Abstract
Plant species confirmation is a crucial step in using native plant species for forest restoration. To enhance this, a hybrid method of DNA barcoding and high-resolution melting analysis (Bar-HRM) was investigated in this study. In total, 12 native plant species samples were collected from forest restoration sites in Nan, a province in Northern Thailand. Simulation HRM analysis was performed to find the most appropriate region for in vitro Bar-HRM analysis. After that, in vitro Bar-HRM was carried out to validate the performance of native plant species. Results from both simulation and in vitro analyses revealed that the nuclear ribosomal internal transcribed spacer (ITS) region can be used as a primer set that can clearly discriminate native plant species in this study. With our study, Bar-HRM was proved of use in native plant species confirmation, even if that species had no molecular data available. In this context, Bar-HRM would be useful for the identification of native plant species used in tropical forest restoration not only in Thailand but also in any areas with similar plant groups.
1. Introduction
Tropical forests are the habitat of approximately two-thirds of our world’s flora and fauna, yet this type of forest covers less than 7% of the earth’s land surface [1,2]. Southeast Asia was reported to be home to around 15% of the world’s tropical forests [3]. There has been an unprecedented loss of tropical forests in the region because of population growth, infrastructure development, agricultural expansion, illegal logging, and uncontrolled forest fires [4,5,6]. The tropical forests of Southeast Asia are under immense pressure. This tropical forest region has lost a large proportion of its original forest cover and is now a deforestation hotspot [7]. Over the past two decades, Southeast Asia has lost 61 million hectares of forest; the annual loss was 4 million hectares a year on average from 2010 to 2019 [8]. Deforestation is a major problem in the region, with Indonesia in the lead, followed by other hotspots, such as Cambodia, Malaysia, Vietnam, and Thailand. The forest area in Thailand dramatically dropped from 70% in 1950 to 31% in 2018 [9]. Although all parts of Thailand are facing the same situation, the worst-affected area is the northern part of Thailand [10,11].
Over the past few decades, many countries have recognized the problem and put effort into forest conservation and restoration. One commonly used reforestation strategy is planting or seeding of native or introduced species [12,13]. Two main strategies of plantations have been established, which are monoculture and mixed-species plantations. Monocultures (a single species with the same genotype with almost no variation) have been used extensively in the tropics to reforest and are well documented in forest research. However, monocultural plantations have also been reported to have several negative social and environmental impacts despite their economic benefits [14,15]. Monoculture forests may only provide short-term economic benefits, with lower ecosystem service and biodiversity conservation benefits [16,17]. In contrast, mixed-species plantations based on native trees have been found to be more productive and sustainable plantation systems over monocultures [18,19,20]. Thus, mixed-species plantations using indigenous species are increasingly being considered for sustainable reforestation and used worldwide to restore disturbed and degraded areas [18,21,22,23,24]. Many native species have proven ability to grow well in deforested sites and have higher growth rates than introduced species [25]. Perhaps the key to the success of mixed-species plantations using native plants is for a system to rely on accurate species identification and an efficient propagation approach [25,26,27].
Therefore, plant species identification is important for forest conservation and management [28]. However, morphological-based identification normally requires experts and is time consuming. Moreover, several parts of the plants are needed for species-level identification, such as flower, leave, and fruit. So, it is practically impossible to do this when plants are in a vegetative or seedling phase [29]. Having a reliable and fast method for species identification would benefit the mixed-species plantations. Several procedures, such as seed-handling, germination pretreatments, and storage, are involved in the mix-species, and wrong identification of native species stock could be a waste of time and money [30]. Various native plant species were selected to be included in the forest restoration project in Nan province, Thailand, launched by the Forest Restoration Research Unit, Chiang Mai University (FORRU-CMU). The majority of the plant materials generated in the project are seeds and plantlets, which are then used for restoration so that conventional plant species identification techniques are inefficient. Lack of appropriate identification and characterization of the plant materials could lead to failure. Thus, what is required is a method that can be used to identify plant species at an early stage (e.g., seed and plantlet).
The most popular molecular species identification system is the use of DNA barcoding. Two DNA regions, rbcL and matK, were recommended by CBOL Plant Working Group as a universal barcode for plants [31]. In addition, the nuclear ribosomal internal transcribed spacer (ITS) is reported to be a good DNA barcode for species identification in several plant groups [32,33,34]. Recently, the combination of DNA barcoding and high-resolution melting analysis (HRM), called Bar-HRM, has been developed. Bar-HRM analysis has been used as a species authenticating method for herbal and agricultural products [35,36,37,38,39,40,41]. In addition, the method was used to identify a wide range of plant species [32,42,43,44]. Here, the Bar-HRM was evaluated for its performance in species identification of the native plants used for forest restoration in Nan province, Thailand. Results of this study will be useful for native plant species confirmation for tropical forest restoration and conservation.
2. Materials and Methods
2.1. Plant Samples and DNA Extraction
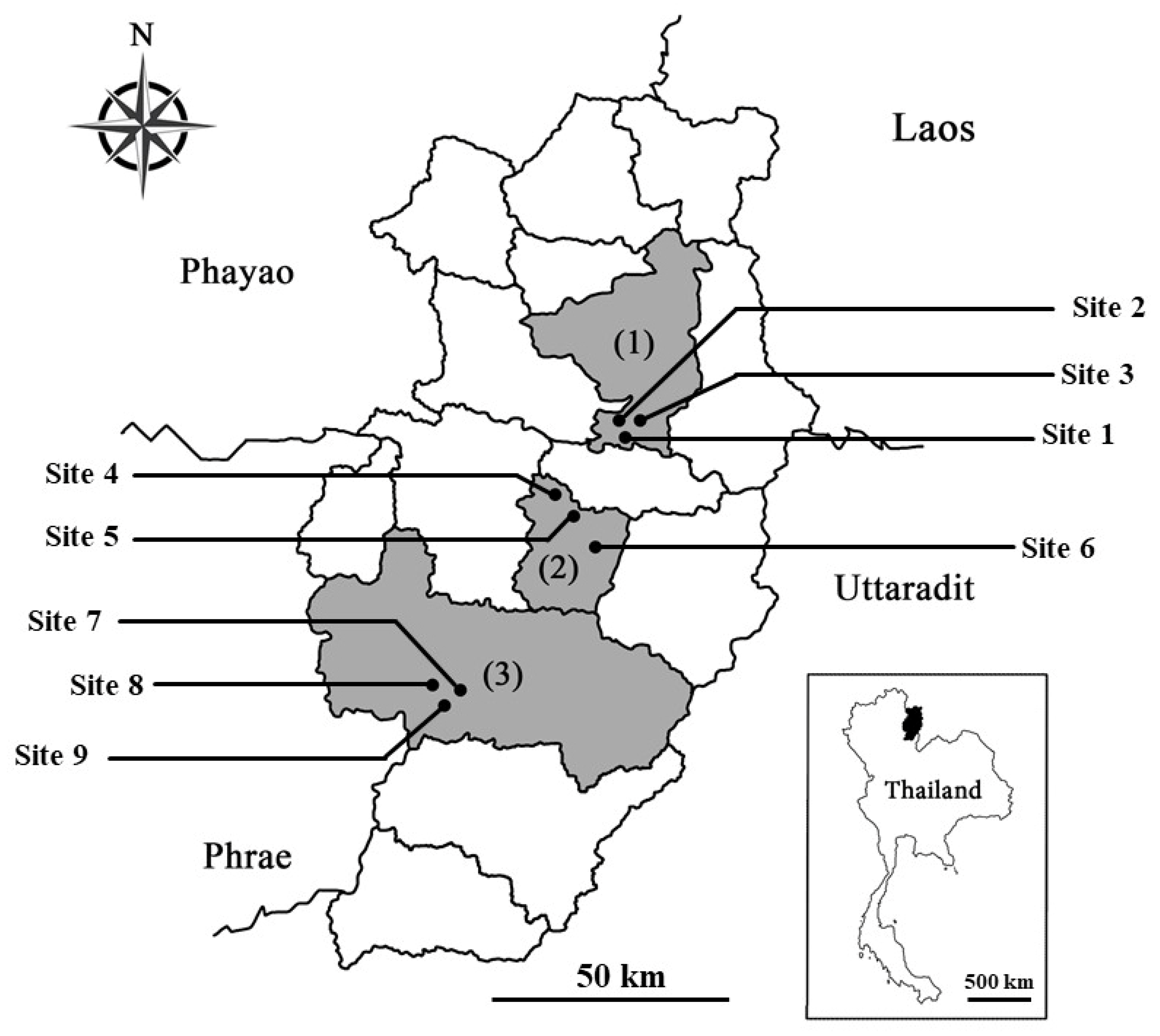
Plant tissue from 12 native species was collected from nine deforested sites in three districts (Pua, Phu Phiang, and Wiang Sa) of Nan province, Northern Thailand (Table 1 and Figure 1). Most of forest in Nan is mainly highland headwater mixed forest, which is an important watershed that feeds Thai people in many vital waterways. Chao Phraya River, the main river of Thailand, partly originates here. The nine selected sites were deforested due to agricultural in the past. Forest restoration is currently in progress here under the “From a bare mountain to a regenerated forest: comparing landscape planting design for forest restoration in NAN province” project, which was launched by the Forest Restoration Research Unit, Chiang Mai University (FORRU-CMU), in 2019.

Table 1.
Twelve native plants samples used in this study.
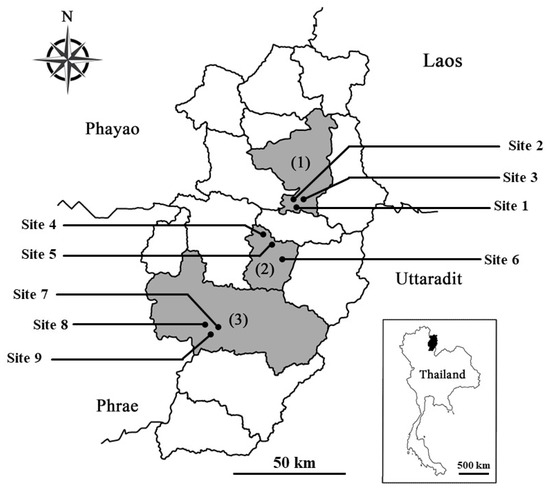
Figure 1.
Locations of the nine sampling sites (black circle) in (1) Pua, (2) Phu Phiang, and (3) Wiang Sa districts in Nan, Northern Thailand.
The plant tissues were ground with liquid nitrogen. DNA from all samples was extracted using the CTAB-chloroform solution protocol [45]. DNA concentrations were determined using the Qubit dsDNA HS Assay (Invitrogen, CA, USA). Various DNA concentrations of the samples were obtained and are recorded in Table 2. Final concentrations of the DNA solutions were adjusted to 120 ng/µL. The DNA solutions were stored at −20 °C for further use.

Table 2.
DNA concentration of each sample.
2.2. Data Mining
To address the most suitable markers for the identification of the tested species (Table 2) based on the Bar-HRM technique, a sequence dataset was constructed for sequence profile analysis. The sequences of the four selected regions (ITS, matK, rbcL, and trnL) of the 12 native species were retrieved from GenBank. Multiple alignments of the obtaining sequences were performed using MEGA 11 [46], and sequence length (bp) and variable sites (%) were recorded.
2.3. Simulated High-Resolution Melting Analysis
The retrieved sequences were 5′ and 3′ trimmed. Six DNA sequences or fragments were selected based on their availability in all selected DNA regions, to test the feasibility of Bar-HRM. To determine the melting profile of each region, simulated HRM analyses were performed using uMELT Quartz (melting prediction software) following the user guide [47]. The species included in the assay were N2, N7-N8, and N10-N12 (Table 3). The melting curves of the four chosen regions of the tested species were compared for their efficiency in species discrimination. The region with the best performance was then used in the next experiment.

Table 3.
Details of sequences retrieved from GenBank.
2.4. In Vitro High-Resolution Melting Analysis
To distinguish the tested species, the melting profile of each species was generated in HRM. The extracted DNA was amplified using the Rotor-Gene Q 5plex HRM system (Qiagen, Hilden, Germany). The reaction mixture for the HRM analysis consisted of a total volume of 10 µL, containing 4 µL of Evagreen HRM Master Mix, 0.2 µL of 10 mM forward primer, 0.2 µL of 10 mM reverse primer, 1 µL of 120 ng DNA, and 3.6 µL of ddH2O. The reaction conditions were as follows: an initial denaturing step at 95 °C for 5 min followed by 40 cycles at 95 °C for 30 s, 57 °C for 30 s, and 72 °C for 20 s. Melting curves were generated after the last extension step. Only the ITS primer pair was used in this experiment. The nucleotides of the forward primer (ITSF) are 5′-GGTGAACCTGCGGAAGGATCATTG-3′ and the reverse primer (ITSR) are 5′-CCGAGATATCCATTGCCGAGAGTC-3′. The temperature for the HRM analysis was increased from 60 to 95 °C at 0.1 °C/s. The negative derivative of the fluorescence (F) over temperature (T) (dF/dT) curve displays the Tm, and the normalized raw curve depicts the decreasing fluorescence vs. the increasing temperature.
3. Results
To find the most suitable DNA region for the identification of the 12 target species, their sequences were retrieved from GenBank. Not all target species have sequences deposited in the database. There are 11 of 12 species of the matK, rbcL, and trnL regions, whereas there are 10 for the ITS (Table 2). The Afzelia xylocarpa (N6) sequence of only the trnL region can be found in the database. However, 10 species (N1-N4 and N7-N12) contain sequences of all four selected regions (Table 3).
The sequence data of the four selected regions (ITS, matK, rbcL, and trnL) were analyzed. ITS led the charts for nucleotide variation (61.56%), followed by matK (41.99%), trnL (40.47%), and rbcL (21.56%), respectively (Table 4).

Table 4.
Characteristics of the analyzed sequences.
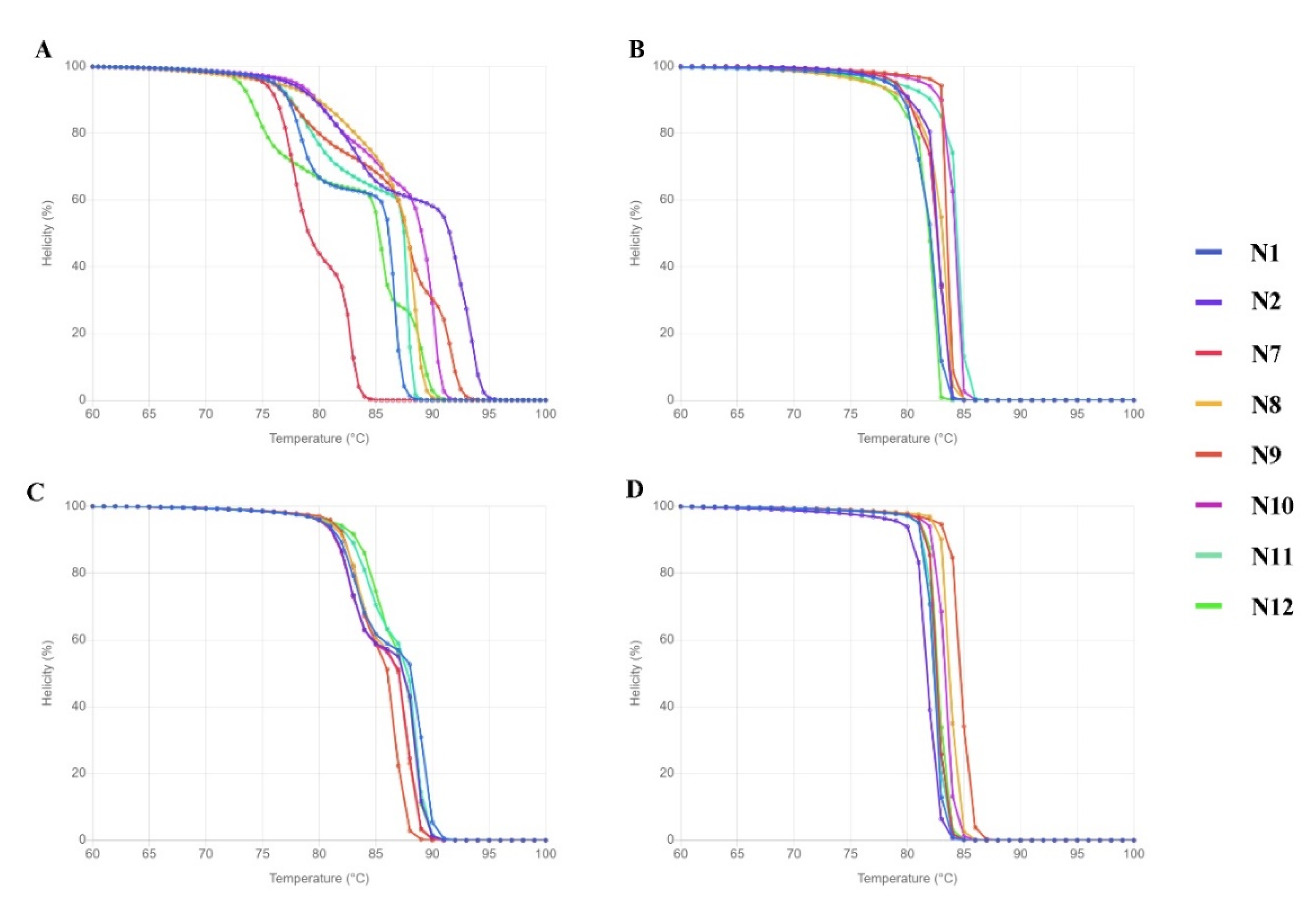
Before conducting an in vitro HRM, the DNA sequences were used in simulation HRM (uMELT Quartz) to predict the melting curves of each species. As can be seen in Figure 2, the melting curves of all the tested species were clearly distinguished only with the ITS primer set.
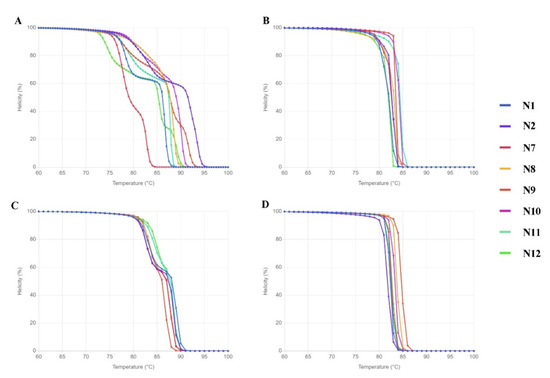
Figure 2.
Melting curve prediction from uMELT Quartz based on DNA sequences from the four selected regions. (A) ITS, (B) matK, (C) rbcL, and (D) trnL.
By containing the highest nucleotide variation (61.56%) and good performance in discriminating tested species in the simulation HRM, ITS is the most suitable for the task and thus only the ITS primer set was taken further to be used in an in vitro HRM assay. Although there were DNA samples of 12 native species (Table 2), only 9 were good enough for the HRM analysis: N1-N4, N6-N8, and N11-N12.
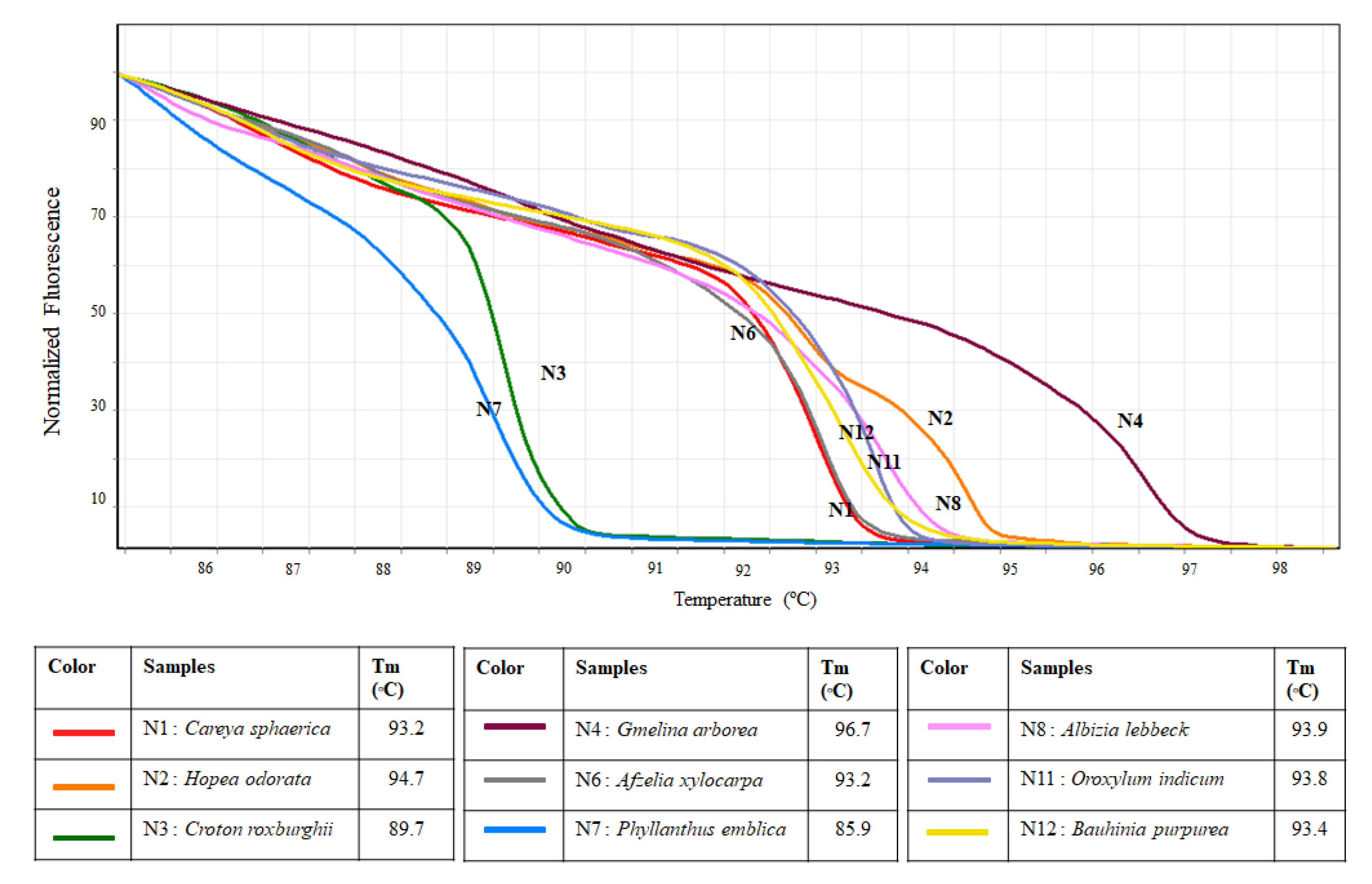
Results from the in vitro HRM analysis are similar to those obtained from the simulation. The ITS primer set can be used in HRM to separate the nine tested species as none of the melting curves of the tested species were similar (Figure 3). Melting temperatures of the tested species were also recorded, which range from 85.9 °C to 96.7 °C (Figure 3).
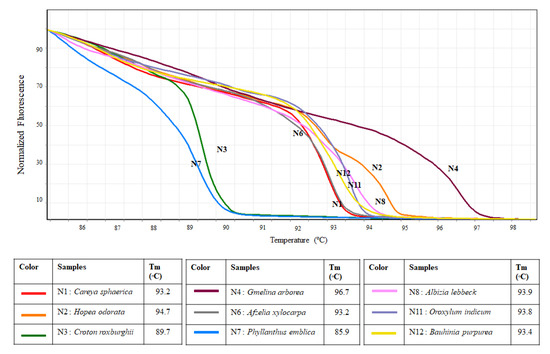
Figure 3.
Melting profiles of nine tested species generated from HRM analysis using ITS primers.
4. Discussion
Although DNA barcoding has been proven useful for the species-level identification of various plant groups, there are some limitations to the method. It is costly and time consuming and not easy to apply routinely in developing countries due to financial constraints for sequencing. The Bar-HRM is a free sequencing approach that is cost-effective for a large-scale study. Bar-HRM analysis has proven to be one good molecular approach for plant identification. Several DNA regions were used with Bar-HRM to identify a wide range of plant groups in previous studies, such as matK for flowering plants [42], trnL for bean crops [35] and nut products [36], rbcL for medicinal products from Acanthaceae species [37], ITS for Euphorbiaceae [38], and Dipterocarpaceae and Fagaceae [44]. It is therefore undoubtable that Bar-HRM would also be successful in identifying the tested species in this study.
Results from data mining and simulation HRM analysis indicate that the ITS is the most suitable region that can clearly discriminate the plant species in this study. As described in previous works, a key to success in HRM analysis is the variation in nucleotides within amplicon (more is better) [44,48]. The ITS as a non-coding region commonly exhibits higher nucleotide variations than coding regions, such as matK and rbcL [32,49].
In the in vitro HRM analysis, only 9 of the 12 samples were good enough for Bar-HRM analysis. Due to both quantity and quality, three DNA samples (Irvingia malayana, Chukrasia velutina, and Spondias mombin) were omitted from the analysis. A main process in HRM analysis involves measuring the fluorescence dye bound to double-stranded DNA in a reaction. Thus, the initial DNA concentration of all samples needs to be the same [50]. In addition, DNA concentration correlates with melting temperature (Tm) [51]. Therefore, if there are any differences in the DNA concentration of the analyzed samples, results of HRM analysis could be wrong or uninterpretable [50].
5. Conclusions
Higher growth rate and greater ecosystem services are the main advantages of native plant species over other plant species for forest restoration. However, use of native plant species in forest restoration involves some time-consuming steps, including species identification. A rapid and reliable identification method is thus required. Bar-HRM has proved to be a rapid and reliable method for plant species identification in this study. The choice of the DNA region plays an important role in the success of Bar-HRM, as discriminated efficiency was found to be varied among the primer pairs selected (ITS, matK, rbcL, and trnL). From the results shown here, a primer set based on the ITS region that exhibits the highest nucleotide variations is suitable for the identification of the tested native plant species. Thus, Bar-HRM with ITS primers could be used to confirm the species of plants used for reforestation.
Author Contributions
Conceptualization, M.O., P.M. and W.P.; methodology, M.O., P.M. and N.S.; formal analysis, M.O. and N.S.; data curation, M.O. and W.P.; writing original draft preparation, M.O. and W.P.; writing review and editing, M.O. and W.P.; funding acquisition, M.O. and W.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Chiang Mai University.
Data Availability Statement
DNA sequences with GenBank accessions are all included in the manuscript.
Acknowledgments
We thank the Forest Restoration Research Unit, Chiang Mai University (FORRU-CMU), for providing and identifying the samples. We acknowledge and are thankful to our colleagues for their help. We are also truly thankful to Lauren R. Clark for English editing.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bierregaard, R.O., Jr.; Lovejoy, T.E.; Kapos, V.; dos Santos, A.A.; Hutchings, R.W. The biological dynamics of tropical rainforest fragments: A prospective comparison of fragments and continuous forest. BioScience 1992, 42, 859–866. [Google Scholar] [CrossRef]
- DeFries, R.; Hansen, A.; Newton, A.C.; Hansen, M.C. Increasing isolation of protected areas in tropical forests over the past twenty years. Ecol. Appl. 2005, 15, 19–26. [Google Scholar] [CrossRef]
- Stibig, H.J.; Achard, F.; Carboni, S.; Rasi, R.; Miettinen, J. Change in tropical forest cover of Southeast Asia from 1990 to 2010. Biogeosciences 2014, 11, 247–258. [Google Scholar] [CrossRef]
- Achard, F.; Eva, H.D.; Stibig, H.J.; Mayaux, P.; Gallego, J.; Richards, T.; Malingreau, J.P. Determination of deforestation rates of the world’s humid tropical forests. Science 2002, 297, 999–1002. [Google Scholar] [CrossRef]
- Sodhi, N.S.; Posa, M.R.C.; Lee, T.M.; Bickford, D.; Koh, L.P.; Brook, B.W. The state and conservation of Southeast Asian biodiversity. Biodivers. Conserv. 2010, 19, 317–328. [Google Scholar] [CrossRef]
- Miettinen, J.; Shi, C.; Liew, S.C. Deforestation rates in insular Southeast Asia between 2000 and 2010. Glob. Chang. Biol. 2011, 17, 2261–2270. [Google Scholar] [CrossRef]
- Estoque, R.C.; Ooba, M.; Avitabile, V.; Hijioka, Y.; DasGupta, R.; Togawa, T.; Murayama, Y. The future of Southeast Asia’s forests. Nat. Commun. 2019, 10, 1829. [Google Scholar] [CrossRef]
- Feng, Y.; Ziegler, A.D.; Elsen, P.R.; Liu, Y.; He, X.; Spracklen, D.V.; Holden, J.; Jiang, X.; Zheng, C.; Zeng, Z. Upward expansion and acceleration of forest clearance in the mountains of Southeast Asia. Nat. Sustain. 2021, 4, 892–899. [Google Scholar] [CrossRef]
- Royal Forest Department. 2018. Forest Area of Thailand 1973–2018. Available online: http://forestinfo.forest.go.th/Content/file/stat2561/Binder1.pdf (accessed on 21 February 2022).
- Delang, C.O. Deforestation in northern Thailand: The result of Hmong farming practices or Thai development strategies? Soc. Nat. Resour. 2002, 15, 483–501. [Google Scholar] [CrossRef]
- Virapongse, A. Smallholders and forest landscape restoration in upland northern Thailand. Int. For. Rev. 2017, 19, 102–119. [Google Scholar] [CrossRef]
- Carnus, J.M.; Parrotta, J.; Brockerhoff, E.; Arbez, M.; Jactel, H.; Kremer, A.; Lamb, D.; O’Hara, K.; Walters, B. Planted forests and biodiversity. J. For. 2006, 104, 65–77. [Google Scholar]
- Nghiem, N.; Tran, H. The biodiversity benefits and opportunity costs of plantation forest management: A modelling case study of Pinus radiata in New Zealand. Forests 2016, 7, 297. [Google Scholar] [CrossRef]
- Erskine, P.D.; Lamb, D.; Bristow, M. Tree species diversity and ecosystem function: Can tropical multi-species plantations generate greater productivity? For. Ecol. Manag. 2006, 233, 205–210. [Google Scholar] [CrossRef]
- Alem, S.; Pavlis, J.; Urban, J.; Kucera, J. Pure and mixed plantations of Eucalyptus camaldulensis and Cupressus lusitanica: Their growth interactions and effect on diversity and density of undergrowth woody plants in relation to light. Open J. For. 2015, 5, 375–386. [Google Scholar]
- Moore, S.E.; Allen, H.L. Plantation forestry. In Maintaining Biodiversity in Forest Ecosystems; Hunter, M.L., Ed.; Cambridge University Press: New York, NY, USA, 1999; pp. 400–433. [Google Scholar]
- Baral, H.; Guariguata, M.R.; Keenan, R.J. A proposed framework for assessing ecosystem goods and services from planted forests. Ecosyst. Serv. 2016, 22, 260–268. [Google Scholar] [CrossRef]
- Manson, D.G.; Schmidt, S.; Bristow, M.; Erskine, P.D.; Vanclay, J.K. Species-site matching in mixed species plantations of native trees in tropical Australia. Agrofor. Syst. 2013, 87, 233–250. [Google Scholar] [CrossRef]
- Potvin, C.; Dutilleul, P. Neighborhood effects and size asymmetric competition in a tree plantation varying in diversity. Ecology 2009, 90, 321–327. [Google Scholar] [CrossRef]
- Petit, B.; Montagnini, F. Growth in pure and mixed plantations of tree species used in reforesting rural areas of the humid region of Costa Rica, Central America. For. Ecol. Manag. 2006, 233, 338–343. [Google Scholar] [CrossRef]
- McNamara, S.; Tinh, D.V.; Erskine, P.D.; Lamb, D.; Yates, D.; Brown, S. Rehabilitating degraded forest land in central Vietnam with mixed native species plantings. For. Ecol. Manag. 2006, 233, 358–365. [Google Scholar] [CrossRef]
- Raman, T.; Mudappa, D.; Kapoor, V. Restoring rainforest fragments: Survival of mixed-native species seedlings under contrasting site conditions in the western Ghats, India. Restor. Ecol. 2009, 17, 137–147. [Google Scholar] [CrossRef]
- Ashton, M.S.; Gunatilleke, C.V.S.; Gunatilleke, I.A.U.N.; Singhakumara, B.M.P.; Gamage, S.; Shibayama, T.; Tomimura, C. Restoration of rain forest beneath pine plantations: A relay floristic model with special application to tropical South Asia. For. Ecol. Manag. 2014, 329, 351–359. [Google Scholar] [CrossRef]
- Elliott, S.; Chairuangsri, S.; Cherdsak, K.; Sangkum, S.; Sinhaseni, K.; Shannon, D.; Nippanon, P.; Manohan, B. Collaboration and conflict-developing forest restoration techniques for Northern Thailand’s upper watersheds whilst meeting the needs of science and communities. Forests 2019, 10, 732. [Google Scholar] [CrossRef]
- Shono, K.; Davies, S.J.; Chua, Y. Performance of 45 native tree species on degraded lands in Singapore. J. Trop. For. Sci. 2007, 19, 25–34. [Google Scholar]
- Meli, P.; Martínez-Ramos, M.; Rey-Benayas, J.M.; Carabias, J. Combining ecological, social, and technical criteria to select species for forest restoration. Appl. Veg. Sci. 2014, 17, 744–753. [Google Scholar] [CrossRef]
- Kremer, K.N.; Bauhus, J. Drivers of native species regeneration in the process of restoring natural forests from mono-specific, even-aged tree plantations: A quantitative review. Restor. Ecol. 2020, 28, 1074–1086. [Google Scholar] [CrossRef]
- Margules, C.R.; Pressey, R.L. Systematic conservation planning. Nature 2000, 405, 243. [Google Scholar] [CrossRef]
- Forman, L.L. Trigonobalanus a new genus of Fagaceae with notes on the classification of the family. Kew Bull. 1964, 17, 381–396. [Google Scholar] [CrossRef]
- Francis, J.F. Collection. In Tropical Tree Seed Manual. Agriculture Handbook Number 721; Vozzo, J.A., Ed.; United States Department of Agriculture Forest Service: Washington, DC, USA, 2003; pp. 119–124. [Google Scholar]
- CBOL Group. A DNA barcode for land plants. Proc. Natl. Acad. Sci. USA 2009, 106, 12794–12797. [Google Scholar] [CrossRef]
- Chen, S.; Yao, H.; Han, J.; Liu, C.; Song, J.; Shi, L.; Zhu, Y.; Ma, X.; Gao, T.; Pang, X.; et al. Validation of the ITS2 Region as a Novel DNA Barcode for Identifying Medicinal Plant Species. PLoS ONE 2010, 5, e8613. [Google Scholar] [CrossRef]
- Hollingsworth, P.M. Refining the DNA barcode for land plants. Proc. Natl. Acad. Sci. USA 2011, 108, 19451–19452. [Google Scholar] [CrossRef]
- China Plant BOL Group; Li, D.-Z.; Gao, L.-M.; Li, H.-T.; Wang, H.; Ge, X.-J.; Liu, J.-Q.; Chen, Z.-D.; Zhou, S.-L.; Chen, S.-L.; et al. Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc. Natl. Acad. Sci. USA 2011, 108, 19641–19646. [Google Scholar] [PubMed]
- Madesis, P.; Ganopoulos, I.; Anagnostis, A.; Tsaftaris, A. The application of Bar-HRM (Barcode DNA-High Resolution Melting) analysis for authenticity testing and quantitative detection of bean crops (Leguminosae) without prior DNA purification. Food Control 2012, 25, 576–582. [Google Scholar] [CrossRef]
- Madesis, P.; Ganopoulos, I.; Bosmali, I.; Tsaftaris, A. Barcode High Resolution Melting analysis for forensic uses in nuts: A case study on allergenic hazelnuts (Corylus avellana). Food Res. Int. 2013, 50, 351–360. [Google Scholar] [CrossRef]
- Osathanunkul, M.; Madesis, P.; de Boer, H. Bar-HRM for Authentication of Plant-Based Medicines: Evaluation of Three Medicinal Products Derived from Acanthaceae Species. PLoS ONE 2015, 10, e0128476. [Google Scholar] [CrossRef]
- Osathanunkul, M.; Suwannapoom, C.; Ounjai, S.; Rora, J.A.; Madesis, P.; de Boer, H. Refining DNA barcoding coupled high resolution Melting for discirmination of 12 closely related Croton species. PLoS ONE 2015, 10, e0138888. [Google Scholar] [CrossRef]
- Osathanunkul, M.; Suwannapoom, C.; Khamyong, N.; Pintakum, D.; Lamphun, S.; Triwitayakorn, K.; Osathanunkul, K.; Madesis, P. Hybrid analysis (barcode-high resolution melting) for authentication of Thai herbal products, Andrographis paniculata (Burm.f.) Wall. ex Nees. Pharmacogn. Mag. 2016, 12, 71–75. [Google Scholar] [CrossRef][Green Version]
- Singtonat, S.; Osathanunkul, M. Fast and reliable detection of toxic Crotalaria spectabilis Roth. in Thunbergia laurifolia Lindl. herbal products using DNA barcoding coupled with HRM analysis. BMC Complement. Altern. Med. 2015, 15, 162. [Google Scholar] [CrossRef]
- Ganopoulos, I.; Madesis, P.; Darzentas, N.; Argiriou, A.; Tsaftaris, A. Barcode High Resolution Melting (Bar-HRM) analysis for detection and quantification of PDO “Fava Santorinis” (Lathyrus clymenum) adulterants. Food Chem. 2012, 133, 505–512. [Google Scholar] [CrossRef]
- Lahaye, R.; Van der Bank, M.; Bogarin, D.; Warner, J.; Pupulin, F. DNA barcoding the floras of biodiversity hotspots. Proc. Natl. Acad. Sci. USA 2008, 105, 2923–2928. [Google Scholar] [CrossRef]
- Ballin, N.Z.; Onaindia, J.O.; Jawad, H.; Fernandez-Carazo, R.; Maquet, A. High-resolution melting of multiple barcode amplicons for plant species authentication. Food Control 2019, 105, 141–150. [Google Scholar] [CrossRef]
- Osathanunkul, M.; Madesis, P. The identification of several Dipterocarpaceae and Fagaceae trees by barcode DNA coupled with high-resolution melting analysis. Forests 2021, 12, 1466. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure from small quantities of fresh leaf tissues. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Dwight, Z.; Palais, R.; Wittwer, C.T. uMELT: Prediction of high-resolution melting curves and dynamic melting profiles of PCR products in a rich web application. Bioinformatics 2011, 27, 1019–1020. [Google Scholar] [CrossRef]
- Wittwer, C.T.; Reed, G.H.; Gundry, C.N.; Vandersteen, J.G.; Pryor, R.J. High-Resolution Genotyping by Amplicon Melting Analysis Using LCGreen. Clin. Chem. 2003, 49, 853–860. [Google Scholar] [CrossRef]
- Kress, W.J.; Erickson, D.L. A two-locus global DNA barcode for land plants: The coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS ONE 2007, 2, e508. [Google Scholar] [CrossRef]
- Slomka, M.; Sobalska-Kwapis, M.; Wachulec, M.; Bartosz, G.; Strapagiel, D. High Resolution Melting (HRM) for high-throughput genotyping—Limitations and caveats in practical case studies. Int. J. Mol. Sci. 2017, 18, 2316. [Google Scholar] [CrossRef]
- Ng, J.W.S.; Holt, D.C.; Andersson, P.; Giffard, P.M. DNA concentration can specify DNA melting point in a High-Resolution Melting analysis master mix. Clin. Chem. 2014, 60, 414–416. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).