Abstract
In recent years, Castanopsis carlesii, a keystone species in southern China’s forest ecosystems with high ecological and economic importance, has faced growing challenges from severe nut rot diseases. Gnomoniopsis (Gnomoniaceae, Diaporthales, Sordariomycetes, Ascomycota) represents a significant fungal genus that causes leaf spots, branch cankers, and fruit rot diseases. In this study, rotten nuts of C. carlesii were collected from Fujian Province, and fungal isolates were obtained using the tissue isolation method. Morphological characterization and molecular phylogenetic analysis, based on the combined sequences of the internal transcribed spacer region of rDNA (ITS), the translation elongation factor 1-alpha (tef1) gene, and the partial beta-tubulin (tub2) gene were used to identify these isolates. As a result, new isolates from diseased nuts of C. carlesii formed a distinct clade with Gnomoniopsis, and morphologically differentiated from the other species; hence, G. flava sp. nov. is proposed herein. Furthermore, pathogenicity tests involving three isolates of G. flava were conducted on healthy nuts of C. carlesii, confirming its role as the causal agent of this new plant disease. This study not only advances our understanding of species diversity within Gnomoniopsis but also lays the groundwork for developing control strategies for C. carlesii nut rot disease.
1. Introduction
Castanopsis carlesii (Hemsl.) Hayata is a fagaceous species, commonly known as the Carles’ chinkapin, which is widely distributed in subtropical regions of East Asia [1,2,3]. This evergreen tree is an important component of mixed broadleaf forests and plays a significant ecological role in its native habitat. C. carlesii is valued not only for its ecological importance but also for its economic and cultural significance [1,2,3]. The wood is hard and durable, making it suitable for construction, furniture, and tool handles. In traditional medicine, various parts of the tree, including the bark and leaves, have been used to treat ailments such as diarrhea and inflammation [2,4]. Additionally, the tree produces small, edible nuts that are consumed by wildlife and local communities [2,3,4]. However, the availability of these nuts can be affected by plant diseases, which threaten the survival of C. carlesii populations.
The fungal genus Gnomoniopsis Berl., classified within the family Gnomoniaceae G. Winter of the order Diaporthales Nannf., comprises several species known to act as nut pathogens of Fagaceae Dumort. Hosts [5,6,7,8]. Among these, G. smithogilvyi L.A. Shuttlew., E.C.Y. Liew and D.I. Guest has been identified as the primary causative agent of nut rot in Castanea sativa Mill. across Europe and Oceania [7,8,9,10,11,12], while G. daii C.M. Tian and N. Jiang is responsible for nut rot in C. mollissima Blume in China [6]. Beyond their impact on nuts, species within Gnomoniopsis are also known to induce a range of plant diseases, including leaf spots, branch cankers, and wilting, highlighting their broader ecological and agricultural significance [13,14,15,16].
Gnomoniopsis was initially established as a subgenus within Gnomonia Ces. and De Not. to accommodate species characterized by ascospores that develop additional septa [17,18]. However, it was later reduced to a synonym of Gnomonia when the presence of additional septa was deemed an unreliable characteristic [18]. The genus name Gnomoniopsis was subsequently revived following molecular phylogenetic analyses, which re-evaluated the generic boundaries within the family Gnomoniaceae [17,19]. Gnomoniopsis proved phylogenetically distinct from Gnomonia [17]. Meanwhile, the type species Gnomoniopsis chamaemori (Fr.) Berl. and six additional species were included within the genus [17].
Morphologically, members of Gnomoniopsis are characterized by the presence of small, black perithecia that are typically immersed within the host tissue [17,18,19]. These perithecia are solitary, lacking a stroma or loosely aggregated in a minimal stroma [17,18,19]. Each perithecium features a single neck, which can be centrally, marginally, or laterally positioned [17,18,19]. The asci are oval to fusiform in shape and contain eight ascospores with a visible apical ring [17,18,19]. The ascospores are one-septate, oval to fusiform [17,18,19]. For the anamorphic stage, species within Gnomoniopsis produce pycnidia characterized by a wide ostiole [14,15,20]. These pycnidia generate conidia that vary in shape, ranging from oval, oblong, and globose to femur-shaped [14,15,17,18,19,20].
Recent studies integrating molecular phylogeny and morphological analyses have unveiled several new species within the genus Gnomoniopsis, including G. castanopsidis (N. Jiang, G. lithocarpi Shi Wang, Zhao X. Zhang, X.Y. Liu, and X.G. Zhang), among others [20,21,22,23]. Interestingly, most of these species were discovered in association with hosts from the families Fagaceae and Rosaceae Juss., suggesting that host-specific adaptation may serve as a primary driving force in the speciation of Gnomoniopsis [21,24,25,26,27].
In recent years, C. carlesii has been significantly affected by severe nut rot disease, which not only hinders the natural regeneration of this tree species but also reduces the edible quality of its seeds. The primary objectives of this study were to identify the causal pathogen based on the morphological and molecular methods and evaluate its pathogenicity in accordance with Koch’s postulates.
2. Materials and Methods
2.1. Sample Collection and Isolation
Mature nuts of Castanopsis carlesii were gathered from beneath the trees across various locales in Fujian Province, China, where the tree naturally proliferates. The tissues of infected nuts turned brown, with white mycelium emerging on the surface of the discolored areas (Figure 1). The diseased nuts were carefully encased in paper bags and expedited to the laboratory for the purpose of fungal isolation.

Figure 1.
Symptoms of nut rot disease of Castanopsis carlesii.
Upon arrival, the nuts were subjected to thorough cleansing under running tap water and subsequently underwent surface sterilization by immersion in a 75% ethanol solution for three minutes. Post-sterilization, the nuts were rinsed in sterile distilled water three times and dried using sterile absorbent cotton. A rigorous inspection ensued to cull any nuts exhibiting damage, with only those presenting an unblemished exterior being chosen for further analysis. Each selected nut was bisected to reveal its internal constitution; kernels displaying a spectrum from white to yellow were deemed healthy, whereas those manifesting a brown hue were classified as rotten. A tally was conducted to enumerate both the diseased and healthy nuts.
From the interface of discolored and healthy tissues, diminutive segments measuring 0.2 cm × 0.2 cm were excised and meticulously placed onto the surface of PDA (potato dextrose agar) plates, which were composed of 200 g of potatoes, 20 g of dextrose, and 20 g of agar per liter. After two days, hyphal tips were excised and transferred to fresh PDA plates to procure uncontaminated isolates under the scrutiny of a dissecting stereomicroscope with the aid of a sterile needle. The resultant cultures were cataloged and preserved at the China Forestry Culture Collection Center (CFCC), and corresponding specimens were archived in the herbarium of the Chinese Academy of Forestry (CAF).
2.2. Morphological Identification and Characterization
The morphological characteristics of the Gnomoniopsis species identified in this study were meticulously analyzed based on the fruiting bodies developed on PDA plates. The conidiomata were carefully examined and sectioned using a precision double-edged blade under the observation of a Zeiss Discovery V8 stereomicroscope (Jena, Germany). The microscopic structures, including conidiophores, conidiogenous cells, and conidia, were scrutinized and captured using an Olympus BX51 microscope (Tokyo, Japan). For spore measurement, a random selection of 50 conidia was made. The findings are delineated with the maximum and minimum dimensions provided in parentheses, accompanied by the range expressed as the mean ± standard deviation. Observations and documentation of the colony characteristics on PDA plates were conducted after periods of 1 and 2 weeks.
2.3. Sequence Data
Fungal DNA was extracted from 7-day-old colonies grown on PDA plates using the CTAB method [28]. Polymerase chain reaction (PCR) was performed to amplify the internal transcribed spacer region of rDNA (ITS), the translation elongation factor 1-alpha (tef1) gene, and the partial beta-tubulin (tub2) gene, utilizing the primer pairs ITS5/ITS4, 688F/EF2, and T1/Bt2b, respectively [29,30,31,32]. The PCR protocol consisted of an initial denaturation step at 94 °C for 5 min, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 48 °C (for ITS) or 54 °C (for tef1 and tub2) for 50 s, and extension at 72 °C for 1 min, with a final elongation step at 72 °C for 7 min. The PCR amplification products were visualized and analyzed through electrophoresis on 2% agarose gels and DNA sequencing was carried out by Sangon Biotech Company Limited (Beijing, China).
2.4. Phylogenetic Analyses
The sequences generated in this study were assembled using Seqman v. 7.1.0 (DNASTAR Inc., Madison, WI, USA) and subsequently deposited in GenBank (Table 1). Reference sequences were selected from recent studies on Gnomoniopsis (Table 1). Sequence alignments for the three loci ITS, tef1, and tub2 were performed using MAFFT v. 7 and manually refined in MEGA v. 7.0.21 [33].

Table 1.
Strains and GenBank accession numbers used in this study.
Phylogenetic analyses were conducted on a combined dataset of the three loci using both maximum likelihood (ML) and Bayesian inference (BI) approaches. For the ML analysis, the GTR substitution model was employed, and 1000 bootstrap replicates were performed via the CIPRES Science Gateway portal (https://www.phylo.org/; accessed on 6 February 2025) using RAxML-HPC BlackBox v. 8.2.10 [34,35]. For the BI analysis, partition-specific evolutionary models were selected using MrModeltest v. 2.3 based on the Akaike Information Criterion (AIC). Markov Chain Monte Carlo (MCMC) simulations were executed in MrBayes v. 3.1.2, running for 10 million generations with two independent chains initiated from random trees. Convergence was confirmed by an average standard deviation of split frequencies below 0.01, and trees were sampled every 1000 generations [36]. The first 25% of the sampled trees were discarded as burn-in, and posterior probabilities (BPP) were calculated from the remaining trees. Bootstrap support (BS) values in the ML analysis were derived from 1000 replicates. The resulting phylogenetic trees were visualized and annotated using FigTree v. 1.4.4.
2.5. Pathogenicity Trials
Three isolates of Gnomoniopsis flava (CFCC 71563, CFCC 71566, and CFCC 71567) were selected for inoculation experiments. A total of 200 healthy nuts of Castanopsis carlesii were shelled and surface-sterilized by immersing them in a 75% ethanol solution for three minutes. The isolates were cultured on PDA plates for 20 days to obtain conidia masses. The conidial suspension was prepared by filtering the conidial masses through sterile gauze and adjusting the concentration to 106 conidia/mL using sterile water. Subsequently, 5 µL of the conidial suspension was applied to the center of each chestnut, while 5 µL of sterile water was used as a control. Each isolate was inoculated onto 50 nuts, which were then placed in transparent plastic bags to maintain high humidity and incubated at 25 °C in the dark for 5 days. This setup included 50 replicates (50 nuts) per treatment. After incubation, re-isolations were conducted from the inoculated nuts and identified based on morphological characteristics combined with ITS sequence analysis.
3. Results
3.1. Phylogenetic Analysis
The combined dataset, comprising ITS, tef1, and tub2 sequences from 66 strains, was analyzed with Apiognomonia errabunda (AR 2813) designated as the outgroup taxon. The final alignment spanned 1881 characters (ITS: 461; tef1: 990; tub2: 430), including gaps. Maximum likelihood (ML) analysis yielded an optimal tree with a likelihood value of −14,687.13, featuring 811 distinct alignment patterns and 20.83% undetermined characters or gaps. The estimated base frequencies were A = 0.220472, C = 0.276547, G = 0.242728, and T = 0.260253. Substitution rates were calculated as follows: AC = 1.313857, AG = 3.276239, AT = 1.451147, CG = 0.818668, CT = 4.522579, and GT = 1.000000. The gamma distribution shape parameter (α) was 0.271083. For Bayesian inference (BI), the most suitable substitution models for each locus, determined using MrModeltest, were TN+F+I+G4 for ITS, TIM2e+I+G4 for tef1, and TIM3e+I+G4 for tub2. The BI results were consistent with the ML tree topology. Branches in Figure 1 are annotated with ML bootstrap support values (BS) ≥ 50% and Bayesian posterior probabilities (BPP) ≥ 0.90. Notably, three isolates from this study formed a distinct clade close to Gnomoniopsis daii, G. mengyinensis, G. silvicola, G. diaoluoshanensis, and G. yunnanensis, representing a novel species designated as Gnomoniopsis flava sp. nov. (Figure 2).
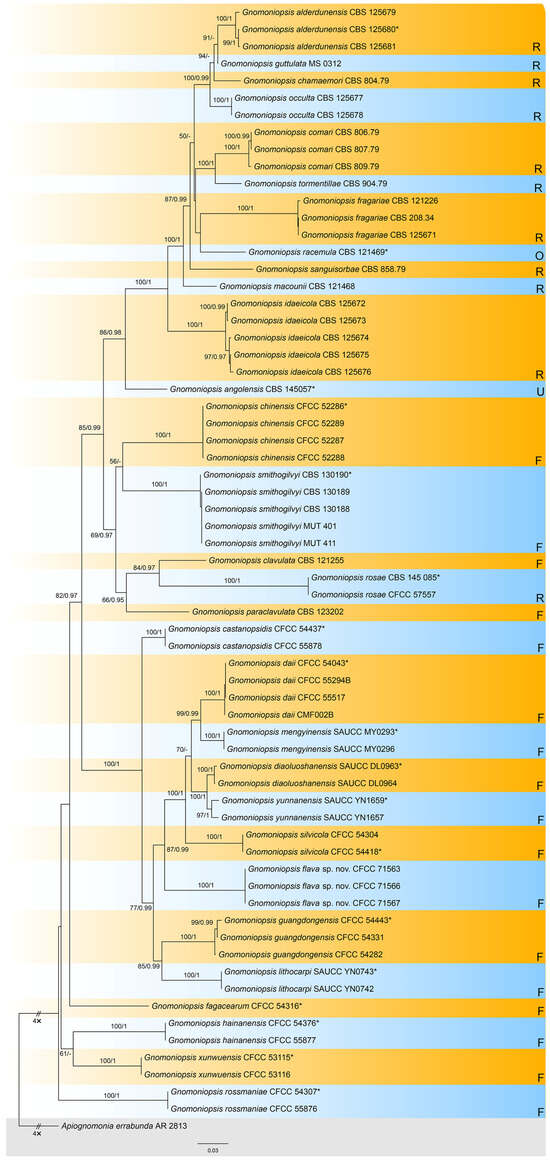
Figure 2.
Phylogram of Gnomoniopsis resulting from a maximum likelihood analysis based on the combined dataset of ITS, tef1, and tub2. Numbers above the branches indicate ML bootstraps (left, ML BS ≥ 50%) and Bayesian posterior probabilities (right, BPP ≥ 0.90). The tree is rooted with Apiognomonia errabunda (AR 2813). The ex-type strains are indicated by an asterisk (*). Fungal species are annotated according to their host families: those from Rosaceae are marked with R, from Fagaceae with F, from Onagraceae with O, and from unknown hosts with U.
3.2. Taxonomy
Gnomoniopsis flava Ning Jiang, sp. nov.
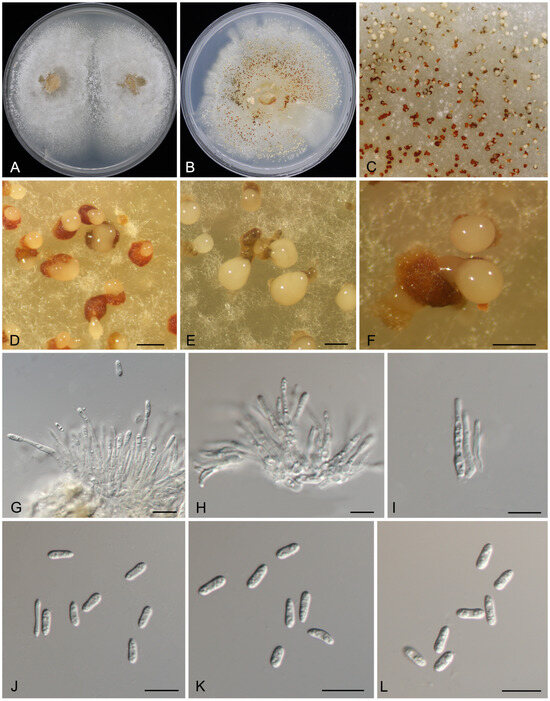
Figure 3.
Morphology of Gnomoniopsis flava (CFCC 71563). (A) Colony on PDA after 10 d. (B) Colony on PDA after 20 d. (C–F) Conidiomata formed on PDA. (G–I) Conidiogenous cells with attached conidia. (J–L) Conidia. Scale bars: (D–F) = 200 μm; (G–L) = 10 μm.
MycoBank: MB857963
Etymology: named after the color of the conidial masses.
Description: Causing nut rot disease of Castanopsis carlesii. Colonies on PDA flat, spreading, with moderate aerial mycelium and undulating margin, initially white, forming honey irregular center area after 10 d, reaching 90 mm diam after 10 d at 25 °C, forming abundant conidiomata with buff conidial masses after 20 d. Conidiomata pycnidial, aggregated or solitary, erumpent, globose to pulvinate, hazel or scarlet, 200–450 μm diam. Conidiophores indistinct, often reduced to conidiogenous cells. Conidiogenous cells hyaline, smooth, multi-guttulate, cylindrical to ampulliform, attenuate towards apex, phialidic, 11.5–26 × 1.5–2.5 μm. Conidia aseptate, hyaline, smooth, multi-guttulate, cylindrical, straight or slightly curved, base truncate, (5.5–)6.5–8(–9) × (2–)2.5–3 μm ( = 7.3 × 2.6 μm), L/W = 2.4–3.3.
Materials examined: CHINA, Fujian Province, Longyan City, Changting County, Tingzhou Town, on a rotten nut of Castanopsis carlesii, 16 July 2024, Ai-Ning Li, (CAF800142, holotype), ex-type culture CFCC 71563; Fujian Province, Longyan City, Changting County, Nanshan Town, on a rotten nut of C. carlesii, 14 July 2024, Ai-Ning Li, culture CFCC 71566; Fujian Province, Longyan City, Changting County, Hetian Town, on a rotten nut of C. carlesii, 15 July 2024, Ai-Ning Li, culture CFCC 71567.
Notes: Gnomoniopsis flava, isolated from rotten nuts of C. carlesii, is phylogenetically closely related to several other Gnomoniopsis species: G. daii from rotten nuts and leaf spots of Castanea mollissima, G. mengyinensis from diseased leaves of Castanea mollissima, G. silvicola from leaf spots of various fagaceous hosts, G. diaoluoshanensis from diseased leaves of C. chinensis, and G. yunnanensis from leaf spots of Castanea mollissima (Figure 1). All six Gnomoniopsis species infect hosts within the host family Fagaceae and are distributed across China. Morphologically, G. flava exhibits longer conidiogenous cells compared to G. diaoluoshanensis, G. mengyinensis, G. silvicola, and wider conidiogenous cells than G. yunnanensis, while its conidiogenous cells are similar in size to those of G. daii (11.5–26 × 1.5–2.5 μm in G. flava vs. 5–18 × 1.5–2.5 μm in G. daii vs. 8–12 × 1–2 μm in G. diaoluoshanensis vs. 8–11.5 × 1.3–2.2 μm in G. mengyinensis vs. 7–15 × 1.5–2.5 μm in G. silvicola vs. 9–18 × 0.5–1.6 μm in G. yunnanensis). Additionally, G. flava produces larger conidia than the other five species (6.5–8 × 2.5–3 μm in G. flava vs. 5.5–7 × 2–3.5 μm in G. daii vs. 3.8–7 × 1.2–2 μm in G. diaoluoshanensis vs. 4.5–6.5 × 1.8–2.8 μm in G. mengyinensis vs. 4.5–5.3 × 2.2–2.6 μm in G. silvicola vs. 4.1–5.5 × 1.3–2 μm in G. yunnanensis) [6,14,20,21].
3.3. Pathogenicity Test
Five days post-incubation, the nuts of Castanopsis carlesii were examined for kernel browning by carefully removing their shells. The fungal isolates CFCC 71563, CFCC 71566, and CFCC 71567 caused rot symptoms in 38, 34, and 33 nuts, respectively (Figure 4). In contrast, no symptoms were observed in nuts treated with sterile water. Re-isolation procedures were performed, and the fungi were confirmed to be Gnomoniopsis flava based on consistent colony and conidia morphology, as well as identical ITS sequences.
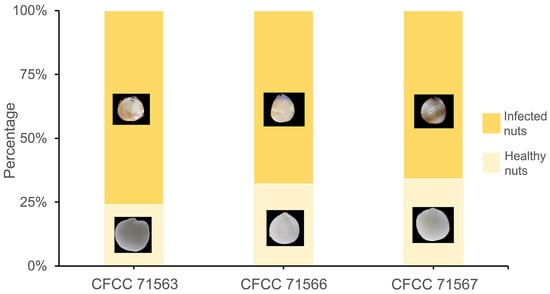
Figure 4.
Percentage of infected and healthy nuts in Castanopsis carlesii five days post-incubation.
4. Discussion
Members of the genus Gnomoniopsis are usually plant pathogens, responsible for diseases such as nut rot, branch canker, and leaf spot worldwide [7,14,19,20]. Among these, the most well-known pathogen is G. smithogilvyi, also referred to as G. castaneae, which causes severe chestnut rot in Castanea sativa, primarily affecting regions in Oceania and Europe [7,8,9,10,16,37,38,39]. Similarly, G. daii has been reported to cause significant chestnut rot in China, leading to production losses of up to 20% [6]. Unlike the widely recognized Castanea species, Castanopsis carlesii is a relatively rare nut-bearing tree within the genus Castanopsis, valued for its unique flavor and cultural significance among local communities. In this study, we identified a newly emerging nut rot disease in Fujian, China, and characterized the causative fungal pathogen as a novel species, which we named Gnomoniopsis flava. Naming and reporting this new disease and its associated pathogen are crucial steps for advancing future disease management strategies.
Generally, species within the genus Gnomoniopsis exhibit a preference for hosts in the families Rosaceae and Fagaceae, with the exception of G. racemula, which was discovered in the USA and inhabits Chamerion angustifolium (Onagraceae) [17]. Additionally, the host of Gnomoniopsis angolensis, identified in Angola, remains unknown [40]. As illustrated in Figure 1, a total of 30 species are currently recognized within Gnomoniopsis, with 11 species (36.67%) associated with Rosaceae and 17 species (56.67%) associated with Fagaceae. This distribution indicates host specificity and specialization may be the primary drivers of speciation within the genus Gnomoniopsis, like its relative Ophiognomonia [27].
Among the 17 Gnomoniopsis species associated with Fagaceae, 14 species (82.35%) were discovered in China, with the exceptions being G. clavulata and G. paraclavulata from the USA, and G. smithogilvyi from Oceania and Europe [17]. This suggests that China may be the potential origin site for Gnomoniopsis species inhabiting Fagaceae hosts. In contrast, of the 11 species associated with Rosaceae, only 1 species (9.10%), G. rosae, is distributed in China [41]. This distribution pattern indicates that the potential origin site for Gnomoniopsis species inhabiting Rosaceae hosts is likely to be Europe or the USA.
Traditionally, the identification of Gnomoniopsis species has relied primarily on teleomorph characteristics and molecular phylogeny [17,19]. However, recently discovered species have been described based on their anamorphic stages [20,21,22]. Notably, the shape and size of conidiogenous cells and conidia have proven to be useful diagnostic features when combined with multigene phylogenetic analysis [20,21,22,41]. In this study, a new anamorphic species is identified and described. Given the diversity of Fagaceae hosts and their wide distribution across China, it is anticipated that future studies will uncover additional novel species within this genus.
5. Conclusions
In this study, we investigated nut rot disease affecting Castanopsis carlesii in Fujian Province, China, and identified the fungal isolates as a novel species within the genus Gnomoniopsis, which we named G. flava sp. nov. Additionally, we conducted pathogenicity tests on C. carlesii, confirming that G. flava is the causative agent of nut rot. These findings provide a scientific foundation for developing future disease control strategies.
Author Contributions
Conceptualization, Y.L. and A.L.; methodology, N.J.; software, N.J.; validation, Y.L. and A.L.; formal analysis, Y.L.; investigation, Y.L. and A.L.; resources, Y.L. and A.L.; data curation, Y.L. and A.L.; writing—original draft preparation, Y.L., A.L. and N.J.; writing—review and editing, N.J.; visualization, N.J.; supervision, N.J.; project administration, N.J.; funding acquisition, N.J. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Microbial Resource Center of the Ministry of Science and Technology of the People’s Republic of China (NMRC-2024-7).
Data Availability Statement
All sequence data are available in NCBI GenBank listed in Table 1.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Cheng, Y.P.; Hwang, S.Y.; Lin, T.P. Potential refugia in Taiwan revealed by the phylogeographical study of Castanopsis carlesii Hayata (Fagaceae). Mol. Ecol. 2005, 14, 2075–2085. [Google Scholar]
- Zhong, X.; Zhang, L.; Zhang, J.; He, L.; Sun, R. Maxent modeling for predicting the potential geographical distribution of Castanopsis carlesii under various climate change scenarios in China. Forests 2023, 14, 1397. [Google Scholar] [CrossRef]
- Li, X.; Liu, X.; Xie, J.; Zhang, Q.; Yang, Z.; Schindlbacher, A.; Yang, Y. Contribution of above ground litterfall and roots to the soil CO2 efflux of two sub-tropical Cunninghamia lanceolata and Castanopsis carlesii forests. Agric. For. Meteorol. 2021, 311, 108671. [Google Scholar] [CrossRef]
- Yang, B.Y.; Wang, Y.F.; Li, G.Q.; He, R.J.; Huang, Y.L. Genus Castanopsis: A review on phytochemistry and biological activities. Fitoterapia 2024, 179, 106216. [Google Scholar] [PubMed]
- Visentin, I.; Gentile, S.; Valentino, D.; Gonthier, P.; Tamietti, G.; Cardinale, F. Gnomoniopsis castanea sp. nov. (Gnomoniaceae, Diaporthales) as the causal agent of nut rot in sweet chestnut. J. Plant Pathol. 2012, 94, 411–419. [Google Scholar]
- Jiang, N.; Tian, C.M. An emerging pathogen from rotted chestnut in China: Gnomoniopsis daii sp. nov. Forests 2019, 10, 1016. [Google Scholar] [CrossRef]
- Shuttleworth, L.A.; Liew, E.C.Y.; Guest, D.I. Survey of the incidence of chestnut rot in south-eastern Australia. Australas. Plant Pathol. 2013, 42, 63–72. [Google Scholar]
- Shuttleworth, L.A.; Guest, D.I. The infection process of chestnut rot, an important disease caused by Gnomoniopsis smithogilvyi (Gnomoniaceae, Diaporthales) in Oceania and Europe. Australas. Plant Pathol. 2017, 46, 397–405. [Google Scholar]
- Crous, P.W.; Summerell, B.A.; Shivas, R.G.; Burgess, T.I.; Decock, C.A.; Dreyer, L.L.; Granke, L.L.; Guest, D.I.; Hardy, G.E.S.J.; Hausbeck, M.K.; et al. Fungal Planet Description Sheets: 107–127. Persoonia 2012, 28, 138–182. [Google Scholar]
- Shuttleworth, L.A.; Walker, D.M.; Guest, D.I. The chestnut pathogen Gnomoniopsis smithogilvyi (Gnomoniaceae, Diaporthales) and its synonyms. Mycotaxon 2016, 130, 929–940. [Google Scholar]
- Sakalidis, M.L.; Medina-Mora, C.M.; Kolp, M.; Fulbright, D.W. First report of Gnomoniopsis smithogilvyi causing chestnut brown rot on chestnut fruit in Michigan. Plant Dis. 2019, 103, 2134. [Google Scholar]
- Dennert, F.G.; Broggini, G.A.; Gessler, C.; Storari, M. Gnomoniopsis castanea is the main agent of chestnut nut rot in Switzerland. Phytopathol. Mediterr. 2015, 54, 199–211. [Google Scholar]
- Pasche, S.; Calmin, G.; Auderset, G.; Crovadore, J.; Pelleteret, P.; Mauch-Mani, B.; Barja, F.; Paul, B.; Jermini, M.; Lefort, F. Gnomoniopsis smithogilvyi causes chestnut canker symptoms in Castanea sativa shoots in Switzerland. Fungal Genet. Biol. 2016, 87, 9–21. [Google Scholar] [PubMed]
- Jiang, N.; Fan, X.L.; Tian, C.M. Identification and characterization of leaf-inhabiting Fungi from Castanea plantations in China. J. Fungi 2021, 7, 64. [Google Scholar] [CrossRef] [PubMed]
- Jiang, N.; Liang, L.Y.; Tian, C.M. Gnomoniopsis chinensis (Gnomoniaceae, Diaporthales), a new fungus causing canker of Chinese chestnut in Hebei Province, China. MycoKeys 2020, 67, 19–32. [Google Scholar] [PubMed]
- Linaldeddu, B.T.; Deidda, A.; Scanu, B.; Franceschini, A.; Alves, A.; Abdollahzadeh, J.; Phillips, A.J.L. Phylogeny, morphology and pathogenicity of Botryosphaeriaceae, Diatrypaceae and Gnomoniaceae associated with branch diseases of hazelnut in Sardinia (Italy). Eur. J. Plant Pathol. 2016, 146, 259–279. [Google Scholar] [CrossRef]
- Sogonov, M.V.; Castlebury, L.A.; Rossman, A.Y.; Mejía, L.C.; White, J.F. Leaf-inhabiting genera of the Gnomoniaceae, Diaporthales. Stud. Mycol. 2008, 62, 1–77. [Google Scholar]
- Barr, M.E. The Diaporthales in North America with emphasis on Gnomonia and its segregates. Mycologia 1978, 7, 43–184. [Google Scholar]
- Walker, D.M.; Castlebury, L.A.; Rossman, A.Y.; Sogonov, M.V.; White, J.F. Systematics of genus Gnomoniopsis (Gnomoniaceae, Diaporthales) based on a three-gene phylogeny, host associations and morphology. Mycologia 2010, 102, 1479–1496. [Google Scholar]
- Wang, S.; Zhang, Z.; Liu, R.; Liu, S.; Liu, X.; Zhang, X. Morphological and phylogenetic analyses reveal four new species of Gnomoniopsis (Gnomoniaceae, diaporthales) from China. J. Fungi 2022, 8, 770. [Google Scholar] [CrossRef]
- Jiang, N.; Voglmayr, H.; Bian, D.R.; Piao, C.G.; Wang, S.K.; Li, Y. Morphology and phylogeny of Gnomoniopsis (Gnomoniaceae, Diporthales) from Fagaceae leaves in China. J. Fungi 2021, 7, 792. [Google Scholar]
- Yang, Q.; Jiang, N.; Tian, C.M. Tree inhabiting gnomoniaceous species from China, with Cryphogonomonia gen. nov. proposed. MycoKeys 2020, 69, 71–89. [Google Scholar]
- Udayanga, D.; Miriyagalla, S.D.; Manamgoda, D.S.; Lewers, K.S.; Gardiennet, A.; Castlebury, L.A. Molecular reassessment of diaporthalean fungi associated with strawberry, including the leaf blight fungus, Paraphomopsis obscurans gen. et comb. nov. (Melanconiellaceae). IMA Fungus 2021, 12, 15. [Google Scholar] [CrossRef]
- Jaklitsch, W.M.; Voglmayr, H. European species of Dendrostoma (Diaporthales). MycoKeys 2019, 59, 1–26. [Google Scholar] [PubMed]
- Mejía, L.C.; Castlebury, L.A.; Rossman, A.Y.; Sogonov, M.V.; White, J.F. Phylogenetic placement and taxonomic review of the genus Cryptosporella and its synonyms Ophiovalsa and Winterella (Gnomoniaceae, Diaporthales). Mycol. Res. 2008, 112, 23–35. [Google Scholar] [CrossRef] [PubMed]
- Mejía, L.C.; Castlebury, L.A.; Rossman, A.Y.; Sogonov, M.V.; White, J.F. A systematic account of the genus Plagiostoma (Gnomoniaceae, Diaporthales) based on morphology, host associations, and a four-gene phylogeny. Stud. Mycol. 2011, 68, 211–235. [Google Scholar]
- Walker, D.M.; Castlebury, L.A.; Rossman, A.Y.; Struwe, L. Host conservatism or host specialization? Patterns of fungal diversification are influenced by host plant specificity in Ophiognomonia (Gnomoniaceae, Diaporthales). Biol. J. Linnean Soc. 2013, 111, 1–16. [Google Scholar]
- Doyle, J.J. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. Guide Methods Appl. 1990, 18, 315–322. [Google Scholar]
- Carbone, I.; Kohn, L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 1999, 91, 553–556. [Google Scholar]
- Glass, N.L.; Donaldson, G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microb. 1995, 61, 1323–1330. [Google Scholar]
- O’Donnell, K.; Cigelnik, E. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol. Phylogenet Evol. 1997, 7, 103–116. [Google Scholar] [PubMed]
- Katoh, K.; Toh, H. Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics 2010, 26, 1899–1900. [Google Scholar] [PubMed]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the Gateway Computing Environments Workshop, GCE, New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar]
- Benigno, A.; Aglietti, C.; Cacciola, S.O.; Moricca, S. Trunk injection delivery of biocontrol strains of Trichoderma spp. effectively suppresses nut rot by Gnomoniopsis castaneae in Chestnut (Castanea sativa Mill.). Biology 2024, 13, 143. [Google Scholar] [CrossRef]
- Galdames, R.E. First report of Gnomoniopsis castaneae associated with branch dieback on chestnut tree (Castanea sativa) in Southern Chile. Plant Dis. 2024, 108, 3186. [Google Scholar]
- Çakar, D. Significance of Gnomoniopsis smithogilvyi as kernel rot of sweet chestnut in Turkey. J. Phytopathol. 2024, 172, e13293. [Google Scholar]
- Crous, P.W.; Luangsa-Ard, J.J.; Wingfield, M.J.; Carnegie, A.J.; Hernández-Restrepo, M.; Lombard, L.; Roux, J.; Barreto, R.W.; Baseia, I.G.; Cano-Lira, J.F.; et al. Fungal Planet description sheets: 785–867. Persoonia 2018, 41, 238–417. [Google Scholar] [CrossRef]
- Jiang, N.; Xue, H.; Piao, C.; Li, Y. Characterization and identification of Gnomoniopsis species. Terr. Ecosyst. Conserv. 2022, 2, 44–52. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).