Abstract
To improve the ecological and economic benefits of Korean pine (Pinus koraiensis), we analysed and evaluated its germplasm resources. This promotes in-depth research and utilisation of germplasm resources, providing excellent genetic resources for Korean pine breeding. We performed genetic analysis based on morphological and physiological traits and nuclear SSR molecular marker data was performed by collecting 314 clones (5 ramets of each clone) of Korean pine from eight (8) locations within the Korean pine range. The core collection underwent testing and evaluation for representativeness using variable rate (VR), coincidence rate (CR), variance difference percentage (VD), mean difference percentage (MD), Shannon index (I), and other indicators. The results indicated significant differences in morphological and physiological traits among the populations. All traits had a coefficient of variation (CV) greater than 10%, except for the water content of the needles (WC), which had an average CV of 17.636%. The populations showed high overall genetic diversity, with the HL (Helong) population exhibiting the highest genetic diversity, with an Ne (number of effective alleles), I, and He (expected heterozygosity) of 3.171, 1.103, and 0.528, respectively. Genetic variation mainly originated from individuals within populations, while the variation between populations was relatively small, at only 3%. The population did not exhibit any distinct subpopulation structures and was mainly derived from two admixed gene pools. Six core sets were obtained using different sampling strategies, and subset 6 was identified as the core collection, consisting of 114 individuals, representing a selection rate of 36.31%. In conclusion, the most appropriate method for constructing the core collection of Korean pines is the M-strategy (maximizing the number of alleles), based on both phenotypic and molecular data. The resulting core collection effectively represents the genetic diversity of the entire population effectively.
1. Introduction
Germplasm resources are essential for genetic research, enabling the exploration and utilisation of economically and ecologically important genes and traits [1]. Therefore, it is crucial to collect, preserve, evaluate, and utilise germplasm resources for breeding improved cultivars and new varieties [2,3]. Germplasm work can also be applied in theoretical studies related to plant origin, evolution, classification, and other aspects. However, the management and utilisation of germplasm resources require significant resources for appropriate assessment, particularly regarding redundant and duplicated germplasm resources [4,5,6]. Due to their large quantity, diverse structure, and incomplete information, the available potential of collected germplasm may not be fully utilised effectively. To tackle these issues, the concept of a “core collection” was further expanded in the early 1980s, introducing additional principles and methodologies for construction [7]. A core collection is a selected subset of the entire germplasm resource, chosen through certain methods, with the objective of representing the genetic diversity of the entire germplasm using the minimum number of resources required. They facilitate the rapid capture of germplasms possessing target traits and promote the effective utilisation of these resources [8,9]. Core collection resources are a novel approach to the utilisation and preservation of germplasm resources. Core collections perform a crucial function in the management and utilisation of genetic resources [10].
Initially, researchers used geographic origin and phenotypic characters to establish core collections, since these characters visually represent plant differences and are easy to measure, e.g., pomegranate Punica granatum L. [11], Ziziphus mauritiana Lam. [12], Prunus persica L. Batsch [13], etc. Nevertheless, there are some dilemmas with the method, such as the loss, incompleteness, and unreliability of germplasm and genetic data, as well as the sensitivity of phenotypic data to environmental conditions. These dilemmas mean that a core collection does not correctly represent the diversity of the original population [14,15]. Microsatellites or simple sequence repeats (SSRs) are tandemly repeated motifs of 1–6 bases found in all prokaryotic and eukaryotic genomes analysed to date [16]. These help to reveal the population structure and are often used as a tool for core collection development, with the advantage of exhibiting high polymorphisms that lead to population-specific alleles. A core collection has been developed using SSR markers in quite a few species, such as Corylus avellana L. [17]. In practice, breeders often use multiple data types to improve the quality of a core collection, which can greatly prevent the loss of key germplasm and improve the accuracy and comprehensiveness of a core collection. To name only a few, Krichen L et al. established a core collection of Prunus armeniaca L. through molecular markers and morphological traits [18]. Sun et al. developed a core collection of Litchi chinensis Sonn. based on genotypic data and agronomic traits [19]. Kumar et al. developed a core collection of Carthamus tinctorius L. using molecular, phenotypic, and geographic diversity [20]. Wang et al. used a combination of molecular markers and different phenotypic data to construct a core collection of Pinus yunnanensis [21].
Korean pine (Pinus koraiensis) is a valuable tree species in Northeastern China, serving as both an excellent source of timber and a valuable edible dry fruit and oilseed tree species [22]. The distribution area of Korean pine includes Northeastern China, the Korean Peninsula, and far southeastern Russia, with intermittent distribution in Honshu and Shikoku, Japan. In China, it is mainly found in Changbaishan, Zhangguangcailing, Laoyeling, Wandashan, and Xiaoxinganling [23]. Korean pine is a tall and straight tree with a perfectly shaped stem, high longevity, good physical quality of wood, and high productivity levels [24]. Additionally, the seeds, resin, and bark of Korean pine hold high economic value [25]. Korean pine is one of the main species of pine nuts and has a similar nutrient composition to other pine family cones such as Pinus pinea, Pinus sibirica, Pinus sinensis, Pinus cembra, Pinus edulis, and Pinus monophylla. These cones have significant food value and are highly beneficial to human health [26]. Since the 1960s, Heilongjiang Province has focused on the production and utilisation of Korean pine resources. The establishment of numerous seed orchards has provided a strong foundation for the development of Korean pine nut resources and the cultivation of nut forests [27]. Breeding research has been conducted based on Korean pine seed source zoning [28], resulting in the accumulation of abundant germplasm resources and a theoretical basis [29,30,31,32]. However, the direction and method of seed orchard construction has mainly been determined by geographical origin and growth traits. The genetic relationships and diversity of germplasm were overlooked. The repetitive and redundant materials in the germplasm resources of seed orchards has impeded the effective conservation of germplasm resources and may have also impede the germplasm resources’ evaluation and efficient use. Therefore, it is necessary to establish a core collection of Korean pine seed orchard germplasms.
The aim of this study is to enhance the effective management and utilisation of Korean pine resources. To achieve this, we constructed a core collection consisting of 314 clones from eight (8) populations, based on 11 SSR markers and nine (9) morphological and physiological traits, using various data types and strategies. The core collection was selected based on an analysis of genetic diversity parameters. The characteristics of the core collection have been described and compared to those of the entire collection. The results of this study will lay the foundation for research on good genes and molecular markers and provide the basis and valuable materials for the effective use and conservation of Korean pine resources. This study is of theoretical importance for the development of conservation strategies and breeding programs for Korean pine germplasm.
2. Materials and Methods
2.1. Plant Material
A total of 314 Korean pine clones were collected from six (6) Korean pine clones seed orchards. The assessment of genetic diversity is the basis of breeding research. Morphological variation is an important component of genetic variation. The clones were grafted from superior trees (refers to individuals with excellent growth, timber, and resistance adaptations in natural or planted forests with similar environmental conditions) from eight (8) Korean pine seed source natural forests in 1975–1980 (Table 1). The above eight (8) natural forests cover the distribution area of Korean pine in China in the same climate zone and their germs have been well preserved. Sampling trees were planted during the installation of orchards using specific grid planting diagrams. The annual and biennial disease-free needles of five ramets from each clone were randomly collected and stored at −20 °C, protected from light.

Table 1.
Information on the origin of clones.
2.2. Evaluated Traits
Needle traits were measured in biennial needles from five (5) ramets of 314 clones (Table 2). CL was measured using a ruler with an accuracy of 0.1 cm and CW was measured using an electronic caliper, respectively. Both FWC and DWC were measured as 15-needle weights using a 10,000-point balance. Three (3) randomized samples were taken from each ramet and replicated. DWC was the weight of fresh needle dried to constant weight. Chl in needles was determined using the acetone–ethanol method [33]. The 1:1 acetone–ethanol solution was used to dissolve chlorophyll and the absorbance of the solution was determined at 665 mm and 649 mm using INESA 722s (Shanghai, China). Chlorophyll content was calculated using the following formula:
where D649 and D665 are the optical density values at the corresponding wavelengths, V is the volume of the extract, and W is the fresh weight of the needles.

Table 2.
Investigated traits information of Korean pine.
2.3. DNA Extraction and Genotyping
DNA of all samples were extracted from 0.2 g annual needles using the DP-320 Plant Genome Extraction Kit (Tiangen Biochemical Technology (Beijing) Co. Ltd., Beijing, China). The integrity of genomic DNA was examined using a 1% agarose gel and DNA concentration and quality were examined using MicroDrop 3000 (Shanghai BIO-DL Science Instrument Co., Ltd., Shanghai, China) after extraction. The concentration of each DNA sample was diluted to 10 ng/mL and stored at −20 °C.
SSR primers from published studies in the field and synthesised by Sangon Biotech Co. Ltd. (Shanghai, China) [29] were used. The information is shown in Table A1. Each forward primer was labelled at the 5′ end with fluorescent dyes FAM, HEX, ROX, and TAMRA, respectively [34]. The PCR system consisted of 2U DNA polymerase (TransGen Biotech Co., Ltd. Beijing, China), 400μM Dntp (TransGen Biotech Co., Ltd. Beijing, China), 2.0μL of 10× buffer (TransGen Biotech Co., Ltd. Beijing, China), 0.5 μmol each of forward and reverse primer (Sangon Biotech Co. Ltd., Shanghai, China), and 10 ng DNA. The PCR procedure was as follows: 94 °C for 3 min, 35 cycles (30 s at 94 °C, 30 s at Tm, and 15 s at 72 °C), with a final extension at 72 °C for 7 min. The above procedures were executed under light-avoidance conditions. PCR products were assayed on an ABI 3730 Genetic Analyzer (Applied Biosystems, Inc., Foster City, CA, USA) by Shagon Biotech Co. Ltd. (Shanghai, China) and diploid genotype data were collected using DataCollection 3.0 software (Applied Biosystems, Inc., Foster City, CA, USA).
2.4. Data Analysis
The mean value, standard deviation (SD), coefficient of variation (CV), and analysis of variance (ANOVA) of the traits were calculated using SPSS 27.0 software (IBM SPSS Inc., Chicago, IL, USA). The diversity index (H) and principal coordinates analysis (PCoA) was performed. Morphological and physiological properties were classified into 10 levels, with values less than −2σ ( is the mean value and σ is the standard deviation) as level 1 and values greater than or equal to +2σ as level 10, where every 0.5σ was classified as a level.
The Shannon-Weaver diversity index (H) of morphological and physiological traits was used, which was calculated as:
where H is the diversity index of a trait; Pi is the number of germplasm copies of the itch level of a trait as a percentage of the total germplasm material; and ln denotes the natural logarithm [35,36].
The concept of Kataring Lindgren’s (1984) equivalent latitude was adopted, which was calculated as:
where Le is equivalent latitude; L is latitude; and A is altitude. When the altitude ≥300 m, a = 140. When the altitude <300 m, a = 140 [37].
GenAIEX6.51b2 (Australian National University, Acton ACT, Australian) was applied to calculate the number of alleles (Na), number of effective alleles (Ne), Shannon index (I), observed heterozygosity (Ho), expected heterozygosity (He), unbiased expected heterozygosity (uHe), and fixation index (F). Analysis of molecular variance (AMOVA), principal coordinate analysis (PCoA), and Nei genetic distances between populations were also performed in GenAIEX 6.51b2 (Australian National University, Acton ACT, Australian) [38]. Genetic structure was analysed using the admixture model in Structure 2.3.4 software (Stanford University, Palo Alto, CA, USA), with the number of populations (K) set from 1 to 10 and each K value was simulated 10 times. MCMC was set to 100,000 times for both the uncounted iterations and the MCMC after uncounted iterations. The results of the operations were imported into the Structure Harvester website (https://taylor0.biology.ucla.edu/structureHarvester/, accessed on 25 January 2024). The optimal K value was selected according to the method of Evanno et al. [39]. Significant deviations from both the Hardy–Weinberg equilibrium (HWE) and linkage disequilibrium (LD) between all pairs of SSR loci were identified using Genepop v4.2 (Laboratiore de Genetique et Environment, Montpellier, France) [40].
To improve the retention of alleles of the original germplasm, we performed random sampling (R-) and simulated annealing sampling (S-) using different sampling strategies, as follows: M-strategy (maximizing the number of alleles), E-strategy (Entropy), and D-strategy (Diversity), respectively. We used a sampling ratio of 1%–99% by PowerMarker 3.25 (NC State University, Raleigh, NC, USA) to determine the optimal sampling method and quantity [41]. M-strategy sampling was performed using PowerCore 1.0 (National Institute of Agricultural Biotechnology, Suwon, Republic of Korea) [42] based on an improved heuristic algorithm that considers different data types. Each program was run independently 10 times. GeneAlEx 6.501 (Australian National University, Canberra, Australia) was used to calculate the genetic parameters (Na, Ne, Ho, He, and I) for each collection. To evaluate the representativeness of the core collection, mean difference percentage (MD), variance difference percentage (VD), coincidence rate (CR), and variable rate (VR) were calculated according to the method of Hu et al. [14]. The Brekin Comprehensive Evaluation Method was used to evaluate the collections, as follows:
where Qi is the comprehensive score, Xij is the average value of an indicator, Xjmax is the optimal value of an indicator, and n is the number of evaluation indicators [43].
3. Results
3.1. Variation of Traits
Table 3 shows the results of genetic variation for nine (9) traits. The coefficient of variation (CV) for each trait was on average 17.64%, with all traits having a CV greater than 10%, except for water content (WC). The highest level of variation was found in Chlb (33.45%). Similarly, the value of H for all traits was greater than 2.00, except for water content. The average H for each trait was 2.052, with the highest H found in CW (2.089). The highest average H of each trait was found in HB (2.015), while all other populations had values less than 2. The CV was less than 20% for all groups, with the highest CV (19.87%) found in SGL and the lowest H and CV in HL, which were 1.851 and 9.170%, respectively. Morphological and physiological traits from different populations in the same orchard tended to behave similarly. There were no significant differences in CL, CW, CL/CW, FWC, or DWC between HL and SCZ. However, there were significant differences in CL, FWC, and DWC between LX and SGL. The given statement indicates that the morphological and physiological traits were influenced not only by genetic factors but also by the environment in which they were cultivated.

Table 3.
Morphological and physiological traits of Korean pine in different populations.
PCoA of the morphological and physiological traits for each group produced comparable results to those of the CV (Figure 1). HL had the smallest distribution range, while SGL, XBH, and WY, which had a large CV, had a large distribution range.
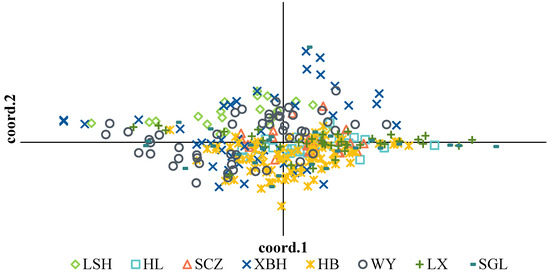
Figure 1.
Principal coordinates analysis (PCoA) of morphological and physiological traits among the eight (8) populations.
3.2. Genetic Diversity
The results of the genetic diversity analysis of the populations revealed that 11 SSR markers identified 93 alleles in 314 Korean pine clones, with an average Ne of 3.067. It is important to note that these results are objective and based solely on the data presented in Table 4. The genetic diversity analysis of populations revealed that 11 SSR markers identified 93 alleles in 314 Korean pine clones, with an average Ne of 3.067. The highest I was HL (1.103) and the I of the total population was 1.153. HL also had the largest Ne (3.171), He (0.528), uHe (0.538), and F (0.169), indicating that HL had the highest level of genetic diversity with heterozygosity deficiency. SGL exhibited the lowest level of genetic diversity, with the smallest Ne of 2.244, I of 0.956, He of 0.482, and uHe of 0.491. The smallest F of −0.002 was observed in HB, which indicates that the HB had the most reasonable mating system and did not have inbreeding. Positive values of FIS indicate that mates are more closely related on average than expected, and also indicate a deficit of heterozygotes relative to Hardy–Weinberg proportions. This is most likely due to the mean values shown in Table 5. This could be an artificial effect from a single locus that shows high linkage disequilibrium.

Table 4.
Genetic diversity of eight (8) Korean pine populations, resulting from 11 loci.
3.3. Analysis of Genetic Structure
The AMOVA results (Table 5) indicate that only 3% of the variation occurred among the eight (8) populations, while 87% of the variation was due to differences among individuals, and the remaining 10% was due to differences among the materials within each population at the significance level of p ≤ 0.001. The Nei genetic distance between populations was unequal (Figure 2), with the largest genetic distance being 0.161 between LSH and HL, and the smallest genetic distance being 0.015 between WY and XBH.
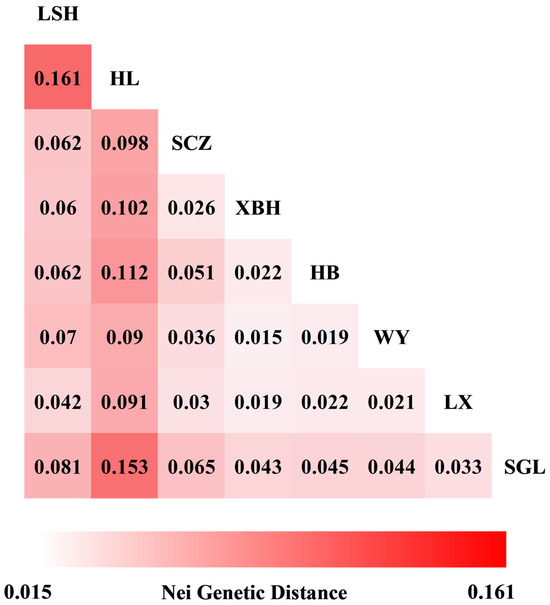
Figure 2.
Pairwise population matrix of Nei genetic distance.

Table 5.
Results of AMOVA of Korean pine populations.
Table 5.
Results of AMOVA of Korean pine populations.
| Source | df | SS | MS | Est. Var. | % |
|---|---|---|---|---|---|
| Among Pops | 7 | 66.152 | 9.450 | 0.084 | 3% |
| Among Indiv. | 306 | 950.221 | 3.105 | 0.298 | 10% |
| Within Indiv. | 314 | 788.000 | 2.510 | 2.510 | 87% |
| Total | 627 | 1804.373 | 2.891 | 100% |
df: degrees of freedom. SS: sum of squares. MS: mean of the squares. Est. Var.: estimated variance of components. %: percentage of total variance contributed by each component. p ≤ 0.001.
Figure 3 displays the results of the genetic structure analysis. The analysis revealed that the germplasm mainly originated from two gene pools, with a maximum at K = 2 in the range of 2–10 for ΔK. Gene pool 1 was the main contributor to LSH and HL, while gene pool 2 was the main contributor to SGL, consistent with the results of the genetic distance analysis. The other populations did not exhibit any significant subpopulation structures and were genetically characterized by a mixed composition of the two gene pools. This finding aligns with the results of AMOVA, indicating that there is only a minor genetic variation between populations, which indicates the frequent gene flow among the populations. This may be related to the small range of distribution and the long evolutionary history of Korean pine.
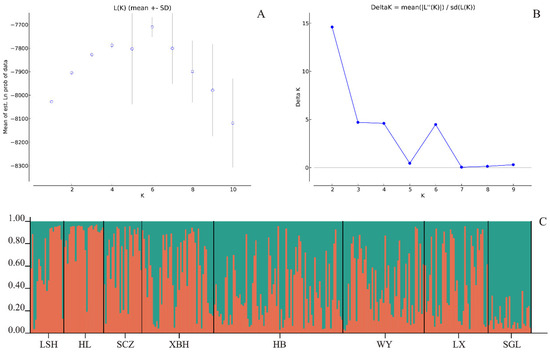
Figure 3.
Results of structure clustering analysis: (A) LnP(D) for each K value; (B) ΔK estimates of the posterior probability distribution of the data for a given K; and (C) estimated population structure and clustering with K = 2.
3.4. Construction of a Core Collection
Figure 4 shows the results of the varying sampling strategies and ratio using PowerMaker 3.25. Regardless of the strategy, R- had smaller values for Na (number of alleles), GE (gene entropy), and GD (gene diversity) than S-. Na increased with the sampling rate in different strategies, reaching a maximum at a sampling ratio of 32% (100) in the E-strategy. The E-strategy repeated the construction 1000 times at a 32% sampling proportion. Based on the frequency of each sample, the 100 samples with the highest frequency were selected as Core set 1. The maximum GE occurred at a sampling rate of 5% (16) in the E-strategy, while the maximum GD occurred at a sampling rate of 3% (9) in the D-strategy. Coresets 2 and 3, containing 16 and 9 individuals, respectively, were constructed in the same manner. Core sets 4, 5, and 6 were constructed using molecular markers, morphological and physiological traits, or both, after 10 independent repetitions using the M-strategy of PowerCore 1.0. The sample sizes for core sets 4, 5, and 6 were 103, 18, and 114, respectively.
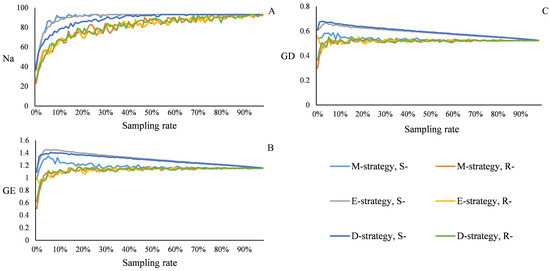
Figure 4.
Relationship between sampling rate and genetic parameters. (A) Number of alleles, (B) GE: gene entropy, and (C) gene diversity in different strategies.
3.5. Confirmation and Evaluation of Core Collection
The study initially selected six (6) core sets. The aim was to identify a core collection that could represent the entire population. To verify the validity of the core sets and identify the core collection, a comprehensive comparison of the six (6) core sets and the entire population was carried out. This was carried out following the principle of restoring the genetic level of the entire population as much as possible (Table 6). The study found that core sets 1, 4, and 6 retained all alleles of the entire population (93). Core set 4 had the smallest MD (0.953%) and the largest value of H (2.038). Coreset 2 had the highest value of Ne (4.479), I (1.453), He (0.568), and Ho (0.668), but its CR and VR were only 52.091% and 77.714%, respectively. The largest CR and VR were observed in set 6 (98.788%) and set 5 (148.626%), respectively. The highest composite score was obtained for core set 6 (2.652), thus identifying set 6 as the core collection. The genetic diversity parameters of core set 6, such as Ne, I, and Ho, were comparable and slightly greater than those of the entire population. The means, variances, CV, and range of morphological and physiological traits of core set 6 were also similar to those of the entire population. Therefore, core set 6 can effectively restore the genetic diversity and morphological and physiological variation of the entire population.

Table 6.
Comparison of six (6) core sets with original germplasm.
The PCoA of the core collection and the entire population revealed that the individuals of the core collection were uniformly distributed in the entire population. Both the morphological and physiological traits and the molecular markers of the core collection accurately represent the entire population. The variation and genetic diversity of the entire population could be restored (Figure 5a). The core collection accounted for 36.31% of the total, comprising 114 individuals from 8 populations (Figure 5b). The inner ring displays the proportion of each population in the core collection, while the outer ring shows the proportion of each population in the entire population. In the core collection, HB accounted for the largest proportion at 20.18%, while SGL accounted for the smallest proportion at 6.14%. However, both proportions were smaller than those in the entire population. The proportions of HL, LX, LSH, SCZ, and XBH in the core collection were larger than those in the entire population. The results above indicate that the HL and other populations exhibit a high level of genetic variation, whereas the SGL and HB populations exhibit a lower level of genetic variation. This finding is consistent with the data.
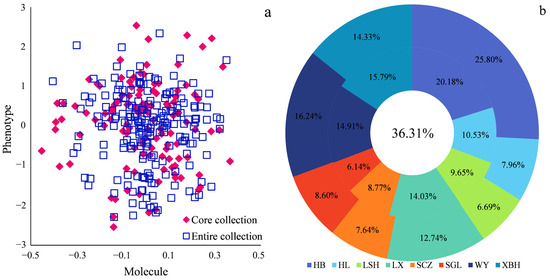
Figure 5.
Evaluation of the core collection: (a) principal component analysis (PCA) plot of the core and raw sets and (b) percentage of core collection in each population.
4. Discussion
Germplasm resources are essential for breeding. It is crucial to understand the genetic diversity and variation of germplasm resources for the conservation and sustainable use of plant resources [44,45]. The greater the range of tree species, the higher the genetic variation, leaf phenology, and physiological variation [46,47]. Among the populations, Chlb exhibited the highest CV. The average CV for each trait was highest in SGL among the populations. The degree of genetic diversity is higher when the CV of quantitative traits in germplasm resources is greater. The results of PCoA indicate that there is a high degree of overlap between different populations from the same orchard. Therefore, it is necessary to use multiple site materials to build the core collection.
Molecular markers are based on DNA polymorphisms, which are not biologically active and are rarely affected by the growth period of the plants or the external environment. They are therefore more suitable than morphological markers for constructing core collections and assessing genetic diversity. SSRs are co-dominant markers that provide richer allelic information than other molecular markers. In this study, Ne and I were higher than those in the study of Feng Fujuan et al. [48]. This study found that the use of SSR markers resulted in a higher efficiency compared to other types of molecular markers. The population with the highest genetic diversity was HL, while the lowest was SGL. The HB population had the lowest F (−0.002), indicating the presence of inbreeding in the seed orchard. All other populations had an F greater than zero, suggesting heterozygous deficiencies. These deficiencies may be related to the size of the original population and the procedures and criteria used to select superior trees. AMOVA revealed that despite the populations having different origins, interpopulation genetic variation accounted for only 3% of the total genetic variation. These results suggest that the interpopulation variation is insignificant and there is no clear genetic differentiation. The most genetically distant relationships are between HL and SGL.
The construction of a core collection typically involves four steps, data collection and organization, grouping of materials, determination of sampling strategy, and testing and evaluation of the core collection [49]. Observing genetic diversity is crucial for conserving germplasm resources and for breeding work. Sets 1, 2, and 3 compared the characteristics of collections constructed using different strategies. Both the E-strategy and M-strategy were able to maintain the Na, but only the E-strategy significantly improved the Ne, I, Ho, and He of the core collection. The D-strategy was able to restore the Ho of the original germplasm by using the least amount of germplasm, making it more suitable for the construction of breeding populations. Collections 4, 5, and 6 compared the quality of the core collection constructed using different data types. The results show that the core collection, constructed using multiple data types not only retained the Na, but also the morphological and physiological traits of the entire population. Previous studies have also demonstrated that constructing a more robust and reliable core collection using multiple data types can significantly prevent the loss of critical germplasm and improve the accuracy and comprehensiveness of the core collection [6]. Genetic analyses suggest that there is no clear genetic structure or significant genetic differentiation between geographic sources of Korean pine clones. Therefore, an appropriate sampling strategy is required to construct a core collection. The construction of a core collection depends on its purpose [17]. In recent years, the M-strategy has been widely used for this purpose. The M-strategy selects materials with high allele abundance and low redundancy by maximizing the number of alleles at each locus. This approach can preserve rare and local alleles [50,51]. The core collection constructed using M-strategy was able to restore more alleles and also restore the level of genetic diversity of the original germplasm under the same sample size.
The study found that the sampling rate of the core collection of Korean pine was at 36.31%, which is higher than that of Schima superba (15.3%), but with a comparable core collection size (115) [52], and lower than that of Juglans regia L. (44.23%) [53]. These results suggest that the sampling rate of the core collection varies depending on the special characteristics. The evaluation of the core collection depends on various factors, including the size and accessibility of the original germplasm, the similarity of the germplasm, the number and relevance of traits investigated, and the sampling strategy [54]. In addition to parameters such as MD, VD, Na, and Ne, the evaluation of morphological and physiological traits has introduced H. The core collection was determined using the Brekin Comprehensive Evaluation Method, which calculates composite scores for multiple traits in superior germplasm [43,55]. This method was also found to be suitable for the comprehensive evaluation of the core set. To compare distributional characteristics between the core collection and the original population, PCoA was used [56]. The distribution of molecular markers and morphological and physiological traits was consistent between the core and original groups, indicating no significant differences between the two. The highest proportion of the core collection was from HB, while the lowest was from SGL, which is related to the population size. The proportion of the core collection in each population of the original population reflects the redundancy of the population. SGL, which has the smallest proportion, exhibits the lowest genetic diversity. The higher proportion observed in LSH may be attributed to its limited population size, in addition to its rich genetic variation.
5. Conclusions
Establishing a core collection of Korean pine based on molecular and phenotypic traits can enhance the efficiency of its genetic resources. This study evaluated 314 clones from eight (8) Korean pine populations in China for genetic diversity using SSR markers and morphological and physiological traits. A core collection of 114 Korean pine clones was constructed, covering all eight (8) populations and representing the diversity of the entire population. The technology has wide-ranging applications in future breeding work. Firstly, it can be used for genomic research and molecular marker development to promote the conservation and utilization of germplasm resources. This includes genetic diversity evaluation, identification and classification of germplasm, genetic map construction, and the development of specific genes. Secondly, it can be used for genome-wide association research, such as the identification of disease-resistant genes and the discovery of superior genes. It also plays an important role in diversifying the genetic basis of germplasm materials and developing a breeding program.
Author Contributions
Investigation, data curation, formal analysis, supervision, validation, writing—original draft, P.Y.; methodology, L.Z.; investigation, J.H. and J.W.; resources, G.S. and Z.H.; software, R.W.; methodology, writing—review and editing, Z.L.; conceptualization, project administration, resources, supervision, writing—review and editing, H.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Key Research and Development Program of China “Research on high-yield and high-efficiency technologies for seed orchards of northern coniferous species”, grant number 2023YFD2200605, the Innovation Project of State Key Laboratory of Tree Genetics and Breeding (Northeast Forestry University) “Research on the Evaluation of Korean Pine Germplasm Resources and Construction Technology of Breeding Population”, grant number 2022A02, and the Heilongjiang Touyan Innovation Team Program.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors on request.
Conflicts of Interest
Authors Guofei Sun and Zhenyu Hu were employed by the Mengjiagang Forest Farm. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The Mengjiagang Forest Farm had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Appendix A

Table A1.
SSR primer information for estimating Genetic diversity in eight (8) populations.
Table A1.
SSR primer information for estimating Genetic diversity in eight (8) populations.
| Locus | Primer Sequence | Motif | Tm (°C) | Size (bp) | Fluorescent Dye |
|---|---|---|---|---|---|
| p49 | F: GAGATGAGCGAATCTGGG | (AAG)7 | 52 | 261 | FAM |
| R: TACAAGTTCCACCTACGG | |||||
| p70 | F: CAACATCGCCAATGACTC | (CTCA)6 | 54 | 294 | FAM |
| R: CCTACCTACGCTCTGCTC | |||||
| p72 | F: TGGGTTACCACCTTTAGC | (GCT)6 | 52 | 193 | HEX |
| R: CAATCAGAGTCTGGAGCA | |||||
| p79 | F: CCACCGCCAAGTCCATTA | (CAA)7 | 55 | 190 | HEX |
| R: GCTTTGTTAGCCGTCCAG | |||||
| p82 | F: GGAAGATGAATCGCAAACC | (GCG)6 | 54 | 280 | ROX |
| R: ACACCCGCCTGAAGAGCA | |||||
| EPD11 | F: GTGGATGCAATGAAGAAAAACAT | (AGG)6 | 60 | 139 | TAM |
| R: ACGAATTGCAAAACTGCATAACT | |||||
| NFPK-34 | F: AACCCACAGAAAGCTGAGGA | (TAA)6 | 60 | 221 | TAM |
| R: CACCCCTGAACAGAGAGGAG | |||||
| P6* | F: TCAAATTACCAGACAATAA | (TA)3 (GT)15 | 55 | 125 | FAM |
| R: GAATTCGCCAATGAAATCA | |||||
| P45* | F: CTTACATTTTGCTGCTTTTC | (TG)16 (AG)17 | 55 | 173 | HEX |
| R: TTGTCAGTTTTAGGTTGGAT | |||||
| P51* | F: CCTAAGAGCAATGTAAAATG | (AG)15 | 55 | 204 | TAM |
| R: AGCTTGACAACGACTAACT | |||||
| P52* | F: CCATCCTTCAAATTTTCCT | (AG)26 | 56 | 138 | ROX |

Table A2.
Genetic diversity of 8 Korean pine populations by each locus.
Table A2.
Genetic diversity of 8 Korean pine populations by each locus.
| Pop | Locus | N | Na | Ne | I | Ho | He | uHe | F |
|---|---|---|---|---|---|---|---|---|---|
| LSH | p49 | 21 | 3 | 2.178 | 0.876 | 0.476 | 0.541 | 0.554 | 0.119 |
| p70 | 21 | 3 | 2.125 | 0.864 | 0.143 | 0.529 | 0.542 | 0.730 | |
| p72 | 21 | 3 | 1.273 | 0.425 | 0.190 | 0.214 | 0.220 | 0.111 | |
| p79 | 21 | 2 | 1.630 | 0.575 | 0.524 | 0.387 | 0.396 | −0.355 | |
| p82 | 21 | 2 | 1.960 | 0.683 | 0.095 | 0.490 | 0.502 | 0.806 | |
| EPD11 | 21 | 2 | 1.960 | 0.683 | 0.667 | 0.490 | 0.502 | −0.361 | |
| NFPK-34 | 21 | 2 | 1.208 | 0.314 | 0.190 | 0.172 | 0.177 | −0.105 | |
| P6* | 21 | 4 | 1.834 | 0.872 | 0.429 | 0.455 | 0.466 | 0.057 | |
| P45* | 21 | 11 | 5.313 | 1.973 | 0.714 | 0.812 | 0.832 | 0.120 | |
| P52* | 21 | 9 | 4.820 | 1.815 | 0.762 | 0.793 | 0.812 | 0.039 | |
| P51* | 21 | 11 | 5.444 | 2.019 | 0.810 | 0.816 | 0.836 | 0.008 | |
| HL | p49 | 25 | 1 | 1.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| p70 | 25 | 3 | 2.023 | 0.844 | 0.560 | 0.506 | 0.516 | −0.108 | |
| p72 | 25 | 3 | 1.390 | 0.546 | 0.240 | 0.281 | 0.287 | 0.145 | |
| p79 | 25 | 4 | 2.887 | 1.173 | 0.120 | 0.654 | 0.667 | 0.816 | |
| p82 | 25 | 2 | 1.471 | 0.500 | 0.400 | 0.320 | 0.327 | −0.250 | |
| EPD11 | 25 | 3 | 1.649 | 0.717 | 0.440 | 0.394 | 0.402 | −0.118 | |
| NFPK-34 | 25 | 2 | 2.000 | 0.693 | 0.200 | 0.500 | 0.510 | 0.600 | |
| P6* | 25 | 8 | 2.747 | 1.409 | 0.680 | 0.636 | 0.649 | −0.069 | |
| P45* | 25 | 12 | 7.440 | 2.196 | 0.800 | 0.866 | 0.883 | 0.076 | |
| P52* | 25 | 9 | 4.513 | 1.821 | 0.520 | 0.778 | 0.794 | 0.332 | |
| P51* | 25 | 12 | 7.764 | 2.232 | 0.640 | 0.871 | 0.889 | 0.265 | |
| SCZ | p49 | 24 | 2 | 1.843 | 0.650 | 0.708 | 0.457 | 0.467 | −0.548 |
| p70 | 24 | 3 | 2.268 | 0.907 | 0.500 | 0.559 | 0.571 | 0.106 | |
| p72 | 24 | 3 | 1.471 | 0.602 | 0.292 | 0.320 | 0.327 | 0.089 | |
| p79 | 24 | 3 | 1.453 | 0.548 | 0.292 | 0.312 | 0.318 | 0.064 | |
| p82 | 24 | 3 | 1.341 | 0.475 | 0.292 | 0.254 | 0.260 | −0.147 | |
| EPD11 | 24 | 3 | 1.607 | 0.688 | 0.375 | 0.378 | 0.386 | 0.007 | |
| NFPK-34 | 24 | 2 | 1.385 | 0.451 | 0.250 | 0.278 | 0.284 | 0.100 | |
| P6* | 24 | 6 | 2.007 | 1.077 | 0.458 | 0.502 | 0.512 | 0.087 | |
| P45* | 24 | 12 | 6.400 | 2.106 | 0.792 | 0.844 | 0.862 | 0.062 | |
| P52* | 24 | 6 | 4.159 | 1.557 | 0.208 | 0.760 | 0.776 | 0.726 | |
| P51* | 24 | 9 | 6.621 | 2.005 | 0.542 | 0.849 | 0.867 | 0.362 | |
| XBH | p49 | 45 | 2 | 1.670 | 0.591 | 0.378 | 0.401 | 0.406 | 0.058 |
| p70 | 45 | 4 | 1.968 | 0.892 | 0.511 | 0.492 | 0.497 | −0.039 | |
| p72 | 45 | 3 | 1.480 | 0.615 | 0.356 | 0.324 | 0.328 | −0.097 | |
| p79 | 45 | 3 | 1.599 | 0.601 | 0.378 | 0.375 | 0.379 | −0.009 | |
| p82 | 45 | 3 | 1.199 | 0.359 | 0.178 | 0.166 | 0.168 | −0.073 | |
| EPD11 | 45 | 5 | 1.946 | 0.955 | 0.444 | 0.486 | 0.492 | 0.086 | |
| NFPK-34 | 45 | 3 | 1.411 | 0.563 | 0.200 | 0.291 | 0.294 | 0.313 | |
| P6* | 45 | 6 | 2.179 | 1.115 | 0.600 | 0.541 | 0.547 | −0.109 | |
| P45* | 45 | 12 | 5.000 | 1.921 | 0.578 | 0.800 | 0.809 | 0.278 | |
| P52* | 45 | 11 | 4.018 | 1.831 | 0.822 | 0.751 | 0.760 | −0.095 | |
| P51* | 45 | 10 | 7.181 | 2.096 | 0.600 | 0.861 | 0.870 | 0.303 | |
| HB | p49 | 81 | 2 | 1.608 | 0.566 | 0.481 | 0.378 | 0.380 | −0.274 |
| p70 | 81 | 4 | 1.433 | 0.612 | 0.222 | 0.302 | 0.304 | 0.264 | |
| p72 | 81 | 3 | 1.522 | 0.635 | 0.370 | 0.343 | 0.345 | −0.080 | |
| p79 | 81 | 7 | 1.804 | 0.801 | 0.519 | 0.446 | 0.449 | −0.163 | |
| p82 | 81 | 3 | 1.160 | 0.285 | 0.148 | 0.138 | 0.139 | −0.073 | |
| EPD11 | 81 | 4 | 2.180 | 0.921 | 0.593 | 0.541 | 0.545 | −0.095 | |
| NFPK-34 | 81 | 4 | 1.429 | 0.541 | 0.309 | 0.300 | 0.302 | −0.029 | |
| P6* | 81 | 9 | 2.184 | 1.256 | 0.543 | 0.542 | 0.545 | −0.002 | |
| P45* | 81 | 18 | 7.062 | 2.308 | 0.728 | 0.858 | 0.864 | 0.151 | |
| P52* | 81 | 9 | 3.670 | 1.632 | 0.679 | 0.728 | 0.732 | 0.067 | |
| P51* | 81 | 11 | 7.397 | 2.165 | 0.679 | 0.865 | 0.870 | 0.215 | |
| WY | p49 | 51 | 2 | 1.613 | 0.568 | 0.314 | 0.380 | 0.384 | 0.174 |
| p70 | 51 | 4 | 1.818 | 0.829 | 0.412 | 0.450 | 0.454 | 0.085 | |
| p72 | 51 | 3 | 1.502 | 0.630 | 0.353 | 0.334 | 0.338 | −0.056 | |
| p79 | 51 | 3 | 1.775 | 0.666 | 0.373 | 0.437 | 0.441 | 0.147 | |
| p82 | 51 | 3 | 1.148 | 0.278 | 0.137 | 0.129 | 0.130 | −0.064 | |
| EPD11 | 51 | 4 | 1.565 | 0.640 | 0.412 | 0.361 | 0.365 | −0.140 | |
| NFPK-34 | 51 | 3 | 1.702 | 0.744 | 0.412 | 0.412 | 0.416 | 0.001 | |
| P6* | 51 | 7 | 2.971 | 1.448 | 0.647 | 0.663 | 0.670 | 0.025 | |
| P45* | 51 | 11 | 6.149 | 2.029 | 0.627 | 0.837 | 0.846 | 0.251 | |
| P52* | 51 | 9 | 4.172 | 1.709 | 0.765 | 0.760 | 0.768 | −0.006 | |
| P51* | 51 | 12 | 7.930 | 2.231 | 0.686 | 0.874 | 0.883 | 0.215 | |
| LX | p49 | 40 | 3 | 1.682 | 0.639 | 0.475 | 0.405 | 0.410 | −0.172 |
| p70 | 40 | 3 | 1.868 | 0.818 | 0.500 | 0.465 | 0.471 | −0.076 | |
| p72 | 40 | 3 | 1.396 | 0.544 | 0.325 | 0.283 | 0.287 | −0.147 | |
| p79 | 40 | 4 | 1.951 | 0.887 | 0.350 | 0.488 | 0.494 | 0.282 | |
| p82 | 40 | 3 | 1.467 | 0.591 | 0.275 | 0.318 | 0.322 | 0.136 | |
| EPD11 | 40 | 4 | 1.996 | 0.881 | 0.475 | 0.499 | 0.505 | 0.048 | |
| NFPK-34 | 40 | 3 | 1.227 | 0.391 | 0.050 | 0.185 | 0.187 | 0.729 | |
| P6* | 40 | 8 | 1.968 | 1.132 | 0.475 | 0.492 | 0.498 | 0.034 | |
| P45* | 40 | 16 | 7.159 | 2.302 | 0.750 | 0.860 | 0.871 | 0.128 | |
| P52* | 40 | 9 | 3.865 | 1.620 | 0.750 | 0.741 | 0.751 | −0.012 | |
| P51* | 40 | 9 | 6.061 | 1.963 | 0.600 | 0.835 | 0.846 | 0.281 | |
| SGL | p49 | 27 | 2 | 1.874 | 0.659 | 0.444 | 0.466 | 0.475 | 0.047 |
| p70 | 27 | 3 | 1.581 | 0.671 | 0.370 | 0.368 | 0.375 | −0.007 | |
| p72 | 27 | 3 | 1.409 | 0.557 | 0.333 | 0.290 | 0.296 | −0.149 | |
| p79 | 27 | 3 | 1.839 | 0.708 | 0.444 | 0.456 | 0.465 | 0.026 | |
| p82 | 27 | 3 | 1.256 | 0.426 | 0.148 | 0.204 | 0.208 | 0.273 | |
| EPD11 | 27 | 4 | 2.363 | 1.066 | 0.778 | 0.577 | 0.588 | −0.348 | |
| NFPK-34 | 27 | 3 | 1.255 | 0.420 | 0.074 | 0.203 | 0.207 | 0.635 | |
| P6* | 27 | 9 | 2.390 | 1.379 | 0.630 | 0.582 | 0.593 | −0.083 | |
| P45* | 27 | 11 | 3.984 | 1.854 | 0.667 | 0.749 | 0.763 | 0.110 | |
| P52* | 27 | 9 | 3.496 | 1.537 | 0.667 | 0.714 | 0.727 | 0.066 | |
| P51* | 27 | 4 | 3.233 | 1.238 | 0.111 | 0.691 | 0.704 | 0.839 |
References
- Liu, Y.; Geng, Y.; Xie, X.; Zhang, P.; Hou, J.; Wang, W. Core collection construction and evaluation of the genetic structure of Glycyrrhiza in China using markers for genomic simple sequence repeats. Genet. Resour. Crop Evol. 2020, 67, 1839–1852. [Google Scholar] [CrossRef]
- Boczkowska, M.; Apiński, B.; Kordulasińska, I.; Dostatny, D.F.; Czembor, J.H. Promoting the Use of Common Oat Genetic Resources through Diversity Analysis and Core Collection Construction. PLoS ONE 2016, 11, e0167855. [Google Scholar] [CrossRef]
- Cuevas, H.E.; Prom, L.K. Evaluation of genetic diversity, agronomic traits, and anthracnose resistance in the NPGS Sudan Sorghum Core collection. BMC Genom. 2020, 21, 88. [Google Scholar] [CrossRef]
- Sun, Y.; Dong, S.; Liu, Q.; Chen, J.; Pan, J.; Zhang, J. Selection of a core collection of Prunus sibirica L. germplasm by a stepwise clustering method using simple sequence repeat markers. PLoS ONE 2021, 16, e0260097. [Google Scholar] [CrossRef]
- Li, N.; Yang, Y.; Xu, F.; Chen, X.; Wei, R.; Li, Z.; Pan, W.; Zhang, W. Genetic Diversity and Population Structure Analysis of Castanopsis hystrix and Construction of a Core Collection Using Phenotypic Traits and Molecular Markers. Genes 2022, 13, 2383. [Google Scholar] [CrossRef]
- Guo, Q.; Liu, J.; Li, J.; Cao, S.; Zhang, Z.; Zhang, J.; Zhang, Y.; Deng, Y.; Niu, D.; Su, L.; et al. Genetic diversity and core collection extraction of Robinia pseudoacacia L. germplasm resources based on phenotype, physiology, and genotyping markers. Ind. Crops Prod. 2022, 178, 114627. [Google Scholar] [CrossRef]
- Frankel, O.H.; Brown, A.H.D. Current plant genetic resources—A critical appraisal. In Crop Genetic Resources: Conservation and Evaluation; Holden, J.H.W., Williams, J.T., Eds.; George Allan and Unwin: London, UK, 1984; Chapter 21; pp. 249–257. [Google Scholar]
- Charmet, G.; Balfourier, F.; Ravel, C.; Denis, J.B. Genotype x environment interactions in a core collection of French perennial ryegrass populations. TAG Theor. Appl. Genetics. Theor. Und Angew. Genet. 1993, 86, 731–736. [Google Scholar] [CrossRef]
- Gepts, P. Plant genetic resources conservation and utilization: The accomplishments and future of a societal insurance policy. Crop Sci. 2006, 46, 2278–2292. [Google Scholar] [CrossRef]
- Gu, R.; Fan, S.; Wei, S.; Li, J.; Zheng, S.; Liu, G. Developments on Core Collections of Plant Genetic Resources: Do We Know Enough? Forests 2023, 14, 926. [Google Scholar] [CrossRef]
- Razi, S.; Soleimani, A.; Zeinalabedini, M.; Vazifeshenas, M.R.; Martínez-Gómez, P.; Kermani, A.M.; Raiszadeh, A.R.; Tayari, M.; Martínez-García, P.J. Development of a Multipurpose Core Collection of New Promising Iranian Pomegranate (Punica granatum L.) Genotypes Based on Morphological and Pomological Traits. Horticulturae 2021, 7, 350. [Google Scholar] [CrossRef]
- Sivalingam, P.N.; Singh, D.; Chauhan, S.; Changal, H.K.; Bhan, C.; Mohapatra, T.; More, T.A.; Sharma, S.K. Establishment of the core collection of Ziziphus mauritiana Lam. from India. Plant Genet. Resour. 2014, 12, 140–142. [Google Scholar] [CrossRef]
- Li, Y.X.; Gao, Q.J.; Li, T.H. Sampling strategy based on fruit characteristics for a primary core collection of peach cultivars. J. Fruit Sci. 2006, 23, 359–364. [Google Scholar] [CrossRef]
- Hu, J.; Zhu, J.; Xu, H.M. Methods of constructing core collections by stepwise clustering with three sampling strategies based on the genotypic values of crops. Theor. Appl. Genet. 2000, 101, 264–268. [Google Scholar] [CrossRef]
- Tanksley, S.D.; Mccouch, S. Seed banks and molecular maps: Unlocking genetic potential from the wild. Science 1997, 277, 1063–1066. [Google Scholar] [CrossRef]
- Zane, L.; Bargelloni, L.; Patarnello, T. Strategies for microsatellite isolation: A review. Mol. Ecol. 2002, 11, 1–16. [Google Scholar] [CrossRef]
- Boccacci, P.; Aramini, M.; Ordidge, M.; Hintum, T.J.L.v.; Marinoni, D.T.; Valentini, N.; Sarraquigne, J.-P.; Solar, A.; Rovira, M.; Bacchetta, L.; et al. Comparison of selection methods for the establishment of a core collection using SSR markers for hazelnut (Corylus avellana L.) accessions from European germplasm repositories. Tree Genet. Genomes 2021, 17, 48. [Google Scholar] [CrossRef]
- Krichen, L.; Audergon, J.M.; Trifi-Farah, N. Relative efficiency of morphological characters and molecular markers in the establishment of an apricot core collection. Hereditas 2012, 149, 163–172. [Google Scholar] [CrossRef]
- Sun, Q.; Bai, L.; Ke, L.; Xiang, X.; Zhao, J.; Ou, L. Developing a core collection of litchi (Litchi chinensis Sonn.) based on EST-SSR genotype data and agronomic traits. Sci. Hortic. 2012, 146, 29–38. [Google Scholar] [CrossRef]
- Kumar, S.; Ambreen, H.; Variath, M.T.; Rao, A.R.; Agarwal, M.; Kumar, A.; Jagannath, A. Utilization of Molecular, Phenotypic, and Geographical Diversity to Develop Compact Composite Core Collection in the Oilseed Crop, Safflower (Carthamus tinctorius L.) through Maximization Strategy. Front. Plant Sci. 2016, 7, 1554. [Google Scholar] [CrossRef]
- Wang, X.; Cao, Z.; Gao, C.; Li, K. Strategy for the construction of a core collection for Pinus yunnanensis Franch. to optimize timber based on combined phenotype and molecular marker data. Genet. Resour. Crop Evol. 2021, 68, 3219–3240. [Google Scholar] [CrossRef]
- Zhen, Z.; Hanguo, Z.; Chi, M.; Lei, Z. Transcriptome Sequencing Analysis and Development of EST-SSR Markers for Pinus koraiensis. Sci. Silvae Sin. 2015, 51, 114–120. [Google Scholar] [CrossRef]
- Jianlu, M.; Liwen, Z.; Jingwen, L.; Dong, C. Geographic Distribution of Pinus koraiensis in The World. J. NorthEast For. Univ. 1992, 20, 9. [Google Scholar] [CrossRef]
- Zhang, Q.; Fan, S.-H.; Shen, H.-L. Research and Development on the Growth Environment of the Young Tree of Pinus koraiensis in Pinus koraiensis-Broadleaved Mixed Forest. For. Res. 2003, 16, 216–224. [Google Scholar] [CrossRef]
- Nergiz, C.; Dönmez, İ. Chemical composition and nutritive value of Pinus pinea L. seeds. Food Chem. 2004, 86, 365–368. [Google Scholar] [CrossRef]
- Yang, J.; Liu, R.H.; Halim, L. Antioxidant and antiproliferative activities of common edible nut seeds. LWT-Food Sci. Technol. 2009, 42, 1–8. [Google Scholar] [CrossRef]
- Zhen, Z.; Lei, Z.; Han-Guo, Z.; Ling, L.; Ying, J. Variations in Nutrition Compositions and Morphology Characteristics of Pine-Nuts in Korean Pine (Pinus koraiensis) Seed Orchard of Tieli. Bull. Bot. Res. 2014, 34, 356–363. [Google Scholar] [CrossRef]
- De’an, X.; Shuwen, Y.; Chuanping, Y.; Qingyou, L.; Guifeng, L.; Peigao, Z. A Study on The Provenance Test of Pinus koraiensis (Ⅰ) Preliminary Division of The Provenance. J. Northeast. For. Univ. 1991, 19, 7. [Google Scholar] [CrossRef]
- Yan, P.; Xie, Z.; Feng, K.; Qiu, X.; Zhang, L.; Zhang, H. Genetic diversity analysis and fingerprint construction of Korean pine (Pinus koraiensis) clonal seed orchard. Front. Plant Sci. 2023, 13, 1079571. [Google Scholar] [CrossRef]
- Zhen, Z.; Zhang, H.-G.; Zhang, L. Age Variations in Productivity and Family Selection of Open-Pollinated Families of Korean Pine (Pinus koraiensis). Bull. Bot. Res. 2016, 36, 305–309. [Google Scholar] [CrossRef]
- Qianping, T.; Limei, Y.; Lei, Z.; Yu, J.; Wenshu, M.; Jiali, Z.; Hanguo, Z. Genetic Diversity Analysis on Indivitual of Pinus koraiensis in Seed Orchard Based on ISSR-PCR. For. Sci. Technol. 2020, 45, 4. [Google Scholar] [CrossRef]
- Pingyu, Y.; Peng, W.; Weiman, Y.; Hanguo, Z. Analysis of Seeding Characters of Korean Pine Seed Orchard and Selection of Excellent Clones. For. Eng. 2020, 36, 11. [Google Scholar] [CrossRef]
- Qi-ping, H.; Ying, C. Comparision on Different Extaction Techniques about Chlorophyll and Determanation of Chlorophyll Content of Common Plants in Campus. Heilongjiang Agric. Sci. 2015, 38, 117–120. [Google Scholar] [CrossRef]
- Schuelke, M. An economic method for the fluorescent labeling of PCR fragments. Nat. Biotechnol. 2000, 18, 233–234. [Google Scholar] [CrossRef]
- Cun, C.; Chang-jun, D.; Qin-jun, H.; Zheng-hong, L.; Jing, Z.; Ning, L.; Bo, L.; Xiao-hua, S. Construction of Phenotypic Core Collection of Populus deltoides. For. Res. 2021, 34, 1–11. [Google Scholar] [CrossRef]
- Guo, W.-L.; Li, Y.-L.; Zhao, F.-C.; Tie, J.; Liao, F.-Y.; Zhong, S.-Y.; Lin, C.-M.; Ye, W.-F. Phenotypic Genetic Diversity of Pinus elliottii*P.caribaea Morelet var. hondurensis Clones. Bull. Bot. Res. 2019, 39, 259–266. [Google Scholar] [CrossRef]
- Chuanping, Y.; Shuwen, Y.; De’an, X.; Guifeng, L.; Qingyou, L.; Hongsheng, Z.; Peigao, Z. Study on The Geographic Variation Rule and Pattern of Growth Characters of Larix olgensis. J. Northeast. For. Univ. 1991, 19, 10. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. Genalex 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Raymond, M.; Rousset, F. GENEPOP (Version 1.2): Population Genetics Software for Exact Tests and Ecumenicism. J. Hered. 1995, 68, 248–249. [Google Scholar] [CrossRef]
- Liu, K.; Muse, S.V. PowerMarker: An integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef]
- Kim, K.-W.; Chung, H.-K.; Cho, G.-T.; Ma, K.-H.; Chandrabalan, D.; Gwag, J.-G.; Kim, T.-S.; Cho, E.-G.; Park, Y.-J. PowerCore: A program applying the advanced M strategy with a heuristic search for establishing core sets. Bioinformatics 2007, 23, 2155–2162. [Google Scholar] [CrossRef]
- Linfeng, Z.; Jianliang, G. Variance analyses of the growth and morphological traits of Chinese fir excellent clones. J. Cent. South Univ. For. Technol. 2022, 42, 8–15. [Google Scholar] [CrossRef]
- Basey, A.; Fant, J.B.; Kramer, A. Producing native plant materials for restoration: 10 rules to collect and maintain genetic diversity. Nativ. Plants J. 2015, 16, 37–53. [Google Scholar] [CrossRef]
- Turchetto, C.; Segatto, A.L.c.A.; Mäder, G.; Rodrigues, D.M.; Bonatto, S.L.; Freitas, L.B. High levels of genetic diversity and population structure in an endemic and rare species: Implications for conservation. Aob Plants 2016, 8, plw002. [Google Scholar] [CrossRef]
- Rongbo, J.; Jun, L.; Jingmin, J.; Chao, K.; Yanjie, L. Geographical Variation in Main Phenotypic Traits and Seedling Traits of Machilus thunbergii. J. Northeast. For. Univ. 2011, 39, 9–11. [Google Scholar]
- Yang, X.X.; Leng, P.S.; Zheng, J.; Hu, Z.H.; Liu, X.Y.; Yang, X.H.; Dou, D.Q. Variation of phenotypic traits of seed and seedling of Syringa reticulata subsp. amurensis from different provenances and their correlations with geographic-climatic factors. J. Plant Resour. Environ. 2016, 25, 80–89. [Google Scholar] [CrossRef]
- Fujuan, F.; Dongdong, Z.; Shijie, H. Genetic Diversity of Superior Clones from Pinus koraiensis Seed Orchard. J. Northeast. For. Univ. 2007, 35, 9–11. [Google Scholar]
- Odong, T.L.; Jansen, J.; van Eeuwijk, F.A.; van Hintum, T.J.L. Quality of core collections for effective utilisation of genetic resources review, discussion and interpretation. Theor. Appl. Genet. 2013, 126, 289–305. [Google Scholar] [CrossRef]
- Marita, J.M.; Rodriguez, J.M.; Nienhuis, J. Development of an algorithm identifying maximally diverse core collections. Genet. Resour. Crop Evol. 2000, 47, 515–526. [Google Scholar] [CrossRef]
- Schoen, D.J.; Brown, A.H. Conservation of allelic richness in wild crop relatives is aided by assessment of genetic markers. Proc. Natl. Acad. Sci. USA 1993, 90, 10623–10627. [Google Scholar] [CrossRef]
- Hanbo, Y.; Rui, Z.; Bangshun, W.; Zhaoyou, X.; Zhichun, Z. Construction of Core Collection of Schima superba Based on SSR Molecular Markers. Sci. Silvae Sin. 2017, 53, 37–46. [Google Scholar] [CrossRef]
- Mahmoodi, R.; Dadpour, M.R.; Hassani, D.; Zeinalabedini, M.; Vendramin, E.; Leslie, C.A. Composite core set construction and diversity analysis of Iranian walnut germplasm using molecular markers and phenotypic traits. PLoS ONE 2021, 16, e0248623. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.-C.; Zhang, H.-L.; Chao, Y.-S. Studies on the Sampling Strategy for Primary Core Collection of Chinese Ingenious Rice. Acta Agron. Sin. 2003, 29, 20–24. [Google Scholar]
- Chen, B.; Zhang, J.; Liu, G.; Li, S.; Gao, Y.; Li, H.; Li, T. Selection of Excellent Families and Evaluation of Selection Method for Pulpwood Half-sibling Families of Betula platyphylla. Bull. Bot. Res. 2023, 43, 690–699. [Google Scholar] [CrossRef]
- Huang, C.Q.; Long, T.; Bai, C.J.; Wang, W.Q.; Tang, J.; Liu, G.D. Establishment of a core collection of Cynodon based on morphological data. Trop. Grassl. Forrajes Trop. 2020, 8, 203–213. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).