molecular Pattern (PAMP) recognition, peripheral infected cells produced IL1β, TNFα, and LPS inflammatory molecules, which induced NFκB and CEBP signaling pathways and increased circulating G-CSF. This directly stimulates the proliferation of granulocytic progenitor cells as receptors for G-CSF, which is mainly expressed in granulocytic precursors and mature neutrophils [82]. The key role of G-CSF in the resolution of bacterial infections has been shown in various mouse models of bacterial infections [82] including L. monocytogenes and Pseudomonas aeruginosa. Thus, granulopoïesis increases at the expense of lymphoid progenitor cells [83] through the induction of HSPC (hematopoietic stem and progenitor cell) compartment reactivation [84]. Hall et al. [80] recapitulated this “demand-driven” granulopoïesis model in a zebrafish model of infection by Salmonella. Following intracerebral injection, the depletion of neutrophils at 1 day post-infection (dpi) is followed by emergency granulopoïesis by 2 dpi at the expense of lymphoid cell progenitors. This was associated with an increased number of HSC at 1 dpi in a detailed CEBP-driven, Nos2a-dependent manner [80]. In a recent work, Willis et al. also demonstrated HSC-driven granulopoïesis in response to Shigella flexneri IC injection in zebrafish larvae [85]. Hematopoietic Stem and Progenitor Cell (HSPC) cell death in the context of uncontrolled bacterial burden, and a strong increase of inflammatory cytokine expression (TNFα, IFN 1–2, and IL1) has also been reported [86]. Altogether, these data have established that bacterial infections can increase hematopoiesis via cytokine signaling.
The impact of viral infections on hematopoiesis is less documented, either via direct infection or through IFN-dependent activation. Viral invasions in host organisms face distinct defense mechanisms compared to bacteria, suggesting that their impact on hematopoiesis may rely on different mechanisms. Virus recognition by cellular sensors triggers well-defined signaling pathways and the transcription of pro-inflammatory cytokines, including type I IFN. Then, type I IFN response induced the synthesis of many ISG that count antiviral effector proteins and regulators of immunity. This local and systemic inflammation triggers adaptive immunity and other mechanisms of antiviral defense. Neutrophils, monocytes, and NK cells are recruited for the clearance of deadly infected cells, while lymphocytes contribute to controlling infection in adaptive immunity.
In human, HSPC express Pattern Recognition Receptors (PRR) receptors such as virus-specific Toll-like receptors [87,88] and NOD-like receptors [89]. Their activation leads to HSC cycling and HSPC differentiation, which is preferentially toward mature myeloid cells [87,88,90]. In addition, the direct infection of stem cells, progenitor cells, or stromal cells of a hematopoietic microenvironment have been reported, affecting hematopoiesis via cell death induction or the establishment of viral latency [56]. Systemic acute VSV infection triggers IFN response in bone marrow and HSC, which might evolve in HSC cell cycle entry upon substantial IFN I response. A similar observation was reported in an IFNAR knock-out mouse model with uncontrolled virus replication, which was in favor of an IFNAR-independent HSC activation [91]. Acute infection with murine cytomegalovirus (MCMV) also increased type I IFN expression in HSC and bone marrow (as compared to low systemic levels), and activated HSC through IFNAR-dependent and independent mechanisms. Chronic infection with MCMV was associated with the long-term impairment of long-term HSC [91]. In keeping with this, the chronic infection of human bone marrow progenitor cells has been reported for Human Herpes Virus (HHV)-6, which may be transmitted to differentiated cells, thus using HSC as a source of virus dissemination [92,93]. This was also described for the HHV-8 virus targeting CD34+ hematopoietic stem cells, HHV-7, and HHV-8 infections, leading to the alteration of survival and differentiation of progenitors [92]. However, stem cells have been found to be highly resistant to viral infection across species, due to the intrinsic expression of many ISGs [94]. An additional level of hematopoiesis regulation relies on miRNAs, which are expressed in hematopoietic lineages.
Thus, HSPC fate can be modified by IFN response directly or indirectly via changes in tissue microenvironment stimulation. Several studies have reported the impacts of type I IFN on HSC proliferation, possibly leading to exhaustion through the reduction of quiescent HSCs [95]. In fact, while the chronic activation of the type I IFN in HSCs impairs their function, acute IFNα treatment activates dormant HSC proliferation. However, Poly I:C treatment mediated the return of HSC to quiescence, thus preventing apoptosis and DNA damage [96,97]. The activation of the hematopoietic system by IFN, which is induced after viral infection, has not been extensively studied, although it might be related to several bone marrow pathologies such as aplastic anemia, pancytopenia, hemophagocytic lymphohistiocytosis, lymphoproliferative disorders, and malignancies.
In fish, the modulation of hematopoiesis by viral infection is not well understood either. Viruses targeting hematopoietic precursors have been reported in zebrafish, including Infectious Hematopoietic Necrosis Virus (IHNV) and Infectious Pancreatic Necrosis Virus (IPNV). Infections in adults showed virus persistence over 10 days and cytotoxicity in hematopoietic precursors in the pronephros (the functional equivalent of mammalian bone marrow) with the decrease of terminally differentiated red blood cells, and recovery involving an expansion of proerythroblast [98]. A detailed description of the innate response to IHNV in zebrafish embryos showed that rapid ongoing infection targets endothelial cells. The infection led to 100% mortality between 65–96 Hours post-infection (hpi), which is probably due to late and inefficient host immune response activation; the induction of type I IFN was late from 30 to 48 hpi [99]. In contrast, Chikungunya Virus (CHIKV) replication targets many cell subtypes. The acute phase of infection was associated with an increase in the neutrophils that produce type I IFN, which is critical for survival and probably has an impact on hematopoiesis. Interestingly, this increase is NOS2a-dependent, which is similar to the granulopoiesis induced by Salmonella in [74]. The virus also gains access to the brain, where it persists for longer [100]. Infection of the zebrafish embryo with IHNV or CHIKV leads to different innate responses [101], which may constitute relevant models to evaluate the impact of viral-induced inflammation on hematopoiesis and the priming of adaptive immune responses.
In a model of zebrafish hematopoietic transplant, repeated intra-peritoneal ip injections of the PolyI:C in donors before transplant led to a faster and more efficient colonization of recipient kidney marrow by HSPCs; however, the difference with controls was not significant 28 d post-transplant, and the survival rate was not modified. PolyI:C injections of recipients before transplant did not affect the colonization. While competitive transplants and imaging explorations will be useful to obtain further insights, as proposed by the author, this work shows that valuable in vivo data about the impact of inflammation on HSPC can be produced in zebrafish embryos ([102]; see also https://spiral.imperial.ac.uk/handle/10044/1/55871).
5. Conclusions
Inflammation induced by type I IFN, or independently by viral infection, has critical impacts on the activation of adaptive immunity and hematopoiesis. It also leads to pathologies via multiple signaling pathways that become activated, comprising pro-inflammatory cytokines and fever, NF κB, ubiquitination, proteasome, apoptosis, etc. We believe that fish provide interesting alternative models that can present new insights about these mechanisms, which remain poorly understood. Since it has been demonstrated that the activation of the TLR3 pathway can even facilitate nuclear reprogramming in induced pluripotent stem cells [121], innate antiviral mechanisms appear increasingly important for the control of cell differentiation. The fast development of gene editing technologies that can be used in virtually all species will provide fish immunologists and virologists with exquisite tools to dissect these mechanisms. Besides the interest of highly conserved—and hence likely essential—mechanisms, such studies may also have important impacts through vaccination in aquaculture. Importantly, zebrafish have recently emerged as a promising model for drug screening, and genome editing is likely to provide useful models in this field in the coming years.
Author Contributions
C.L., P.B., and B.C. drafted and edited the manuscript.
Funding
This research was funded by institutional grants from the Institut National de la Recherche Agronomique and marine Scotland, and by the grants ANR-16-CE20-0002-01 FishRNAVax and BBSRC/NERC (NE/P010946/1).
Acknowledgments
The authors thank Jean-Pierre Levraud for discussions and for his comments on the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Robertsen, B.; Bergan, V.; Røkenes, T.; Larsen, R.; Albuquerque, A. Atlantic salmon interferon genes: Cloning, sequence analysis, expression, and biological activity. J. Interferon Cytokine Res. 2003, 23, 601–612. [Google Scholar] [CrossRef]
- Altmann, S.M.; Mellon, M.T.; Distel, D.L.; Kim, C.H. Molecular and functional analysis of an interferon gene from the zebrafish, Danio rerio. J. Virol. 2003, 77, 1992–2002. [Google Scholar] [CrossRef]
- Lutfalla, G.; Roest Crollius, H.; Stange-Thomann, N.; Jaillon, O.; Mogensen, K.; Monneron, D. Comparative genomic analysis reveals independent expansion of a lineage-specific gene family in vertebrates: The class II cytokine receptors and their ligands in mammals and fish. BMC Genom. 2003, 4, 29. [Google Scholar] [CrossRef]
- Hamming, O.J.; Lutfalla, G.; Levraud, J.-P.; Hartmann, R. Crystal structure of Zebrafish interferons I and II reveals conservation of type I interferon structure in vertebrates. J. Virol. 2011, 85, 8181–8187. [Google Scholar] [CrossRef] [PubMed]
- Boudinot, P.; Langevin, C.; Secombes, C.J.; Levraud, J.P. The Peculiar Characteristics of Fish Type I Interferons. Viruses 2016, 8, 298. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Gorgoglione, B.; Taylor, N.G.; Summathed, T.; Lee, P.T.; Panigrahi, A.; Genet, C.; Chen, Y.M.; Chen, T.Y.; Ul Hassan, M.; et al. Salmonids have an extraordinary complex type I IFN system: Characterization of the IFN locus in rainbow trout oncorhynchus mykiss reveals two novel IFN subgroups. J. Immunol. 2014, 193, 2273–2286. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Tafalla, C.; Truckle, J.; Secombes, C.J. Identification of a second group of type I IFNs in fish sheds light on IFN evolution in vertebrates. J. Immunol. 2007, 179, 3859–3871. [Google Scholar] [CrossRef] [PubMed]
- Svingerud, T.; Solstad, T.; Sun, B.; Nyrud, M.L.J.; Kileng, Ø.; Greiner-Tollersrud, L.; Robertsen, B. Atlantic salmon type I IFN subtypes show differences in antiviral activity and cell-dependent expression: Evidence for high IFNb/IFNc-producing cells in fish lymphoid tissues. J. Immunol. 2012, 189, 5912–5923. [Google Scholar] [CrossRef]
- López-Muñoz, A.; Roca, F.J.; Sepulcre, M.P.; Meseguer, J.; Mulero, V. Zebrafish larvae are unable to mount a protective antiviral response against waterborne infection by spring viremia of carp virus. Dev. Comp. Immunol. 2010, 34, 546–552. [Google Scholar] [CrossRef]
- Chen, S.N.; Zou, P.F.; Nie, P. Retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs) in fish: Current knowledge and future perspectives. Immunology 2017, 151, 16–25. [Google Scholar] [CrossRef]
- Robertsen, B. The role of type I interferons in innate and adaptive immunity against viruses in Atlantic salmon. Dev. Comp. Immunol. 2018, 80, 41–52. [Google Scholar] [CrossRef]
- Secombes, C.J.; Zou, J. Evolution of Interferons and Interferon Receptors. Front. Immunol. 2017, 8, 209. [Google Scholar] [CrossRef] [PubMed]
- Poynter, S.J.; DeWitte-Orr, S.J. Fish interferon-stimulated genes: The antiviral effectors. Dev. Comp. Immunol. 2016, 65, 218–225. [Google Scholar] [CrossRef] [PubMed]
- Lorenzen, E.; Einer-Jensen, K.; Martinussen, T.; Lapatra, S.E.; Lorenzen, N. Feature dna vaccination of rainbow trout against viral hemorrhagic septicemia virus: A dose–response and time–course study. J. Aquat. Anim. Health 2000, 12, 167–180. [Google Scholar] [CrossRef]
- Evensen, O.; Leong, J.A. DNA vaccines against viral diseases of farmed fish. Fish Shellfish Immunol. 2013, 35, 1751–1758. [Google Scholar] [CrossRef]
- Collins, C.; Lorenzen, N.; Collet, B. DNA vaccination for finfish aquaculture. Fish Shellfish Immunol. 2019, 85, 106–125. [Google Scholar] [CrossRef]
- Lazarte, J.M.S.; Kim, Y.R.; Lee, J.S.; Im, S.P.; Kim, S.W.; Jung, J.W.; Kim, J.; Lee, W.J.; Jung, T.S. Enhancement of glycoprotein-based DNA vaccine for viral hemorrhagic septicemia virus (VHSV) via addition of the molecular adjuvant, DDX41. Fish Shellfish Immunol. 2017, 62, 356–365. [Google Scholar] [CrossRef]
- Kim, C.H.; Johnson, M.C.; Drennan, J.D.; Simon, B.E.; Thomann, E.; Leong, J.A. DNA vaccines encoding viral glycoproteins induce nonspecific immunity and Mx protein synthesis in fish. J. Virol. 2000, 74, 7048–7054. [Google Scholar] [CrossRef]
- McLauchlan, P.E.; Collet, B.; Ingerslev, E.; Secombes, C.J.; Lorenzen, N.; Ellis, A.E. DNA vaccination against viral haemorrhagic septicaemia (VHS) in rainbow trout: Size, dose, route of injection and duration of protection-early protection correlates with Mx expression. Fish Shellfish Immunol. 2003, 15, 39–50. [Google Scholar] [CrossRef]
- Ishikawa, H.; Ma, Z.; Barber, G.N. STING regulates intracellular DNAmediated, type I interferon-dependent innate immunity. Nature 2009, 461, 788–792. [Google Scholar] [CrossRef] [PubMed]
- Le Bon, A.; Tough, D.F. Type I interferon as a stimulus for cross-priming. Cytokine Growth Factor Rev. 2008, 19, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Le Bon, A.; Tough, D.F. Links between innate and adaptive immunity via type I interferon. Curr. Opin. Immunol. 2002, 14, 432–436. [Google Scholar] [CrossRef]
- Kulkarni, R.R.; Rasheed, M.A.; Bhaumik, S.K.; Ranjan, P.; Cao, W.; Davis, C.; Marisetti, K.; Thomas, S.; Gangappa, S.; Sambhara, S.; et al. Activation of the RIG-I pathway during influenza vaccination enhances the germinal center reaction, promotes T follicular helper cell induction, and provides a dose-sparing effect and protective immunity. J. Virol. 2014, 88, 13990–14001. [Google Scholar] [CrossRef] [PubMed]
- Carroll, E.C.; Jin, L.; Mori, A.; Munoz-Wolf, N.; Oleszycka, E.; Moran, H.B.T.; Mansouri, S.; McEntee, C.P.; Lambe, E.; Agger, E.M.; et al. The Vaccine Adjuvant Chitosan Promotes Cellular Immunity via DNA Sensor cGAS-STING-Dependent Induction of Type I Interferons. Immunity 2016, 44, 597–608. [Google Scholar] [CrossRef]
- Kastenmuller, K.; Wille-Reece, U.; Lindsay, R.W.; Trager, L.R.; Darrah, P.A.; Flynn, B.J.; Becker, M.R.; Udey, M.C.; Clausen, B.E.; Igyarto, B.Z.; et al. Protective T cell immunity in mice following protein-TLR7/8 agonist-conjugate immunization requires aggregation, type I IFN, and multiple DC subsets. J. Clin. Investig. 2011, 121, 1782–1796. [Google Scholar] [CrossRef]
- Lauksund, S.; Svingerud, T.; Bergan, V.; Robertsen, B. Atlantic salmon IPS-1 mediates induction of IFNa1 and activation of NF- k B and localizes to mitochondria. Dev. Comp. Immunol. 2009, 33, 1196–1204. [Google Scholar] [CrossRef] [PubMed]
- Biacchesi, S.; LeBerre, M.; Lamoureux, A.; Louise, Y.; Lauret, E.; Boudinot, P.; Bremont, M. Mitochondrial antiviral signaling protein plays a major role in induction of the fish innate immune response against RNA and DNA viruses. J. Virol. 2009, 83, 7815–7827. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Chang, M.; Nie, P.; Secombes, C.J. Origin and evolution of the RIG-I like RNA helicase gene family. BMC Evol. Biol. 2009, 9, 85. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.J.; Sun, B.; Robertsen, B. Adjuvant activity of fish type I interferon shown in a virus DNA vaccination model. Vaccine 2015, 33, 2442–2448. [Google Scholar] [CrossRef] [PubMed]
- Sobhkhez, M.; Krasnov, A.; Chang, C.J.; Robertsen, B. Transcriptome analysis of plasmid-induced genes sheds light on the role of type I IFN as adjuvant in DNA vaccine against infectious salmon anemia virus. PLoS ONE 2017, 12, e0188456. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.J.; Robertsen, C.; Sun, B.; Robertsen, B. Protection of Atlantic salmon against virus infection by intramuscular injection of IFNc expression plasmid. Vaccine 2014, 32, 4695–4702. [Google Scholar] [CrossRef]
- Robertsen, B.; Chang, C.J.; Bratland, L. IFN-adjuvanted DNA vaccine against infectious salmon anemia virus: Antibody kinetics and longevity of IFN expression. Fish Shellfish Immunol. 2016, 54, 328–332. [Google Scholar] [CrossRef]
- Osokine, I.; Snell, L.M.; Cunningham, C.R.; Yamada, D.H.; Wilson, E.B.; Elsaesser, H.J.; de la Torre, J.C.; Brooks, D. Type I interferon suppresses de novo virus-specific CD4 Th1 immunity during an established persistent viral infection. Proc. Natl. Acad. Sci. USA 2014, 111, 7409–7414. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.-X.; Zou, J.; Nie, P.; Huang, B.; Yu, Z.; Collet, B.; Secombes, C.J. Intracellular interferons in fish: A unique means to combat viral infection. PLoS Pathog. 2013, 9, e1003736. [Google Scholar] [CrossRef]
- Cao, Y.; Zhang, Q.; Xu, L.; Li, S.; Wang, D.; Zhao, J.; Liu, H.; Feng, J.; Lu, T. Effects of different cytokines on immune responses of rainbow trout in a virus DNA vaccination model. Oncotarget 2017, 8, 112222–112235. [Google Scholar] [CrossRef] [PubMed]
- Prchal, M.; Pilz, A.; Simma, O.; Lingnau, K.; von Gabain, A.; Strobl, B.; Muller, M.; Decker, T. Type I interferons as mediators of immune adjuvants for T- and B cell-dependent acquired immunity. Vaccine 2009, 27 (Suppl. 6), G17–G20. [Google Scholar] [CrossRef] [PubMed]
- Le Bon, A.; Schiavoni, G.; D’Agostino, G.; Gresser, I.; Belardelli, F.; Tough, D.F. Type i interferons potently enhance humoral immunity and can promote isotype switching by stimulating dendritic cells in vivo. Immunity 2001, 14, 461–470. [Google Scholar] [CrossRef]
- Kurath, G.; Garver, K.A.; Corbeil, S.; Elliott, D.G.; Anderson, E.D.; LaPatra, S.E. Protective immunity and lack of histopathological damage two years after DNA vaccination against infectious hematopoietic necrosis virus in trout. Vaccine 2006, 24, 345–354. [Google Scholar] [CrossRef]
- Xu, L.; Zhao, J.; Liu, M.; Kurath, G.; Ren, G.; Lapatra, S.E.; Yin, J.; Liu, H.; Feng, J.; Lu, T. A effective DNA vaccine against diverse genotype J infectious hematopoietic necrosis virus strains prevalent in China. Vaccine 2017, 35, 2420–2426. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.J.; Gu, J.; Robertsen, B. Protective effect and antibody response of DNA vaccine against salmonid alphavirus 3 (SAV3) in Atlantic salmon. J. Fish. Dis. 2017, 40, 1775–1781. [Google Scholar] [CrossRef]
- Boudinot, P.; Blanco, M.; de Kinkelin, P.; Benmansour, A. Combined DNA immunization with the glycoprotein gene of viral hemorrhagic septicemia virus and infectious hematopoietic necrosis virus induces double-specific protective immunity and nonspecific response in rainbow trout. Virology 1998, 249, 297–306. [Google Scholar] [CrossRef]
- Lorenzen, E.; Lorenzen, N.; Einer-Jensen, K.; Brudeseth, B.; Evensen, O. Time course study of in situ expression of antigens following DNA-vaccination against VHS in rainbow trout (Oncorhynchus mykiss Walbaum) fry. Fish Shellfish Immunol. 2005, 19, 27–41. [Google Scholar] [CrossRef] [PubMed]
- Stan, A.C.; Casares, S.; Brumeanu, T.D.; Klinman, D.M.; Bona, C.A. CpG motifs of DNA vaccines induce the expression of chemokines and MHC class II molecules on myocytes. Eur. J. Immunol. 2001, 31, 301–310. [Google Scholar] [CrossRef]
- Joffre, O.P.; Segura, E.; Savina, A.; Amigorena, S. Cross-presentation by dendritic cells. Nat. Rev. Immunol. 2012, 12, 557–569. [Google Scholar] [CrossRef]
- Rizza, P.; Moretti, F.; Belardelli, F. Recent advances on the immunomodulatory effects of IFN-alpha: Implications for cancer immunotherapy and autoimmunity. Autoimmunity 2010, 43, 204–209. [Google Scholar] [CrossRef] [PubMed]
- Lugo-Villarino, G.; Balla, K.M.; Stachura, D.L.; Banuelos, K.; Werneck, M.B.; Traver, D. Identification of dendritic antigen-presenting cells in the zebrafish. Proc. Natl. Acad. Sci. USA 2010, 107, 15850–15855. [Google Scholar] [CrossRef] [PubMed]
- Granja, A.G.; Leal, E.; Pignatelli, J.; Castro, R.; Abos, B.; Kato, G.; Fischer, U.; Tafalla, C. Identification of Teleost Skin CD8alpha+ Dendritic-like Cells, Representing a Potential Common Ancestor for Mammalian Cross-Presenting Dendritic Cells. J. Immunol. 2015, 195, 1825–1837. [Google Scholar] [CrossRef] [PubMed]
- Resseguier, J.; Delaune, E.; Coolen, A.L.; Levraud, J.P.; Boudinot, P.; Le Guellec, D.; Verrier, B. Specific and Efficient Uptake of Surfactant-Free Poly(Lactic Acid) Nanovaccine Vehicles by Mucosal Dendritic Cells in Adult Zebrafish after Bath Immersion. Front. Immunol. 2017, 8, 190. [Google Scholar] [CrossRef]
- Wagstaffe, H.R.; Mooney, J.P.; Riley, E.M.; Goodier, M.R. Vaccinating for natural killer cell effector functions. Clin. Transl. Immunol. 2018, 7, e1010. [Google Scholar] [CrossRef] [PubMed]
- Garver, K.A.; Conway, C.M.; Elliott, D.G.; Kurath, G. Analysis of DNA-vaccinated fish reveals viral antigen in muscle, kidney and thymus, and transient histopathologic changes. Mar. Biotechnol. 2005, 7, 540–553. [Google Scholar] [CrossRef]
- Fu, X.; Li, N.; Lin, Q.; Guo, H.; Zhang, D.; Liu, L.; Wu, S. Protective immunity against infectious spleen and kidney necrosis virus induced by immunization with DNA plasmid containing mcp gene in Chinese perch Siniperca chuatsi. Fish Shellfish Immunol. 2014, 40, 259–266. [Google Scholar] [CrossRef]
- Jung, M.H.; Nikapitiya, C.; Jung, S.J. DNA vaccine encoding myristoylated membrane protein (MMP) of rock bream iridovirus (RBIV) induces protective immunity in rock bream (Oplegnathus fasciatus). Vaccine 2018, 36, 802–810. [Google Scholar] [CrossRef] [PubMed]
- Adamek, M.; Rakus, K.L.; Brogden, G.; Matras, M.; Chyb, J.; Hirono, I.; Kondo, H.; Aoki, T.; Irnazarow, I.; Steinhagen, D. Interaction between type I interferon and Cyprinid herpesvirus 3 in two genetic lines of common carp Cyprinus carpio. Dis. Aquat. Organ. 2014, 111, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, P.; Rakus, K.; van Beurden, S.J.; Westphal, A.H.; Davison, A.J.; Gatherer, D.; Vanderplasschen, A.F. IL-10 encoded by viruses: A remarkable example of independent acquisition of a cellular gene by viruses and its subsequent evolution in the viral genome. J. Gen. Virol. 2014, 95 Pt 2, 245–262. [Google Scholar] [CrossRef]
- Lin, Q.; Dong, C.; Cooper, M.D. Impairment of T and B cell development by treatment with a type I interferon. J. Exp. Med. 1998, 187, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Pascutti, M.F.; Erkelens, M.N.; Nolte, M.A. Impact of Viral Infections on Hematopoiesis: From Beneficial to Detrimental Effects on Bone Marrow Output. Front. Immunol. 2016, 7, 364. [Google Scholar] [CrossRef] [PubMed]
- Langevin, C.; Aleksejeva, E.; Passoni, G.; Palha, N.; Levraud, J.P.; Boudinot, P. The antiviral innate immune response in fish: Evolution and conservation of the IFN system. J. Mol. Biol. 2013, 425, 4904–4920. [Google Scholar] [CrossRef] [PubMed]
- Ablain, J.; Zon, L.I. Of fish and men: Using zebrafish to fight human diseases. Trends Cell Biol. 2013, 23, 584–586. [Google Scholar] [CrossRef] [PubMed]
- Davidson, A.J.; Zon, L.I. The ‘definitive’ (and ‘primitive’) guide to zebrafish hematopoiesis. Oncogene 2004, 23, 7233–7246. [Google Scholar] [CrossRef] [PubMed]
- De Jong, J.L.; Zon, L.I. Use of the zebrafish system to study primitive and definitive hematopoiesis. Annu. Rev. Genet. 2005, 39, 481–501. [Google Scholar] [CrossRef] [PubMed]
- Clements, W.K.; Traver, D. Signalling pathways that control vertebrate haematopoietic stem cell specification. Nat. Rev. Immunol. 2013, 13, 336–348. [Google Scholar] [CrossRef]
- Kulkeaw, K.; Sugiyama, D. Zebrafish erythropoiesis and the utility of fish as models of anemia. Stem Cell Res. 2012, 3, 55. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, J.Y.; Chi, N.C.; Santoso, B.; Teng, S.; Stainier, D.Y.; Traver, D. Haematopoietic stem cells derive directly from aortic endothelium during development. Nature 2010, 464, 108–111. [Google Scholar] [CrossRef]
- Kissa, K.; Herbomel, P. Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature 2010, 464, 112–115. [Google Scholar] [CrossRef]
- Hess, I.; Boehm, T. Intravital imaging of thymopoiesis reveals dynamic lympho-epithelial interactions. Immunity 2012, 36, 298–309. [Google Scholar] [CrossRef]
- Renaud, O.; Herbomel, P.; Kissa, K. Studying cell behavior in whole zebrafish embryos by confocal live imaging: Application to hematopoietic stem cells. Nat Protoc 2011, 6, 1897–1904. [Google Scholar] [CrossRef] [PubMed]
- Kwan, W.; North, T.E. Netting Novel Regulators of Hematopoiesis and Hematologic Malignancies in Zebrafish. Curr. Top. Dev. Biol. 2017, 124, 125–160. [Google Scholar]
- Eckfeldt, C.E.; Mendenhall, E.M.; Flynn, C.M.; Wang, T.F.; Pickart, M.A.; Grindle, S.M.; Ekker, S.C.; Verfaillie, C.M. Functional analysis of human hematopoietic stem cell gene expression using zebrafish. PLoS Biol. 2005, 3, e254. [Google Scholar] [CrossRef]
- Kobayashi, I.; Ono, H.; Moritomo, T.; Kano, K.; Nakanishi, T.; Suda, T. Comparative gene expression analysis of zebrafish and mammals identifies common regulators in hematopoietic stem cells. Blood 2010, 115, e1–e9. [Google Scholar] [CrossRef] [PubMed]
- Manesia, J.K.; Franch, M.; Tabas-Madrid, D.; Nogales-Cadenas, R.; Vanwelden, T.; Van Den Bosch, E.; Xu, Z.; Pascual-Montano, A.; Khurana, S.; Verfaillie, C.M. Distinct Molecular Signature of Murine Fetal Liver and Adult Hematopoietic Stem Cells Identify Novel Regulators of Hematopoietic Stem Cell Function. Stem Cells Dev. 2017, 26, 573–584. [Google Scholar] [CrossRef] [PubMed]
- McKinney-Freeman, S.; Cahan, P.; Li, H.; Lacadie, S.A.; Huang, H.T.; Curran, M.; Loewer, S.; Naveiras, O.; Kathrein, K.L.; Konantz, M.; et al. The transcriptional landscape of hematopoietic stem cell ontogeny. Cell Stem Cell 2012, 11, 701–714. [Google Scholar] [CrossRef] [PubMed]
- Esain, V.; Cortes, M.; North, T.E. Enumerating Hematopoietic Stem and Progenitor Cells in Zebrafish Embryos. Methods Mol. Biol. 2016, 1451, 191–206. [Google Scholar]
- North, T.E.; Goessling, W.; Walkley, C.R.; Lengerke, C.; Kopani, K.R.; Lord, A.M.; Weber, G.J.; Bowman, T.V.; Jang, I.H.; Grosser, T.; et al. Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature 2007, 447, 1007–1011. [Google Scholar] [CrossRef]
- Hall, C.; Crosier, P.; Crosier, K. Inflammatory cytokines provide both infection-responsive and developmental signals for blood development: Lessons from the zebrafish. Mol. Immunol. 2016, 69, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Pietras, E.M. Inflammation: A key regulator of hematopoietic stem cell fate in health and disease. Blood 2017, 130, 1693–1698. [Google Scholar] [CrossRef] [PubMed]
- Travnickova, J.; Tran Chau, V.; Julien, E.; Mateos-Langerak, J.; Gonzalez, C.; Lelievre, E.; Lutfalla, G.; Tavian, M.; Kissa, K. Primitive macrophages control HSPC mobilization and definitive haematopoiesis. Nat. Commun. 2015, 6, 6227. [Google Scholar] [CrossRef]
- Espin-Palazon, R.; Stachura, D.L.; Campbell, C.A.; Garcia-Moreno, D.; Del Cid, N.; Kim, A.D.; Candel, S.; Meseguer, J.; Mulero, V.; Traver, D. Proinflammatory signaling regulates hematopoietic stem cell emergence. Cell 2014, 159, 1070–1085. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Esain, V.; Teng, L.; Xu, J.; Kwan, W.; Frost, I.M.; Yzaguirre, A.D.; Cai, X.; Cortes, M.; Maijenburg, M.W.; et al. Inflammatory signaling regulates embryonic hematopoietic stem and progenitor cell production. Genes Dev. 2014, 28, 2597–2612. [Google Scholar] [CrossRef]
- Sawamiphak, S.; Kontarakis, Z.; Stainier, D.Y. Interferon gamma signaling positively regulates hematopoietic stem cell emergence. Dev. Cell 2014, 31, 640–653. [Google Scholar] [CrossRef]
- Hall, C.J.; Flores, M.V.; Oehlers, S.H.; Sanderson, L.E.; Lam, E.Y.; Crosier, K.E.; Crosier, P.S. Infection-responsive expansion of the hematopoietic stem and progenitor cell compartment in zebrafish is dependent upon inducible nitric oxide. Cell Stem Cell 2012, 10, 198–209. [Google Scholar] [CrossRef]
- Takizawa, H.; Boettcher, S.; Manz, M.G. Demand-adapted regulation of early hematopoiesis in infection and inflammation. Blood 2012, 119, 2991–3002. [Google Scholar] [CrossRef]
- Panopoulos, A.D.; Watowich, S.S. Granulocyte colony-stimulating factor: Molecular mechanisms of action during steady state and ’emergency’ hematopoiesis. Cytokine 2008, 42, 277–288. [Google Scholar] [CrossRef]
- Manz, M.G.; Boettcher, S. Emergency granulopoiesis. Nat. Rev. Immunol. 2014, 14, 302–314. [Google Scholar] [CrossRef]
- Baldridge, M.T.; King, K.Y.; Boles, N.C.; Weksberg, D.C.; Goodell, M.A. Quiescent haematopoietic stem cells are activated by IFN-gamma in response to chronic infection. Nature 2010, 465, 793–797. [Google Scholar] [CrossRef] [PubMed]
- Willis, A.R.; Torraca, V.; Gomes, M.C.; Shelley, J.; Mazon-Moya, M.; Filloux, A.; Lo Celso, C.; Mostowy, S. Shigella-Induced Emergency Granulopoiesis Protects Zebrafish Larvae from Secondary Infection. MBio 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Sheng, Z.; Mao, X.; Li, C.; Chen, J.; Zhang, J.; Huang, H.; Ruan, H.; Luo, L.; Li, L. Systemic inoculation of Escherichia coli causes emergency myelopoiesis in zebrafish larval caudal hematopoietic tissue. Sci. Rep. 2016, 6, 36853. [Google Scholar] [CrossRef] [PubMed]
- Sioud, M.; Floisand, Y.; Forfang, L.; Lund-Johansen, F. Signaling through toll-like receptor 7/8 induces the differentiation of human bone marrow CD34+ progenitor cells along the myeloid lineage. J. Mol. Biol. 2006, 364, 945–954. [Google Scholar] [CrossRef] [PubMed]
- Nagai, Y.; Garrett, K.P.; Ohta, S.; Bahrun, U.; Kouro, T.; Akira, S.; Takatsu, K.; Kincade, P.W. Toll-like receptors on hematopoietic progenitor cells stimulate innate immune system replenishment. Immunity 2006, 24, 801–812. [Google Scholar] [CrossRef] [PubMed]
- Sioud, M.; Floisand, Y. NOD2/CARD15 on bone marrow CD34+ hematopoietic cells mediates induction of cytokines and cell differentiation. J. Leukoc. Biol. 2009, 85, 939–946. [Google Scholar] [CrossRef]
- Granick, J.L.; Simon, S.I.; Borjesson, D.L. Hematopoietic stem and progenitor cells as effectors in innate immunity. Bone Marrow Res. 2012, 2012, 165107. [Google Scholar] [CrossRef] [PubMed]
- Hirche, C.; Frenz, T.; Haas, S.F.; Doring, M.; Borst, K.; Tegtmeyer, P.K.; Brizic, I.; Jordan, S.; Keyser, K.; Chhatbar, C.; et al. Systemic Virus Infections Differentially Modulate Cell Cycle State and Functionality of Long-Term Hematopoietic Stem Cells In Vivo. Cell Rep. 2017, 19, 2345–2356. [Google Scholar] [CrossRef]
- Kolb-Maurer, A.; Goebel, W. Susceptibility of hematopoietic stem cells to pathogens: Role in virus/bacteria tropism and pathogenesis. Fems Microbiol. Lett. 2003, 226, 203–207. [Google Scholar] [CrossRef]
- Luppi, M.; Barozzi, P.; Morris, C.; Maiorana, A.; Garber, R.; Bonacorsi, G.; Donelli, A.; Marasca, R.; Tabilio, A.; Torelli, G. Human herpesvirus 6 latently infects early bone marrow progenitors in vivo. J. Virol. 1999, 73, 754–759. [Google Scholar] [PubMed]
- Wu, X.; Dao Thi, V.L.; Huang, Y.; Billerbeck, E.; Saha, D.; Hoffmann, H.-H.; Wang, Y.; Vale Silva, L.; Sarbanes, S.; Sun, T.; et al. Intrinsic Immunity Shapes Viral Resistance of Stem Cells. Cell 2017, 172, 423–438. [Google Scholar] [CrossRef]
- Sato, T.; Onai, N.; Yoshihara, H.; Arai, F.; Suda, T.; Ohteki, T. Interferon regulatory factor-2 protects quiescent hematopoietic stem cells from type I interferon-dependent exhaustion. Nat. Med. 2009, 15, 696–700. [Google Scholar] [CrossRef]
- Pietras, E.M.; Lakshminarasimhan, R.; Techner, J.M.; Fong, S.; Flach, J.; Binnewies, M.; Passegue, E. Re-entry into quiescence protects hematopoietic stem cells from the killing effect of chronic exposure to type I interferons. J. Exp. Med. 2014, 211, 245–262. [Google Scholar] [CrossRef] [PubMed]
- Essers, M.A.; Offner, S.; Blanco-Bose, W.E.; Waibler, Z.; Kalinke, U.; Duchosal, M.A.; Trumpp, A. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature 2009, 458, 904–908. [Google Scholar] [CrossRef]
- LaPatra, S.E.; Barone, L.; Jones, G.R.; Zon, L.I. Effects of infectious hematopoietic necrosis virus and infectious pancreatic necrosis virus infection on hematopoietic precursors of the zebrafish. Blood Cells Mol. Dis. 2000, 26, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, M.; Palha, N.; Torhy, C.; Briolat, V.; Colucci-Guyon, E.; Bremont, M.; Herbomel, P.; Boudinot, P.; Levraud, J.P. Whole-body analysis of a viral infection: Vascular endothelium is a primary target of infectious hematopoietic necrosis virus in zebrafish larvae. PLoS Pathog. 2011, 7, e1001269. [Google Scholar] [CrossRef] [PubMed]
- Palha, N.; Guivel-Benhassine, F.; Briolat, V.; Lutfalla, G.; Sourisseau, M.; Ellett, F.; Wang, C.H.; Lieschke, G.J.; Herbomel, P.; Schwartz, O.; et al. Real-time whole-body visualization of Chikungunya Virus infection and host interferon response in zebrafish. PLoS Pathog. 2013, 9, e1003619. [Google Scholar] [CrossRef]
- Briolat, V.; Jouneau, L.; Carvalho, R.; Palha, N.; Langevin, C.; Herbomel, P.; Schwartz, O.; Spaink, H.P.; Levraud, J.P.; Boudinot, P. Contrasted innate responses to two viruses in zebrafish: Insights into the ancestral repertoire of vertebrate IFN-stimulated genes. J. Immunol. 2014, 192, 4328–4341. [Google Scholar] [CrossRef] [PubMed]
- McBrien, M. The Effect of Poly I:C Induced Inflammation on Hematopoietic Stem and Progenitor Cell Behaviour in the Zebrafish Hematopoietic Transplant Model. Ph.D. Thesis, Imperial College, London, UK, 2017. [Google Scholar]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Ge, W. Genome editing in fishes and their applications. Gen. Comp. Endocrinol. 2018, 257, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Cai, X.; Zhang, D.; Xu, C.; Xiao, W. Zebrafish foxo3b Negatively Regulates Antiviral Response through Suppressing the Transactivity of irf3 and irf7. J. Immunol. 2016, 197, 4736–4749. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Song, Y.; He, L.; Wan, X.; Lai, L.; Dai, F.; Liu, Y.; Wang, Q. MicroRNA-223 Promotes Type I Interferon Production in Antiviral Innate Immunity by Targeting Forkhead Box Protein O3 (FOXO3). J. Biol. Chem. 2016, 291, 14706–14716. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, J.A.; Kim, E.H.; Plisch, E.H.; Suresh, M. FOXO3 regulates the CD8 T cell response to a chronic viral infection. J. Virol. 2012, 86, 9025–9034. [Google Scholar] [CrossRef] [PubMed]
- Chakrapani, V.; Patra, S.K.; Panda, R.P.; Rasal, K.D.; Jayasankar, P.; Barman, H.K. Establishing targeted carp TLR22 gene disruption via homologous recombination using CRISPR/Cas9. Dev. Comp. Immunol. 2016, 61, 242–247. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.H.; Li, W.; Fan, Y.D.; Liu, Q.L.; Zeng, L.B.; Xiao, T.Y. Tlr22 structure and expression characteristic of barbel chub, Squaliobarbus curriculus provides insights into antiviral immunity against infection with grass carp reovirus. Fish Shellfish Immunol. 2017, 66, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, B.R.; Dikhit, M.R.; Bhoi, G.K.; Maharana, J.; Lenka, S.K.; Dubey, P.K.; Tiwari, D.K. Understanding the distinguishable structural and functional features in zebrafish TLR3 and TLR22, and their binding modes with fish dsRNA viruses: An exploratory structural model analysis. Amino Acids 2015, 47, 381–400. [Google Scholar] [CrossRef] [PubMed]
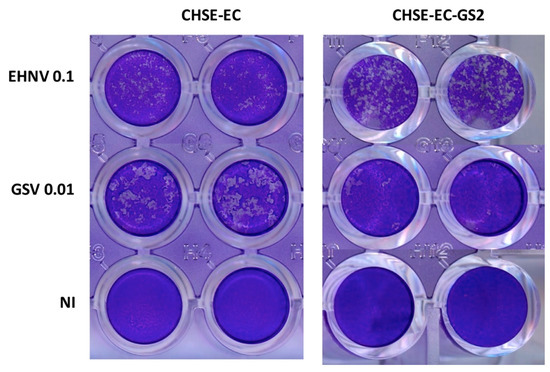
- Collet, B.; Collins, C.; Lester, K. Engineered cell lines for fish health research. Dev. Comp. Immunol. 2018, 80, 34–40. [Google Scholar] [CrossRef]
- Dehler, C.E.; Boudinot, P.; Martin, S.A.; Collet, B. Development of an Efficient Genome Editing Method by CRISPR/Cas9 in a Fish Cell Line. Mar. Biotechnol. 2016, 18, 449–452. [Google Scholar] [CrossRef]
- Kim, M.S.; Shin, M.J.; Kim, K.H. Increase of viral hemorrhagic septicemia virus growth by knockout of IRF9 gene in Epithelioma papulosum cyprini cells. Fish Shellfish Immunol. 2018, 83, 443–448. [Google Scholar] [CrossRef]
- Kim, M.S.; Kim, K.H. CRISPR/Cas9-mediated knockout of HIF-1alpha gene in epithelioma papulosum cyprini (EPC) cells inhibited apoptosis and viral hemorrhagic septicemia virus (VHSV) growth. Arch. Virol. 2018, 163, 3395–3402. [Google Scholar] [CrossRef] [PubMed]
- Schindler, C.; Shuai, K.; Prezioso, V.R.; Darnell, J.E., Jr. Interferon-dependent tyrosine phosphorylation of a latent cytoplasmic transcription factor. Science 1992, 257, 809–813. [Google Scholar] [CrossRef]
- Pasquier, J.; Cabau, C.; Nguyen, T.; Jouanno, E.; Severac, D.; Braasch, I.; Journot, L.; Pontarotti, P.; Klopp, C.; Postlethwait, J.H.; et al. Gene evolution and gene expression after whole genome duplication in fish: The PhyloFish database. BMC Genom. 2016, 17, 368. [Google Scholar] [CrossRef] [PubMed]
- Hasan, M.; Koch, J.; Rakheja, D.; Pattnaik, A.K.; Brugarolas, J.; Dozmorov, I.; Levine, B.; Wakeland, E.K.; Lee-Kirsch, M.A.; Yan, N. Trex1 regulates lysosomal biogenesis and interferon-independent activation of antiviral genes. Nat. Immunol. 2013, 14, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Noyce, R.S.; Collins, S.E.; Mossman, K.L. Identification of a novel pathway essential for the immediate-early, interferon-independent antiviral response to enveloped virions. J. Virol. 2006, 80, 226–235. [Google Scholar] [CrossRef]
- Verrier, E.R.; Dorson, M.; Mauger, S.; Torhy, C.; Ciobotaru, C.; Hervet, C.; Dechamp, N.; Genet, C.; Boudinot, P.; Quillet, E. Resistance to a rhabdovirus (VHSV) in rainbow trout: Identification of a major QTL related to innate mechanisms. PLoS ONE 2013, 8, e55302. [Google Scholar] [CrossRef] [PubMed]
- Verrier, E.R.; Langevin, C.; Tohry, C.; Houel, A.; Ducrocq, V.; Benmansour, A.; Quillet, E.; Boudinot, P. Genetic resistance to rhabdovirus infection in teleost fish is paralleled to the derived cell resistance status. PLoS ONE 2012, 7, e33935. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Sayed, N.; Hunter, A.; Au, K.F.; Wong, W.H.; Mocarski, E.S.; Pera, R.R.; Yakubov, E.; Cooke, J.P. Activation of innate immunity is required for efficient nuclear reprogramming. Cell 2012, 151, 547–558. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).