Abstract
Anomoloma is a cosmopolitan poroid wood-decaying genus, belonging to the Amylocorticiales. During a study on polypores, two new species of Anomoloma were found in Eurasia, and they are described as A. denticulatum and A. eurasiaticum. To examine the phylogenetic relationships among species of Anomoloma, we analyzed nuclear ribosomal sequence data from the ITS regions and the LSU gene. The result demonstrates that A. denticulatum and A. eurasiaticum are independent species that belong to the Anomoloma genus. Both new species share the principal characteristics of the genus, but Anomoloma denticulatum is characterized by extensive white rhizomorphs spreading under the whole fruiting body, angular pores measuring 1–2 per mm, distinctly lacerate to dentate dissepiments and basidiospores of 3.5–4.3 × 2–2.5 μm. Anomoloma eurasiaticum is characterized by bearing plenty of large crystals on the mycelia and growth on Picea in high altitude areas. A key to the accepted species of Anomoloma worldwide is provided.
1. Introduction
Wood-inhabiting fungi are a phylogenetically diverse assemblage of macromycetes and comprise important components of woodlands and some other terrestrial ecosystems. Through their effective enzymatic systems causing wood decomposition, they play a crucial role in the global carbon and nutrient cycles [1,2]. Among the most significant groups of wood-inhabiting macromycetes are polypore fungi. The characterization of additional species in this group would improve our understanding of the decayer fungal community as a whole.
Anomoloma Niemelä & K.H. Larss. is a cosmopolitan wood-inhabiting fungal genus and its species cause a white rot in their wood substrata [3,4]. The genus is typified by Anomoloma albolutescens (Romell) Niemelä & K.H. Larss, and resides in the order Amylocorticiales, and the family Amylocorticiaceae. Anomoloma is distinguished from other similar genera by resupinate, soft, white, cream, or yellow basidiocarps with rhizomorphs, a monomitic hyphal structure with clamp connections, smooth and amyloid basidiospores, and causing a white rot on very rotten angiosperm and gymnosperm wood [3,4,5]. Previously, Anomoloma was treated as Anomoporia Pouzar due to their similar microscopic characteristics [6]. However, Anomoloma was separated from Anomoporia based on the wood decay type and molecular evidence, the former causing a white rot, while the latter causes a brown rot [5]. Four species were accepted when Anomoloma was established, and two species from subtropical China were described recently [4]. Currently, Anomoloma accommodates six species worldwide: A. albolutescens, A. flavissimum (Niemelä) Niemelä & K.H. Larss., A. myceliosum (Niemelä) Niemelä & K.H. Larss., A. rhizosum Y.C. Dai & Niemelä, A. luteoalbum J. Song & B.K. Cui and A. submyceliosum J. Song & B.K. Cui.
Anomoloma has simple morphological structures; sometimes it is difficult to find characteristics to distinguish the species in the genus. Molecular phylogenies supply solid evidence to define species of the genus [4], a similar phenomenon has also been found in other polypores [7,8].
In the course of the exploration of polypores in the East Himalayan area of China, four white, and resupinate specimens were collected from Yunnan and Sichuan Provinces, and their morphological features, taxonomic affinities and phylogenetic relationships were analyzed. Combining these with samples collected from Europe and North America, the taxonomy and phylogeny of the genus here are updated. Two species are confirmed as new within Anomoloma, and they are simultaneously described and illustrated. In addition, an identification key to the accepted species of Anomoloma is provided.
2. Materials and Methods
2.1. Morphological Studies
The analyzed materials were preserved in the herbarium of the Beijing Forestry University (BJFC) in Beijing, China and the National Museum Prague of the Czech Republic (PRM). The macromorphology of the materials was determined according to our photographs in situ and the measurements of voucher materials. The micromorphological study followed Li et al. [9]. For the data on basidiospores, 5% of measurements treated at extreme measured values were excluded and are shown in parentheses. L = arithmetic average length of basidiospores, W = arithmetic average width of spores, Q = variation in the L/W ratios between the voucher specimens examined, n (a/b) = number of spores (a) measured from a given number (b) of specimens. Color terms refer to Petersen [10]. The herbarium code refer to Thiers [11].
2.2. DNA Extraction and Sequencing
A phylogeny based on the combined ITS and LSU DNA sequence data was analyzed with a total of 44 specimens including 20 species of Anomoloma related genera. A CTAB rapid plant genome extraction kit (Aidlab Biotechnologies Co., Ltd., Beijing, China) was applied in obtaining PCR products from dried materials, following the manufacturer’s instructions with some adjustments [12]. To generate PCR amplicons, the following primer pair of ITS5 and ITS4 was used for the ITS (the internal transcribed spacer) region: ITS1, partial sequence; the 5.8S ribosomal RNA gene, complete sequence; and ITS2, partial sequence [13]. The primer pair for LR0R and LR7 was used for the LSU region (large subunit of nuclear ribosomal RNA gene) and included the 28S ribosomal RNA gene following Hopple and Vilgalys [14]. The PCR thermoprofile for different DNA sequences used in this study followed those used in Song et al. [4] The PCR procedure for ITS was as follows: initial denaturation at 95 °C for 3 min, followed by 34 cycles at 94 °C for 40 s, 54 °C for ITS and 72 °C for 1 min, and a final extension of 72 °C for 10 min. The PCR procedure for LSU was as follows: initial denaturation at 94 °C for 1 min, followed by 34 cycles at 94 °C for 30 s, 50 °C for 1 min and 72 °C for 1.5 min, and a final extension of 72 °C for 10 min. The PCR products were purified at the Beijing Genomics Institute (BGI), China. The purified products were then sequenced on an ABI-3730-XL DNA Analyzer (Applied Biosystems, Foster City, CA, USA). The quality of sequence was analysed following Nilsson et al. [15]. All new sequences were submitted to GenBank [16] and the related sequences of representative species were downloaded from GenBank, which belong to the target genus Anomoloma and allied genera (Table 1).

Table 1.
Taxa, voucher specimens and GenBank accession numbers of sequences used in the phylogeny of Anomoloma.
2.3. Phylogenetic Analysis
The obtained sequences were aligned by BioEdit 7.0.5.3 [19]. After automatic alignment using Clustal X1.83 [20], the alignment was manually adjusted. The sequence alignment and the tree file were uploaded to TreeBase (submission ID 29582). The sequence of Hypochniciellum subillaqueatum (Litsch.) Hjortstam downloaded from GenBank was used as an outgroup in the phylogenetic reconstruction of Anomoloma (Figure 1).
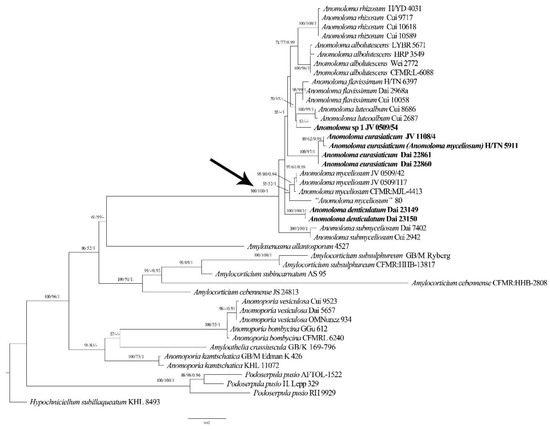
Figure 1.
Maximum likelihood phylogenetic tree of Anomoloma species on the basis of ITS + LSU dataset. Branches are labeled with bootstrap values (MP/ML) >50%, and posterior probabilities (BI) > 0.90 respectively. New taxa are in bold. An arrow pointed the Anomoloma clade.
For the ultimate phylogenetic analyses, maximum parsimony (MP), maximum likelihood (ML) and Bayesian inference (BI) methods were utilized based on the ITS region and LSU sequence data. The three phylogenetic analysis algorithms outputted similar topologies to each other. Thus, the topology of the ML analysis is displayed as a representation with statistical values from the above three algorithms. The most parsimonious phylogenies were based on the combined ITS and LSU data, and their combinability was evaluated with the incongruence length difference (ILD) test [21] performed in PAUP* 4.0b10 [22]. The phylogenetic analysis approaches followed Zhao et al. [23]. The parsimony tree construction procedure was performed in PAUP* version 4.0b10 [22]. Trees were inferred using 100 replicates of the random stepwise addition of sequences and the tree-bisection reconnection (TBR) branch-swapping algorithm, with all characters given equal weight. The branch supports for all parsimony analyses were estimated by performing 1000 bootstrap (BT) replicates [24]. The descriptive tree statistics tree length (TL), consistency index (CI), retention index (RI), rescaled consistency index (RC), and homoplasy index (HI) were calculated for each maximum parsimonious tree (MPT) generated.
In the ML and BI analysis, jModeltest v.2.17 [25] was utilized to screen the best-fitting substitution model. Four unique partitions were established, and GTR + I + G was the selected substitution model for each partition. RaxmlGUI 1.2 [26,27] was applied in the ML analysis. All parameters in the ML analysis were kept at default settings. Statistical support values were obtained from non-parametric bootstrapping with 1000 replicates.
For the BI analysis implemented in MrBayes 3.2.6 [28], there were two independent runs—each of which had four chains for 2,000,000 generations sampling from the posterior distribution every 1000th generation to check that the PSRF (potential scale reduction factors) were reasonably close to 1.0 for all parameters indicative of chain convergence. The first 25% of the sampled trees were discarded as burn-in, while the remaining trees were used to obtain the Bayesian posterior probabilities (BPPs) of the clades.
The trees were shown in TreeView [29]. Branches that gained a bootstrap support for MP (BS) and ML(BP) values and BPPs (Bayesian posterior probabilities for BI) simultaneously of no less than 75% (BS/BP) and 0.95 (BPPs) were considered as significantly supported.
3. Results
3.1. Molecular Phylogeny
The combined ITS and LSU dataset contained 44 sequences representing 20 taxa. The dataset had an aligned length of 2083 characters including gaps (724 characters for ITS, 1359 characters for LSU), of which 1479 characters were constant, 197 were variable and parsimony-uninformative, and 407 were parsimony-informative. The MP analysis yielded 90 trees (TL = 1112, CI = 0.719, RI = 0.855, RC = 0.615, HI = 0.281). The best-fitting substitution model for the combined ITS + LSU dataset was evaluated and applied in the Bayesian analysis was GTR + I + G. The Bayesian analyses exported a nearly identical topology to the ML analyses with an average standard deviation of split frequencies = 0.009428. Both the BT values (≥50%) and BPPs (≥0.90) are displayed on the branches of the ML tree (Figure 1).
The resulting phylogenetic tree resolved a strongly supported Anomoloma clade and the newly sequenced taxa are nested in this clade. Judging from the molecular phylogenies, two new lineages of Anomoloma with robust support were identified. In addition, four samples of Anomoloma eurasiaticum formed an independent lineage with a robust support. JV 1108/4 and H/TN 5911 share the same ITS sequences, and the ITS sequences of Dai 22860 and Dai 22861 are identical, only two base pairs differences are found in the ITS regions between the former two and the latter two samples.
3.2. Taxonomy

Figure 2.
A basidiocarp of Anomoloma denticulatum (Dai 23150). Scale bar = 1.0 cm.
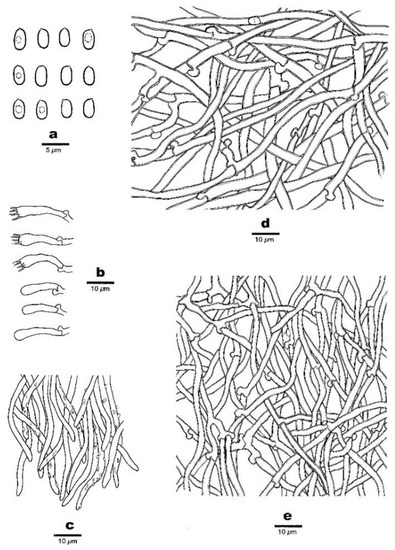
Figure 3.
Micro-morphology of Anomoloma denticulatum (Dai 23150). (a) Basidiospores. (b) Basidia and basidioles. (c) Hyphae at dissepiment edge. (d) A section of subiculum. (e) A section of trama.
MycoBank: MB843478.
Diagnosis—Anomoloma denticulatum is characterized by a white pore surface with extensive white rhizomorphs spread under the whole fruiting body, angular pores measuring 1–2 per mm, distinctly lacerate to dentate dissepiments, ellipsoid basidiospores of 3.5–4.3 × 2–2.5 μm, and growth on decayed wood of Abies in the East Himalayan area.
Holotype—China, Sichuan, Luding County, Hailuogou National Forest Park, E 101°59′, N 29°35′, alt. 3050 m, on rotten wood of Abies, 8 October 2021, Yu-Cheng Dai, Dai 23150 (BJFC 037721).
Etymology—Denticulatum (Lat.): refer to the lacerate to dentate dissepiments of the species.
Description
Fruiting body—The basidiocarps are resupinate, annual, felty, soft corky, have no special odor or taste when fresh and dry, and are 11 cm × 4 cm and 0.2 mm thick. The sterile margin is distinct, snow-white, about 1 cm, radiciform and threadlike, with white rhizomorphs which extensively spread under the whole fruiting body, are visible in the poroid surface, and penetrate into rotten wood. The pores are cream when fresh and dry, angular, and 1–2(–3)/mm; dissepiments are rather thin, distinctly lacerate to dentate. The subiculum is white when fresh and dry, felty, and about 0.1 mm thick. The tubes are white to cream when fresh and dry, soft corky, and about 0.1 mm long.
Hyphal structure—The hyphal system is monomitic; the clamped generative hyphae are neither amyloid nor dextrinoid in Melzer’s reagent, and faintly cyanophilous in Cotton Blue; the tissues are unchanged in 5% potassium hydroxide.
Subiculum—The hyaline generative hyphae are slightly thick-walled with a wide lumen, smooth, frequently branched, flexuous, loosely interwoven, and 3–5 μm in diameter.
Tubes—The hyaline generative hyphae are thin- to slightly thick-walled with a wide lumen, frequently branched, flexuous, loosely interwoven, and 1.5–3 μm in diameter. Hyphae at the dissepiment edge are sometimes encrusted by fine and hyaline crystals. The basidia are clavate, bearing 4-sterigmata with a basal clamp connection, and 15–27 × 3.5–5 μm; the basidioles are of a similar shape but shorter.
Basidiospores—The basidiospores are oblong-ellipsoid to ellipsoid, hyaline, thin- to slightly thick-walled, smooth, amyloid in Melzer’s reagent, acyanophilous, and (3.4−)3.5–4.3(−4.5) × 2–2.5(−2.7) μm, L = 3.84 μm, W = 2.15 μm, Q = 1.75–1.82 (n = 60/2).
Additional specimen (paratype) studied—China, Sichuan, Luding County, Hailuogou National Forest Park, E 101°59′, N 29°35′, alt. 3050 m, on rotten wood of Abies, 8 October 2021, Yu-Cheng Dai, Dai 23150 (BJFC 037720).

Figure 4.
A basidiocarp of Anomoloma eurasiaticum (Dai 22860). Scale bar = 1.0 cm.
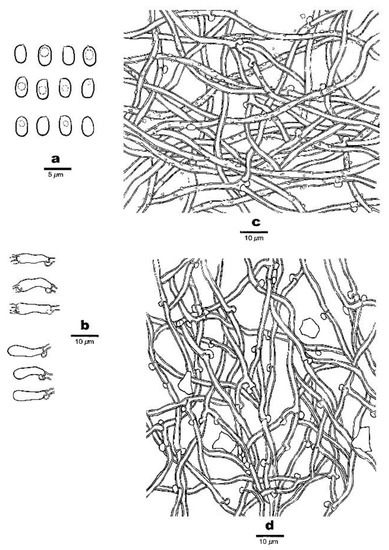
Figure 5.
Micro-morphology of Anomoloma eurasiaticum (Dai 22860). (a) Basidiospores. (b) Basidia and basidioles. (c) A section of subiculum. (d) A section of trama.
MycoBank: MB843480.
Diagnosis—Anomoloma eurasiaticum is characterized by a white pore surface with rhizomorphs, angular pores measuring 3–5 per mm, oblong-ellipsoid to ellipsoid basidiospores of 3.2–4 × 2–2.8 μm, plenty of large crystals dispersed in mycelia, and growth on rotten gymnosperm wood in China and Europe.
Holotype—China, Yunnan, Deqin County, Baima Snow Mountain National Nature Reserve, E 98°50′, N 28°41′, alt. 3750 m, on rotten wood of Picea, 5 September 2021, Yu-Cheng Dai, Dai 22860 (BJFC 037433).
Etymology—Eurasiaticum (Lat.): refer to a distribution in Eurasia.
Description
Fruiting body—The basidiocarps are resupinate, annual, felty, soft corky, of no special odor or taste when fresh and dry, 6 cm × 4 cm, and 0.3 mm thick. The sterile margin is distinct, snow-white, thinning out, about 0.5 cm, radiciform and threadlike, with white rhizomorphs which arise from the margin penetrating into rotten wood. The pores are white to cream when fresh, becoming cream to cream buff when dry, angular, and 3–5/mm; dissepiments are thin and slightly lacerate. The subiculum is white when fresh and dry, felty, and about 0.1 mm thick. The tubes are white when fresh, cream when dry, soft corky, and about 0.2 mm long.
Hyphal structure—The hyphal system is monomitic; the clamped generative hyphae are neither amyloid nor dextrinoid in Melzer’s reagent and faintly cyanophilous in Cotton Blue; tissues are unchanged in 5% potassium hydroxide.
Subiculum—The hyaline generative hyphae are slightly thick-walled with a wide lumen, frequently branched, flexuous, loosely interwoven, bearing plenty of fine crystals, and 2–3 μm in diameter.
Tubes—The hyaline generative hyphae are slightly thick-walled with a wide lumen, frequently branched, flexuous, loosely interwoven, 1.5–2.5 μm in diameter. The basidia are long, barrel-shaped, sometimes constricted at the middle, bearing 4-sterigmata and a basal clamp connection, and 13–20 × 3.5–5 μm; the basidioles are short clavate and shorter than basidia. Some large crystals present in the tube trama.
Basidiospores—The basidiospores are oblong-ellipsoid to ellipsoid, hyaline, thin- to slightly thick-walled, smooth, amyloid in Melzer’s reagent, acyanophilous, and (3.1−)3.2–4(−4.1) × 2–2.8(−3) μm, L = 3.49 μm, W = 2.34 μm, Q = 1.42–1.50 (n = 90/3).
Additional specimen (paratype) studied: China, Yunnan, Deqin County, Baima Snow Mountain National Nature Reserve, E 98°50′, N 28°41′, alt. 3750 m, on rotten wood of Picea, 5 September 2021, Yu-Cheng Dai, Dai 22860 (BJFC 037434); Czechia, Hluboka, Libochovka, E 16°14′, N 49°23′, on rotten wood of Picea, August 2011, Josef Vlasák, JV1108/4 (PRM, duplicate, BJFC038577).
4. Discussion
According to the phylogenetic analysis based on the ITS + LSU dataset, the monophyly of the genus Anomoloma was re-confirmed with robust supports (ML/MP = 100, BI = 1.00). Ten lineages representing ten taxa are nested in the Anomoloma clade (Figure 1). Two new species, Anomoloma denticulatum and A. eurasiaticum, form two well-supported linages within the Anomoloma clade. Interestingly the European and North American “A. myceliosum” are nested in two different linages. The type locality of Anomoloma myceliosum is North America, and the samples JV 0509/42 and JV 0509/117 from the USA and CFMR: MJL-4413 from Canada most probably represent the real A. myceliosum. As Vlasák et al. [30] mentioned, the European “Anomoloma myceliosum” is probably a different species from the North American. In our study, the sample H/TN 5911 from Finland previously identified as Anomoloma myceliosum and sample JV1108/4 from Czechia are nested within the lineage of A. eurasiaticum with robust support. Thus, the samples of previously so-called Anomoloma myceliosum in Europe may represent A. eurasiaticum.
Sample 80, treated as “Anomoloma myceliosum”, was collected in Altai in Russia. We failed to obtain the specimen, but it forms an independent linage and is closer to A. myceliosum (Figure 1). However, there is a 15-base-pair difference between the sequence of the Russian sample and the North American sample of Anomoloma myceliosum, which accounts for >2% of the nucleotides in the ITS regions. Thus, it is possible that another species of Anomoloma exists in Central Asia, but for the time being we treat this as “Anomoloma myceliosum”.
Although the sample JV0509/54 from the USA forms an independent lineage within the Anomoloma clade in our phylogeny (Figure 1), it is temporarily treated as Anomoloma sp. 1 here because of the single sample. The taxon is characterized by a yellow pore surface with rhizomorphs spreading along the surface, angular, irregular pores of 3–4 per mm, a lack of cystidia and cystidioles, oblong-ellipsoid to ellipsoid basidiospores of 3.5–4 × 2.2–2.8 μm, and growth on the rotten wood of Tsuga in southeast USA.
On the basis of our study, the main characteristics of Anomoloma species are listed in Table 2.

Table 2.
A morphological comparison of species in Anomoloma.
In our phylogenetic analysis (Figure 1), Anomoloma denticulatum was nested in the Anomoloma clade as an independent lineage. Morphologically, Anomoloma denticulatum is similar to the white-pore species of A. eurasiaticum, A. myceliosum, and A. submyceliosum. However, Anomoloma eurasiaticum can be distinguished from A. denticulatum by its smaller pores (3–5/mm vs. 1–2/mm), slightly wider basidiospores (L = 3.49 μm, W = 2.34 μm, Q = 1.42–1.50 vs. L = 3.84 μm, W = 2.15 μm, Q = 1.78–1.82), and narrower subicular hyphae with minute hyaline crystals (2–3 µm vs. 3–5 µm). Anomoloma myceliosum is different from A. denticulatum in its wider basidiospores (L = 3.48 μm, W = 2.51 μm, Q = 1.34–1.46 vs. L = 3.84 μm, W = 2.15 μm Q = 1.78–1.82) and hyphae producing an oily substance with inconspicuous guttules [5,6]. In addition, the white rhizomorphs of Anomoloma myceliosum arise from the basidiocarp margin [5,6], but the rhizomorphs of A. denticulatum spread under the whole fruiting body. Anomoloma submyceliosum differs from A. denticulatum in its subicular hyphae covered with fine hyaline crystals, and its growth on angiosperm wood in subtropical areas [4].
Referring to the phylogenetic analysis of the ITS + LSU dataset, four samples of Anomoloma eurasiaticum form a well-supported lineage (ML = 100, MP = 97, BI = 1.00) within the Anomoloma clade. Morphologically, Anomoloma eurasiaticum may be recognized as A. myceliosum and A. submyceliosum by sharing a similar white-colored poroid surface. However, Anomoloma myceliosum differs from A. eurasiaticum in its ellipsoid or broadly ovate basidiospores (3–4.1 × 2.2–2.9 µm, Q = 1.34–1.46), bearing fragile, very thin-walled tramal hyphae and an oily hyphae tip with inconspicuous guttules [5,6]. Anomoloma submyceliosum can be differentiated from A. eurasiaticum by its larger pores (2–3/mm vs. 3–5/mm) and wider subicular hyphae (2.2–4.5 µm vs. 2–3 µm), and its growth on angiosperm wood in subtropical areas [4].
Previously, numerous macrofungi have been described from southwest China [31,32], and the present paper confirms that more undescribed wood-inhabiting fungi are present in the montane forests of the East Himalayas.
Other specimens studied: Anomoloma myceliosum. USA, Tennessee, Great Smoky Mt. on Tsuga, IX.2005, Josef Vlasák, JV0509/117 (dupl. BJFC038579); IX.2007, JV0509/42 (dupl. BJFC038581). Anomoloma sp 1. USA, Tennessee, Great Smoky Mt. on Tsuga, IX.2005, Josef Vlasák, JV0509/54 (dupl. BJFC038578).
Key to Accepted Species of Anomoloma
| 1 Pore surface and rhizomorphs white | 5 |
| 1 Pore surface and rhizomorphs yellowish to yellow | 2 |
| 2 Pores 2–4 per mm | A. albolutescens |
| 2 Pores 4–6 per mm | 3 |
| 3 Basidiospores usually >4 μm in length | A. rhizosum |
| 3 Basidiospores usually <4 μm in length | 4 |
| 4 Pore surface bright chrome or sulphur yellow; vesicular cystidia present | A. flavissimum |
| 4 Pore surface cream to yellowish; vesicular cystidia absent | A. luteoalbum |
| 5 Growth on angiosperm wood, occurrence in subtropical area | A. submyceliosum |
| 5 Growth on gymnosperm wood, occurrence in temperate to boreal areas | 6 |
| 6 Basidiospores ellipsoid or broadly ovate, Q = 1.34–1.46 | A. myceliosum |
| 6 Basidiospores oblong-ellipsoid to ellipsoid, Q >1.5 | 7 |
| 7 Pores 1–2 per mm, basidiospores Q = 1.75–1.82 | A. denticulatum |
| 7 Pores 3–5 per mm, basidiospores Q = 1.42–1.51 | A. crystallinum |
Author Contributions
Conceptualization, Y.-C.D. and M.Z.; methodology, M.Z.; performing the experiment, M.Z.; formal analysis, M.Z.; validation, M.Z., Y.-C.D., J.V. and M.G.-N.; resources, Y.-C.D. and J.V.; writing—original draft preparation, M.Z.; writing—review and editing, M.Z., Y.-C.D., J.V. and M.G.-N.; visualization, M.Z.; supervision, Y.W.L., Y.-C.D. and M.G.-N.; project administration, Y.W.L. and Y.-C.D.; funding acquisition, Y.-C.D., M.G.-N., J.V. and Y.W.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the National Natural Science Foundation of China (project nos. 32161143013, 32011540380, U1802231), the institutional support of the Academy Sciences of the Czech Republic RVO: 60077344, the exchange project between Korea and China for Young Woon Lim (National Research Foundation, project no. NRF-2020K2A9A2A06047605), and by the Iran National Science Foundation (project no. 4000655).
Data Availability Statement
Publicly available datasets were analyzed in this study. These data can be found here: (https://www.ncbi.nlm.nih.gov/; https://www.mycobank.org/page/Simple%20names%20search; http://purl.org/phylo/treebase, submission ID 29582) (accessed on 15 March 2022).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Rayner, A.D.M.; Boddy, L. Fungal decomposition of wood: Its biology and ecology. Q. Rev. Biol. 1988, 73, 1796–1803. [Google Scholar] [CrossRef]
- White, N.A. The importance of wood-decay fungi in forest ecosystems: Fungal biotechnology in agriculture, food and environmental applications. In Fungal Biotechnology in Agricultural, Food, and Environmental Applications; Arora, D.K., Bridge, P.D., Bhatnagar, D., Eds.; Marcel Dekker: New York, NY, USA, 2003; pp. 375–392. [Google Scholar]
- Binder, M.; Larsson, K.H.; Matheny, P.B.; Hibbett, D.S. Amylocorticiales ord. nov and Jaapiales ord. nov.: Early diverging clades of Agaricomycetidae dominated by corticioid forms. Mycologia 2010, 102, 865–880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, J.; Liu, X.Y.; Wang, M.; Cui, B.K. Phylogeny and taxonomy of the genus Anomoloma (Amylocorticiales, Basidiomycota). Mycol. Prog. 2016, 15, 11. [Google Scholar] [CrossRef]
- Niemelä, T.; Larsson, K.H.; Dai, Y.C.; Larsson, E. Anomoloma, a new genus separated from Anomoporia on the basis of decay type and nuclear rDNA sequence data. Mycotaxon 2007, 100, 305–317. [Google Scholar]
- Niemelä, T. Five species of Anomoporia, rare polypores of old forests. Ann. Bot. Fenn. 1994, 31, 93–115. [Google Scholar]
- Yuan, Y.; Chen, J.J.; Korhonen, K.; Martin, F.; Dai, Y.C. An updated global species diversity and phylogeny in the forest pathogenic genus Heterobasidion (Basidiomycota, Russulales). Front. Microbiol. 2021, 11, 596393. [Google Scholar] [CrossRef]
- Song, J.; Sun, Y.F.; Ji, X.; Dai, Y.C.; Cui, B.K. Phylogeny and taxonomy of Laetiporus (Basidiomycota, Polyporales) with descriptions of two new species from western China. MycoKeys 2018, 37, 57–71. [Google Scholar] [CrossRef]
- Li, H.J.; Cui, B.K.; Dai, Y.C. Taxonomy and multi-gene phylogeny of Datronia (Polyporales, Basidiomycota). Persoonia 2014, 32, 170–182. [Google Scholar] [CrossRef] [Green Version]
- Petersen, J.H. The Danish Mycological Society’s Colour-Chart; Foreningen til Svampekundskabens Fremme: Greve, Denmark, 1996; pp. 1–6. [Google Scholar]
- Thiers, B. Index Herbariorum: A Global Directory of Public Herbaria and Associated Staff; New York Botanical Garden’s Virtual Herbarium: New York, NY, USA, 2018; Available online: http://sweetgum.nybg.org/science/ih/ (accessed on 15 March 2022).
- Chen, J.J.; Cui, B.K.; Zhou, L.W.; Korhonen, K.; Dai, Y.C. Phylogeny, divergence time estimation, and biogeography of the genus Heterobasidion (Basidiomycota, Russulales). Fungal Divers. 2015, 71, 185–200. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gefand, D.H., Sninsky, J.J., White, M.J.T., Eds.; Academic Press: San Diego, FL, USA, 1990; pp. 315–322. [Google Scholar] [CrossRef]
- Hopple, J.S.J.; Vilgalys, R. Phylogenetic relationships in the mushroom genus Coprinus and dark-spored allies based on sequence data from the nuclear gene coding for the large ribosomal subunit RNA: Divergent domains, outgroups, and monophyly. Mol. Phylogenet. Evol. 1999, 13, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Nilsson, R.H.; Tedersoo, L.; Abarenkov, K.; Ryberg, M.; Kristiansson, E.; Hartmann, M.; Schoch, C.L.; Nylander, J.A.A.; Bergsten, J.; Porter, T.M.; et al. Five simple guidelines for establishing basic authenticity and reliability of newly generated fungal ITS sequences. MycoKeys 2012, 4, 37–63. [Google Scholar] [CrossRef] [Green Version]
- Sayers, E.W.; Cavanaugh, M.; Clark, K.; Pruitt, K.D.; Schoch, C.L.; Sherry, S.T.; Karsch-Mizrachi, I. GenBank. Nucleic Acids Res. 2022, 50, 161–164. Available online: https://www.ncbi.nlm.nih.gov/ (accessed on 15 March 2022). [CrossRef]
- Larsson, K.H.; Larsson, E.; Kljalg, U. High phylogenetic diversity among corticioid homobasidiomycetes. Mycol. Res. 2004, 108, 983–1002. [Google Scholar] [CrossRef] [PubMed]
- Matheny, P.B.; Curtis, J.M.; Hofstetter, V. Major clades of Agaricales: A multi-locus phylogenetic overview. Mycologia 2007, 98, 984–997. [Google Scholar]
- Hall, T.A. Bioedit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [Green Version]
- Farris, J.S.; Mari, K.; Kluge, A.G.; Bult, C. Testing significance of incongruence. Cladistics 1994, 10, 315–319. [Google Scholar] [CrossRef]
- Swofford, D.L. PAUP*: PHYLOGENETIC Analysis Using Parsimony (*and Other Methods); Version 4.0b10; Sinauer Associates: Sunderland, MA, USA, 2002. [Google Scholar] [CrossRef]
- Zhao, C.L.; Chen, H.; Song, J.; Cui, B.K. Phylogeny and taxonomy of the genus Abundisporus (Polyporales, Basidiomycota). Mycol. Prog. 2015, 14, 38. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence intervals on phylogenetics: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [Green Version]
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analysis with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef] [PubMed]
- Silvestro, D.; Michalak, I. RaxmlGUI: A Graph. Front-End RAxML. Org. Divers. Evol. 2012, 12, 335–337. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Hőhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Page, R.D.M. TreeView: Application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. Cabios 1996, 12, 357–358. [Google Scholar] [CrossRef] [Green Version]
- Vlasák, J.; Vampola, P.; Kout, J. New record of Anomoloma myceliosum in the Czech Republic. Mykol. Listy 2012, 119, 1–5. [Google Scholar]
- Dai, Y.C.; Yang, Z.L.; Cui, B.K.; Wu, G.; Yuan, H.S.; Zhou, L.W.; He, S.H.; Ge, Z.W.; Wu, F.; Wei, Y.L.; et al. Diversity and systematics of the important macrofungi in Chinese forests. Mycosystema 2021, 40, 770–805. [Google Scholar] [CrossRef]
- Wang, K.; Chen, S.L.; Dai, Y.C.; Jia, Z.F.; Li, T.H.; Liu, T.Z.; Phurbu, D.; Mamut, R.; Sun, G.Y.; Bau, T.; et al. Over-view of China’s nomenclature novelties of fungi in the new century (2000–2020). Mycosystema 2021, 40, 822–833. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).