Abstract
Research Highlights: This study assesses the genetic diversity and structure of the ice-cream-bean (Inga edulis Mart.; Fabaceae) in wild and cultivated populations from the Peruvian Amazon. This research also highlights the importance of protecting the biodiversity of the forest in the Peruvian Amazon, to preserve the genetic resources of species and allow further genetic improvement. Background and Objectives: Ice-cream-bean is one of the most commonly used species in the Amazon region for its fruits and for shading protection of other species (e.g., cocoa and coffee plantations). Comprehensive studies about the impact of domestication on this species’ genetic diversity are needed, to find the best conservation and improvement strategies. Materials and Methods: In the current study, the genetic structure and diversity were assessed by genotyping 259 trees, sampled in five wild and 22 cultivated I. edulis populations in the Peruvian Amazon, with microsatellite markers. Pod length was measured in wild and cultivated trees. Results: The average pod length in cultivated trees was significantly higher than that in wild trees. The expected genetic diversity and the average number of alleles was higher in the wild compared to the cultivated populations; thus, a loss of genetic diversity was confirmed in the cultivated populations. The cultivated trees in the Loreto region had the highest pod length and lowest allelic richness; nevertheless, the wild populations’ genetic structure was not clearly differentiated (significantly different) from that of the cultivated populations. Conclusions: A loss of genetic diversity was confirmed in the cultivated populations. The species could have been simultaneously domesticated in multiple locations, usually from local origin. The original I. edulis Amazonian germplasm should be maintained. Cultivated populations’ new germplasm influx from wild populations should be undertaken to increase genetic diversity.
1. Introduction
The Peruvian rainforest, due to its large and relatively continuous area of primary forest, is a worldwide biodiversity hotspot, which is suffering intense disturbance and deforestation from human exploitation and global change [1]. Amazonian inhabitants have used these natural resources through millennia and modified the natural environment, but how human management practices resulted in Amazonian forests’ domestication is not known, in particular the germplasm source [2]. Moreover, the species’ gene pool could have been reduced due to farmers’ selection, thus strategies for genetic resource conservation and management are needed [3]. The Peruvian rainforest that remains a large and relatively continuous area of primary forest has major conservation value and is considered a priority in nearly all global biodiversity inventories, due to its biodiversity and the disturbance and deforestation rates [4,5]. Despite the international recognition of its major conservation value resulting from its uniqueness and global importance, the impacts of human activities throughout the region remain poorly understood [5]. Indeed, information about the species’ genetic structure will assist in tree breeding programmes and conservation strategies, in particular in tropical trees, and, also, to study the implications of human impact on genetic resources [6,7].
The genus Inga (Fabaceae) comprises ca. 300 species of neotropical rainforest trees [8], but earlier studies suggest that the diversification of Inga species in Amazonia is recent, during the past 2–10 million years [9,10]. Ice-cream-bean (Inga edulis Mart.; Fabaceae) is a lowland rainforest light-demanding species, distributed in Colombia and tropical South America to the east of the Andes, extending from south to north-western Argentina (Figure 1). The species’ natural altitudinal range is mostly below 750 m a.s.l., though it has been occasionally identified at 1200 m in Roraima, Brazil [8]. The flowers are hermaphrodite and the pollination is provided by hawkmoths, bats and hummingbirds that may carry the pollen grains across large areas [11]. The species is diploid, 2n = 26 [12], and it is believed to be self-incompatible [13]. Fruiting occurs, in three year-old trees, as a long pod containing recalcitrant seeds, covered by a white, fleshy and slightly sweet edible sarcotesta [14]. Seed dispersal is performed by mammals and birds after eating the sarcotesta [11,15]. The species is widely cultivated for its edible fruit throughout South and Central America [16]. It is one of the most widely distributed and economically useful species in the whole Amazon region [14,17]. In Amazonian Peru, the fruits from cultivated trees may exceed 2 m in length and 5–6 cm in diameter. The wild trees have smaller pods than cultivated trees, rarely exceeding 50 cm in length [8]. This fast growing, symbiotic nitrogen-fixing tree, with umbrella-like canopy, is commonly used as a shade tree for cocoa, coffee, coca and tea plantations, in agroforestry systems and in “home grown” multi-purpose cultivation uses [8].
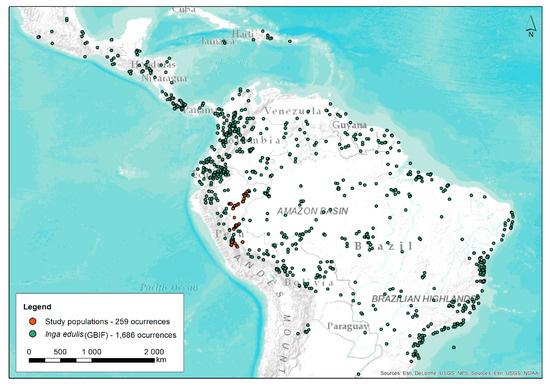
Figure 1.
Distribution map of Inga edulis. The green dots represent the 1686 occurrences, and the red dots the trees sampled in the current study (259 occurrences). GBIF.org (10th October 2018) GIBF Occurrence. Download: https:doi.org/10.15468/dl.ik3uki.
Amazonia was a major centre of crop domestication, with at least 83 native species containing populations domesticated to some degree, which expanded rapidly in the Mid-Holocene [18], including the Inga genus. The historical records for Inga edulis show that this species has been cultivated in Peru for its edible fruit since the pre-Colombian time and has become a commonly used tree species in the Amazon region [19]. The origin of the cultivated populations of I. edulis is uncertain [8]; however, León [17] and Clement [20] claimed West Amazonia as a probable origin. The species’ genetic structure was not studied in detail, yet a reduction of allelic richness in cultivated relative to natural populations was found in I. edulis from the Peruvian Amazon [13,21]. Inga edulis has become a model species to evaluate the maintenance of genetic resources in agroforestry systems, and the putative genetic diversity reduction associated with domestication [22]. More recently, Cruz-Neto et al. [23], using microsatellite markers, observed high levels of genetic diversity within I. vera populations from the Atlantic forest of north-eastern Brazil. They concluded that cultivated populations compared to natural populations displayed reduced genetic diversity. Nevertheless, maintaining high levels of genetic variation within agroforestry trees is important for two main reasons: genetic variation in agricultural landscapes helps farmers to manage their inputs in more efficient ways and because they provide the ability for tree species to adjust to new environments, such as the shifting climate and weather conditions, allowing local adaptation and the migration of better-suited provenances along ecological gradients [3]. In addition, a stronger emphasis on the genetic quality of the trees planted by smallholders is needed, which means paying attention both to domestication and to the systems by which improved germplasm is delivered to farmers for the management of tree genetic resources and the livelihoods of rural communities in the tropics [24].
In the present study, the objectives were to (i) explore differences in pod length between wild trees and cultivated I. edulis trees from different geographical regions in the Peruvian Amazon; (ii) compare the wild and cultivated I. edulis populations’ genetic structure using microsatellite markers; and (iii) determine if the cultivated populations’ genetic structure reflects the different uses and cultivation practices throughout the species’ use history, to help design practical measures to preserve I. edulis genetic resources.
2. Materials and Methods
2.1. Plant Material Sampling
The leaves and mature fruits from 259 individuals and 27 populations of I. edulis were sampled in the Peruvian Amazon, between 2009 and 2012. Each tree was identified according to the morphological aspects detailed by Rollo et al. [25] and, additionally, with the help of locals. Each sampled tree’s young leaves were preserved in micro test-tubes with silica gel for further DNA extraction. One to ten mature pods were sampled per tree, from opposite sides and different heights of the crown, according to the availability of mature fruits on the tree. The pod length was measured from the base to the top of the pod apex. The mature fruits had the following phenological characteristics: seeds from creamy white to purple black up to vivipary; and the sarcotesta membranous creamy to generally flashy white, watery, soft and slightly sweet [8]. A total of 448 mature pods were measured in the 259 trees from the 27 I. edulis populations. We chose to study the pod length, as we can draw a null hypothesis based on the domestication process, since the trees were selected for their pod length (H0: Is the pod length of the domesticated trees higher than that of the wild trees?). Indeed, this trait has economic importance in the species domestication and agroforestry value.
Each sampled tree’s geographical coordinates were recorded, and the minimum distance between any two trees was 200 m. Voucher specimens were kept in the Regional Herbarium of Ucayali IVITA-Pucallpa, Peru, with the code AR1-384. Each population was numbered from 1 to 27 and coded, e.g., 1 SRc, 23 RPw (the two capital letters are taken from the initial letters of the geographic origin of the population, e.g., San Ramón or River Pacaya, and the third letter meaning either c = cultivated—managed by humans or w = wild—growing spontaneously) (Table 1).

Table 1.
The sampling region (Site), population code (Pop.), sample size (N), geographic location (GPS coordinates in WGS84; latitude S and longitude W) and altitude (in metres above sea level) of the 27 sampled Inga edulis populations (cultivated and wild).
Cultivated trees were sampled in 22 geographically different populations, in home gardens and agricultural landscapes surrounding the urban areas in the Selva Central, Ucayali and Loreto regions. Wild trees were sampled in five geographically different populations, in lowland forests; four populations (23 RPw, 24 RSw, 26 MAw, 27 SDw) in protected natural areas in original forest vegetation and one (25 RUw) in secondary forest as described in Rollo et al. [25]. Details on the sampled populations are displayed in Table S1. Finally, no I. edulis wild trees were found in the Selva Central region’s original vegetation, since the species is a lowland rainforest species and has been only occasionally recorded above 750 m [8].
2.2. DNA Extraction and Amplification
Total DNA was extracted from dried young leaves, using the Invitek, Invisorb® Spin Plant Mini Kit following the manufacturer’s instructions. Four microsatellite markers were used to genotype all the individuals, Pel5 [26] and Inga03, Inga08 and Inga33 [21]. For microsatellite detection, each forward primer was fluorescently labelled at the 5′ end (6-FAM, NED or VIC). Amplification conditions were performed according to the conditions described by Rollo et al. [25]. The amplified products were separated on an ABI PRISM 310 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA) and ran according to the manufacturer’s protocol. Fragment sizes were determined using the ROX500 internal size standard and the global southern algorithm implemented by ABI PRISM GeneMapper® software version 4.0 (Applied Biosystems).
2.3. Data Analysis
2.3.1. Morphological Data
The pod length was measured in the 448 pods sampled from the 259 trees of the I. edulis populations from the Peruvian Amazon, and all the individual values were used in the following analysis (not averaged per tree). The pod lengths’ normality (in both cultivated populations originated in Selva Central, Ucayali and Loreto regions and in wild populations) was tested using the Kolmogorov–Smirnov test, and all the groups displayed normal distribution except for the wild trees group. A non-parametric Kruskal–Wallis test (k independent samples) was performed to check for significant differences in the groups’ average followed by the non-parametric Mann–Whitney U post-hoc test [27]. The statistical analyses were performed using the IBM SPSS v.22 statistics software.
2.3.2. Molecular Data
The estimated genetic diversity parameters included the average number of alleles per locus (A), the effective number of alleles (Ne), the number of private alleles (Pa), the mean allelic richness (RS) that uses a rarefaction index to consider differences in sample size [28], the observed (HO) and expected (HE) heterozygosities [29] and the fixation index FIS. Comparisons of the genetic diversity parameters between groups (i.e., cultivated and wild populations) were performed with 10,000 permutations. The estimates were made using Fstat 2.9.3.2 [30] and GenAlEx v. 6.501 [31]. Genepop 4.3 software [32] was used to test the heterozygote deficiency for each population and to compute the average frequency of null alleles.
Genetic variation at the level of populations and groups (i.e., cultivated and wild populations) was investigated with a hierarchical analysis of molecular variance (AMOVA), which partitions the total variance into covariance components due to inter-group differences, inter-populations within groups differences and inter-population differences, in the Arlequin software [33]. Levels of significance were determined by computing 1000 random permutation replicates.
A Bayesian clustering method was performed in the STRUCTURE v.2.3.4. software [34] to infer population genetic structure. The number of genetic clusters (K) was estimated and the individuals sampled from cultivated and wild populations were fractionally assigned to the inferred groups. Afterwards, the allele frequencies were estimated in each of the K groups as well as the proportion of the genome derived from each group for each tree. We applied the model allowing population admixture and correlated allele frequency [34]. However, due to the weak population structure found in the I. edulis populations, we used a model that incorporated a priori sampling location information [35], i.e., a “locprior” model. This improved model has the advantage of allowing cryptic structures to be detected at a lower level of divergence and does not bias towards detecting structures spuriously when none is present, helpful in situations when the standard structure models do not provide a clear signal of structure [35]. Two groups of populations were used as priors, i.e., cultivated and wild populations (see Table 1). The alternative ancestry prior 1/K was used due to unbalanced population sampling [36]. The number of clusters (K) was set from one through twenty-seven and the simulation was run ten times at each K value to confirm the repeatability of the results. Each run comprised a burn-in period of 25,000, followed by 100,000 Markov chain Monte Carlo (MCMC) steps. We used the ΔK distribution statistic of Evanno et al. [37] to determine the most appropriate number of genetic clusters. Hence, the STRUCTURE output data were parsed using the STRUCTURE HARVESTER [38] to determine the optimal K value following the method referred to above. Alignment of cluster assignments across replicate analyses was then conducted in CLUMPP 1.1.2 [39] and subsequently visualized using STRUCTURE PLOT [40]. The results of Bayesian clustering were further mapped in the ArcGIS® Desktop version 10.2 software [41].
3. Results
3.1. Pod Length
From a total of 259 individual trees, 448 fruits were collected and measured: 329 and 119 fruits from 197 cultivated trees and 62 wild trees, respectively. The longest pod (148 cm) was found in 18 EDc, a cultivated population in the Loreto region around El Dorado village (Table S2). The Kruskal–Wallis non-parametric test, using the four groups of populations (regions), showed significant group differences for pod length (Figure 2). The Mann–Whitney U post-hoc test produced three homogeneous groups, which indicated that the pod length average (78 cm) in the Selva Central region was not significantly different (p < 0.05) from the Ucayali region’s 80 cm long average, and both values were significantly different from the Loreto value (90 cm) (Figure 2).
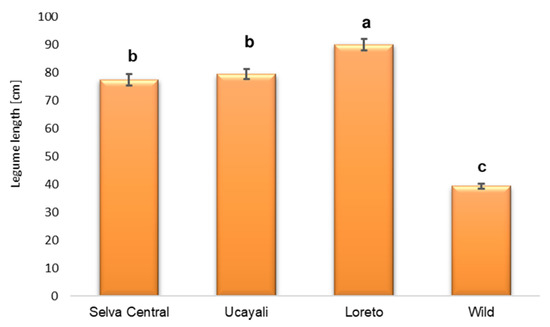
Figure 2.
Pod length comparison among the cultivated and wild populations. From Selva Central, 94 mature pods were collected from 45 trees from 5 populations; from Ucayali, 120 mature pods were collected from 72 trees from 7 populations; and from Loreto, 115 mature pods were collected from 80 trees from 10 populations. In the wild populations, 119 mature pods were collected from 62 trees from 5 populations. Significantly different means are followed by different letters (p < 0.05).
The Selva Central and Ucayali regions could be one group, considering the cultivated trees’ average pod length. The Loreto region’s cultivated trees produced the highest pod length average. The average pod length of 83 ± 1.17 cm (mean ± standard error) in the cultivated trees was significantly higher than the 39 ± 0.95 cm pod length average in the wild trees.
3.2. Genetic Diversity
We identified a total of 71 alleles using the four microsatellite markers after genotyping all the individuals from the 27 populations. The average A was 5.7, the RS was 4.4, the HO was 0.59, and HE was 0.69. The overall inbreeding coefficient FIS was 0.11 (Table 2).

Table 2.
Summary of the genetic diversity of the 27 I. edulis populations. Sample size (N), average number of alleles per locus (A), allelic richness (Rs), effective number of alleles (Ne), expected heterozygosity (HE), observed heterozygosity (HO) and fixation index (FIS) averaged over loci. Sig. refers to the significance resulting from the heterozygote deficiency test (a conservative α value for the test of at least p < 0.01 was used, due to the low number of individuals per population: NS, not significant, ** p < 0.01 and *** p < 0.001, significant). F-null refers to the average estimate of the null frequency over the loci. Standard errors in brackets.
From the results of the current study, the population with the highest and lowest expected heterozygosity possessed the highest and the lowest allelic richness values, in both cultivated and wild populations. The allelic richness parameter correlated well with the populations’ genetic diversity parameters, which is not surprising since the number of individuals sampled per population was unevenly distributed. The population with the highest HE was 2 VRc, 0.83, a cultivated population from the Selva Central region, and the lowest was 21 INc (0.50), a cultivated population from the Loreto region, and they both possessed the highest and the lowest RS, 5.5 and 3.3, respectively. The wild populations 24 RSw and 27 SDw displayed the highest and lowest HE values, 0.79 and 0.60, similarly with the highest and lowest RS values, 5.8 and 4.0. Interestingly, the population with the highest A and Ne was 26 MAw, which could be partially explained by the highest number of sampled individuals (27). The cultivated populations 8 CTc and 13 BRc also had a high number of sampled individuals, which was also reflected in the A and Ne parameters. The average expected genetic diversity is slightly higher in the wild compared to the cultivated populations, 0.72 and 0.69, respectively. The average number of alleles is much higher in the wild (7.4) than in the group of cultivated populations (5.3), but when we consider the allelic richness and effective number of alleles, these differences are reduced (Table 2).
Six cultivated populations out of 22 had significant heterozygote deficiency, but this parameter was not significant in the wild populations (Table 2). The Selva Central group of cultivated populations lacked populations with significant inbreeding coefficient, and the Loreto and the Ucayali regions had only two populations and more than half of the populations with heterozygote deficiency, respectively. Positive and significant FIS values mirror differences between observed and expected heterozygosity, due to putative heterozygosity loss because of non-random mating of the parents. We should also emphasize that the cultivated populations are an assembly of individuals and we should not expect them to be in Hardy–Weinberg Equilibrium. Additionally, the presence of null alleles is an unlikely explanation since the estimated frequency is very low across populations (Table 2). Nevertheless, when we compared the overall inbreeding coefficient from the wild with the cultivated populations, no significant differences were found between them. Conversely, the allelic richness and the observed heterozygosity were significantly lower in the cultivated populations (Table S3).
Seven private alleles (Pa) were identified in three wild populations, the highest Pa per population was found in the 26 MAw population (3) and two in both 23 RPw and 25 RUw. Only one Pa was identified in four different I. edulis cultivated populations (2 VRc, 9 CVc, 14 JHc and 22 MZc). The locus Inga08 had the highest Pa (7 across all populations) and Inga33 and Pel5 only one (data not shown).
The cultivated populations possessed 13 exclusive alleles compared to the wild ones, and only two had a frequency lower than 5%. The regions with the highest number of cultivated populations with exclusive alleles was Selva Central, 80%, followed by Ucayali, 60%, and the Loreto region had the lowest number of populations with those alleles (40%) (data not shown).
3.3. Population Structure
The population genetic structure was investigated by a hierarchical analysis of molecular variance (AMOVA), which revealed that most of the genetic diversity existed within populations (92%). The differentiation between the cultivated and wild group populations (ΦCT = 0.010) was low (~1%), and not significant (p < 0.0958), and the variation among populations within groups was appreciable, ca. 7% (ΦSC = 0.073), and significant (p < 0.0001) (Table 3).

Table 3.
Hierarchical AMOVA between the cultivated and wild population groups, among populations within the cultivated and wild population groups and within I. edulis. populations. df = degrees of freedom; SS = sum of squared deviation; Φ statistics = fixation indexes; P = level of probability of obtaining a more extreme component estimate by chance alone. The significance of the variance components were tested by a permutation test.
The I. edulis genetic structure was further estimated using a Bayesian approach. Using the method of Evanno et al. [37], the most appropriate number of genetic clusters (K) is 2, referred to as red and green (Figure 3; Figure 4 and Figure S1). The red cluster was predominant in the wild populations and in the cultivated populations in the northernmost region (Loreto). Conversely, the green cluster was predominant in the southernmost region (Selva Central). The Ucayali region displayed a mixture of both types of cultivated populations, probably a mixture from the southern and the northern regions (Figure 3; Figure 4).
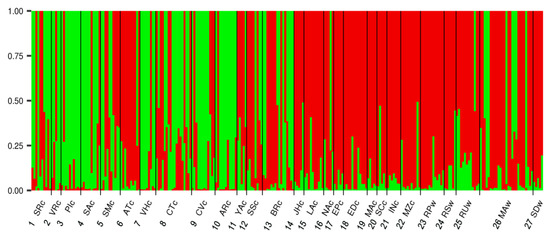
Figure 3.
Proportion of genotype membership q (y-axis) based on STRUCTURE cluster analysis. Plots of proportional group membership for the 259 trees for K = 2. Each tree is represented by a single vertical line, which is divided into different colours based on the genotype affinities to each K cluster (red and green). Divisions between populations are made with black lines.
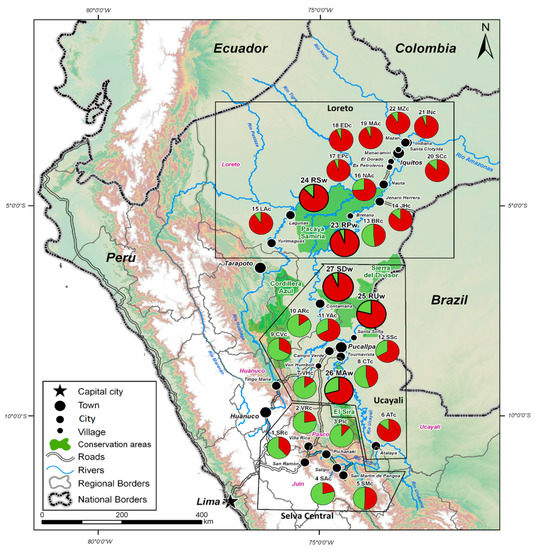
Figure 4.
Inga edulis populations investigated in this study plotted on the map of Peru. Bayesian clustering for K = 2. Populations assigned to two clusters (red and green) corresponding to the I. edulis wild (bigger pie charts outlined in black) and cultivated populations (smaller pie charts).
For K = 2, the highest proportion of red cluster was observed in cultivated populations along the navigable river watersheds in the Loreto and Ucayali regions (e.g., 6 ATc, 11 YAc, 14 JHc, 15 LAc, 16 NAc, 19 MAc, 20 SCc, 21 INc and 22 MZc). Moreover, the green cluster was found to be prevalent in populations cultivated on the Andean foothills and “terra firme” in the Selva Central and Ucayali regions (e.g., 2 VRc, 3 PIc, 4 SAc, 7 VHc and 10 ARc) (Figure 4).
4. Discussion
4.1. Influence of Domestication on Fruit Length
Although the history of cultivation of I. edulis is not well documented, a crop domestication study suggested that humans have domesticated this species over a considerable period of time [20]. Indeed, Amazonia is a major world centre of plant domestication, where selection began in the Late Pleistocene to Early Holocene in peripheral parts of the basin [18]. The origin of cultivated I. edulis trees is uncertain, though probably Amazonian [8]; nevertheless, some authors have suggested it was started by European settlers in west Amazonia [17,20]. Since this tree was cultivated mainly for fruit production, domestication is expected to increase pod length [8,16,20]. To our knowledge, no study has been made comparing both types of populations, cultivated vs. wild, considering this morphologic characteristic (pod length). Certainly, the higher values found in cultivated trees compared to the wild trees clearly support the domestication of I. edulis for food supply. Plant domestication is a long-term process in which natural selection interacts with human selection, driving changes that improve usefulness to humans and adaptations to domesticated landscapes [18].
In the current study, maximum pod length in the wild and the cultivated populations was 73 and 148 cm, respectively, in agreement with Pennington [8]. This author reported that wild trees’ pods rarely exceed 50 cm and cultivated trees could, exceptionally, produce pods exceeding 2 m. The average pod length was higher in the Loreto region’s cultivated trees, compared to Ucayali and Selva Central regions; the smallest fruits were observed in Selva Central. The species’ different cultivation and uses, and differences in ecological conditions, could explain these results. Indeed, in Selva Central, the species was mainly used to shade coffee or cocoa rather than to produce large fruits [42,43]. Farmers were focused mainly on the cash crop yield, rather than the fruit yield provided by shade trees. Additionally, large fruits could be more attractive to uninvited guests, which could then cause damage to the cash crop due to Inga fruit collection. Another supporting argument is the wild I. edulis local name among the Selva Central region inhabitants. The local name for the cultivated I. edulis in Selva Central is “pacay soga”, whereas in the Ucayali and Loreto regions, the name “Guaba” is used for the cultivated type and “guabilla” or “guabilla del monte” for the wild tree (A. Rollo, pers. communication). The difference in local names in these regions might be related to the species abundance, both in the wild and cultivated form. Locals in Selva Central informed us that I. edulis was hard to find in the surrounding wild vegetation; indeed, the species is rarely seen above 750 m [8]. We were also unable to find and sample wild trees in the Selva Central region.
4.2. Genetic Diversity of Wild and Cultivated Populations of I. edulis in the Peruvian Amazon
The overall HE (0.69) was slightly higher than the HO (0.59), inducing an overall inbreeding coefficient index of 11%. In a meta-study for microsatellites and outcrossing species, the author showed a similar value of HE (0.65), but slightly higher HO (0.63) [44].
The results from our study further indicate that all the genetic diversity estimates were lower in the case of the cultivated populations compared to the wild ones, as well as the average inbreeding coefficient. These results confirm a loss of genetic diversity in the cultivated populations, in agreement with the studies by Hollingsworth et al. [21] and Dawson et al. [13] on the same species. These authors concluded that cultivated stands possessed lower total allelic richness than neighbouring wild populations, but the expected genetic diversity remained unchanged, indicating that the process of domestication reduced the number of alleles. Both authors stated that the wild plant material they studied was collected from nearby cultivated populations, in old-growth, primary forest, but due to (i) the long history of the species’ use, (ii) the habits of slash-and-burn in primary forest and (iii) gene flow among nearby stands, the wildness of the trees could be questioned [2,18]. Nevertheless, Dawson et al. [13] observed marked differences in the haplotype composition between natural and cultivated stands. In our case, the wild material was sampled in natural vegetation in protected areas and secondary forest, and unless extensive long-distance gene flow existed, no ambiguities in distinguishing both types existed. In addition, the results regarding pod length clearly distinguish the wild from cultivated material. We found an important effect of the domestication on the natural resources of a species, which is an expected phenomenon when a species is used by humans [23,45]. In some cases, the expected heterozygosity might be higher or similar in the cultivated population than that displayed in the wild population, due to a putative “melting pot” phenomenon in the former populations (introduced alleles from different origins). Nevertheless, the allelic richness and observed heterozygosity found, in our study, in the cultivated populations was lower than in the wild ones, indicating the loss of rare alleles during selection as observed by other authors [23,46]. Some cultivated populations from the current study had a significant heterozygote deficit, particularly in the Ucayali region. The consequences of the inbreeding effect in fruit trees, such as I. edulis, might impact fruit production due to inbreeding depression, which would directly impact farmers’ yield [15,23]. The fact that the species is self-incompatible [13] excludes the possibility of heterozygote deficiency due to self-pollination; probably, related trees were introduced in these populations and the value reflect biparental inbreeding.
4.3. Population Structure
The genetic variance partition in our study (92% of the variance was observed within populations and a low genetic structure, 7%, was detected among populations) is usual in outcrossing tropical forest tree species with high levels of gene flow [6]. The hierarchical AMOVA showed that the Φct between wild and cultivated populations was 1%, yet not significant.
Dawson et al. [13] found low genetic structure, similarly to our results, in I. edulis natural and cultivated stands, with nuclear but not chloroplast microsatellite data. Nevertheless, the authors used only two chloroplast loci, which might have biased the results, since the smaller effective population size of the chloroplast genome makes it more susceptible to genetic drift and species differentiation [47]. Conversely, a high genetic structure was found between natural and cultivated stands of I. vera, and the authors reasoned that the cultivated populations were derived from seeds coming from different mother trees, but a different geographic origin was also possible [23].
In the current study, for K = 2 (Figure 3 and Figure 4), the wild populations displayed identical composition, with the predominant red cluster. The uniform composition of the studied wild material could be due to the relatively recent speciation of the genus [9] and, also, to regional wild populations sampling [8]. The red cluster prevailed in the northern cultivated populations: the Loreto region and along the Ucayali river in the Ucayali region, which could express large population centres occupying the margins of main rivers with extensive trade networks [43]. A tiny green genetic cluster is present in the wild populations and in the cultivated populations of the Loreto region. Conversely, the green cluster is relevant in Selva Central and Ucayali cultivated populations. The green cluster increases in the sub-Andean Selva Central region and in the higher elevated sites from the Ucayali region, where the I. edulis trees were traditionally used on coca fields before the Conquest, and for shading protection of cocoa, coffee and tea plantations after the Spanish settlement [43]. In the Loreto region, the cultivated population 13 BRc possessed a higher proportion of green cluster than others from this region. This population is near Bretaña village, which was named after the Europeans, who arrived from the Andes and the coastal regions of Peru, during the rubber boom at the end of the 19th century [48].
Iquitos, in the Loreto region, is referred to as a crop domestication centre in Amazonia, created as populations expanded, and providing strong evidence that pre-conquest human populations had intensively transformed their plant resources [18,20]. Indeed, the I. edulis domestication was probably achieved by selecting from the local wild population and possibly started in the Loreto region, since the genetic structure of the cultivated populations from this region do not differ much from the wild ones. Moreover, they have bigger pods and lower allelic richness than the other cultivated populations, which could indicate that the selection intensity was higher here. Indeed, some authors claim that the possible origin of I. edulis domestication was in this region, which was also the location for the domestication of other species [18,22]. Additionally, the crop is probably recently domesticated, since when a crop is in an initial process of domestication no clear genetic structuring occurs, as in Brazil nut [22]. The genetic differentiation between the wild and cultivated populations is low and with admixture; the cultivated populations seem to originate from the wild ones. Conversely, the results of Dawson et al. [13] on chloroplast haplotype composition displayed a completely different pattern between natural and cultivated populations. The authors explained these results by a non-local origin of the I. edulis cultivated material. Our results do not support this theory. Instead, we inferred that the cultivated populations had a local germplasm origin, yet without representative sampling, which is expected, since a few trees were probably selected in nearby wild populations. Indeed, a possible genetic drift effect (change in the frequency of the allele in a population due to random sampling of organisms) in the cultivated populations is expected.
4.4. Practical Measures to Maintain I. edulis Genetic Resources
The I. edulis germplasm management should focus on both the wild and the cultivated stands. In the case of wild material, the protection of the original Amazonian vegetation remnants is key to maintaining the species’ genetic resources in the region. In modern-day Amazonia, increasing deforestation for the establishment of pastures has become a global concern due to its impacts on biodiversity [43]. Considering the cultivated stands, the villages and indigenous settlements are the units of interest because they are the domesticated plant population keepers. Consequently, the fate of the village will determine the maintenance of the crop genetic resources. For example, the post-Colombian population collapse that resulted in a loss of village units and corresponded to the loss in human numbers (ca. 90–95% population decline), was quickly reflected in the loss of crop diversity [20,43]. The cultivated populations with low genetic diversity and/or high inbreeding estimates (e.g., 7 VHc, 10 ARc, 12 SSc, 14 JHc, 17 EPc, 18 EDc, 19 MAc and 21 INc) should be supported with new germplasm sources (from wild populations) to eliminate the risk of biparental inbreeding and diversity loss, which might be reflected in the future crop value (inbreeding depression, flower abortion, and crop yield failure).
5. Conclusions
The results of the current study on I. edulis show a significantly higher value for average pod length in cultivated trees than in wild trees. The wide-scale infusion from wild stands into farms could negatively affect fruit size and weaken domestication efforts over time. Additionally, the Loreto region displayed the highest average pod length, as well as having populations with a lower allelic richness when compared to the cultivated populations of other regions.
The cultivated stands in the Selva Central and Ucayali region could, additionally, be a germplasm material source, and thus could provide a long-term safeguard for on-farm conservation since the Loreto region possesses the populations with the lowest values of allelic richness. Hybridization programmes using such a germplasm source and local wild material with backward selection could help increase the crop yield and genetic diversity in the cultivated populations. Additionally, new selection should consider the needs of modern agriculture and forest management practices, as well as global warming.
Supplementary Materials
The following are available online at https://www.mdpi.com/1999-4907/11/6/655/s1, Figure S1. Representation of the ΔK distribution statistic. The method of Evanno et al. [37] was used to determine the most appropriate number of genetic clusters (K); Table S1. Inga edulis 27 populations (cultivated and wild), region (Geographic region), sampling region (Site), population code (Pop.) and sampling details (Sampling description); Table S2. Average fruit length per population. The minimal and maximal pod length values (in brackets) and the number of pods (Nl) per population, in a total of 448 pods. Table S3. Diversity parameters comparison between cultivated and wild populations. Allelic richness (RS), observed heterozygosity (HO) and inbreeding coefficient (FIS). P = probability values for differences between groups for two-sided t-test after 1000 permutations. * = significant test (p < 0.05).
Author Contributions
Conceptualization, A.R., B.L. and M.M.R.; field data collection, A.R., Z.M.C.P., H.V., E.C., S.G.T. and R.M.V.A.; methodology, A.R., M.M.R., R.L.C., C.S.; formal analysis, writing—original draft preparation, M.M.R., A.R., B.M., M.K. and T.H.; writing—review and editing, M.M.R., A.R., B.L., B.M., M.K. and T.H.; funding acquisition, A.R., B.L. and M.M.R. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the Bilateral Project ´Morphological and genetic diversity of indigenous tropical trees in the Amazon—model study of Inga edulis Mart. in Peruvian Amazon´; Czech Academy of Sciences and CONCYTEC, Peru 2011–2012; The Internal Grant Agency of CULS Prague (No. 20185004, No. 20205003); The Scholarship National University of Ucayali, Peru; European Union Lifelong Learning Programme Erasmus Consortium—Practical Placement Scholarship (Certificate No. CZ-01-2009). The Foundation Nadace Nadání Josefa, Marie a Zdeňky Hlávkových, Czech Republic; Supported by the Ministry of Agriculture of the Czech Republic, institutional support MZE-RO0418; Foundation for Science and Technology, Portugal, UIDB/00239/2020 and UIDB/00681/2020 supported MMR.
Acknowledgments
We thank the Servicio Nacional de Áreas Naturales Protegidas and to José Grocio Gil Navarro director of Pacaya Samiria National Reservation for the investigation authorization (N° 004-2012-SERNANP-RNPS-J). Thanks are extended to N. Roque for help with Figure 1, to D. Petrus for help with Figure 4, and to I. Salavessa for editing the English of the manuscript. We want to thank two anonymous reviewers for the very helpful comments and suggestions, which considerably improved the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bodmer, R.; Fang, T.; Antunez, M.; Puertas, P.; Chota, K.; Pittet, M.; Kirkland, M.; Walkey, M.; Rios, C.; Perez-Peña, P.; et al. Impact of Climate Change on Wildlife and Indigenous Communities in Flooded Forests of the Peruvian Amazon. In The Lima Declaration on Biodiversity and Climate Change: Contributions from Science to Policy for Sustainable Development; Rodríguez, L., Anderson, I., Eds.; Secretariat of the Convention on Biological Diversity: Montreal, QC, Canada, 2017; Volume 89, pp. 81–90. [Google Scholar]
- Levis, C.; Flores, B.M.; Moreira, P.A.; Luize, B.G.; Alves, R.P.; Franco-Moraes, J.; Lins, J.; Konings, E.; Peña-Claros, M.; Bongers, F.; et al. How People Domesticated Amazonian Forests. Front. Ecol. Evol. 2018, 5. [Google Scholar] [CrossRef]
- Dawson, I.K.; Lengkeek, A.; Weber, J.C.; Jamnadass, R. Managing genetic variation in tropical trees: Linking knowledge with action in agroforestry ecosystems for improved conservation and enhanced livelihoods. Biodivers. Conserv. 2009, 18, 969–986. [Google Scholar] [CrossRef]
- Brooks, T.M.; Mittermeier, R.A.; da Fonseca, G.A.B.; Gerlach, J.; Hoffmann, M.; Lamoreux, J.F.; Mittermeier, C.G.; Pilgrim, J.D.; Rodrigues, A.S.L. Global biodiversity conservation priorities. Science 2006, 313, 58–61. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, P.J.C.; Asner, G.P.; Knapp, D.E.; Almeyda, A.; Galván-Gildemeister, R.; Keene, S.; Raybin, R.F.; Smith, R.C. Land-use allocation protects the Peruvian Amazon. Science 2007, 317, 1233–1236. [Google Scholar] [CrossRef] [PubMed]
- Finkeldey, R.; Hattemer, H.H. Tropical Forest Genetics; Springer-Verlag: Berlin/Heidelberg, Germany, 2007; p. 315. [Google Scholar] [CrossRef]
- Wee, A.K.S.; Li, C.H.; Dvorak, W.S.; Hong, Y. Genetic diversity in natural populations of Gmelina arborea: Implications for breeding and conservation. New Forests 2012, 43, 411–428. [Google Scholar] [CrossRef]
- Pennington, T.D. The Genus Inga: Botany; Royal Botanic Gardens, Kew: London, UK, 1997; p. 854. [Google Scholar]
- Richardson, J.E.; Pennington, R.T.; Pennington, T.D.; Hollingsworth, P.M. Rapid diversification of a species-rich genus of neotropical rain forest trees. Science 2001, 293, 2242–2245. [Google Scholar] [CrossRef]
- Lavin, M. Floristic and geographical stability of discontinuous seasonally dry tropical forests explains patterns of plant phylogeny and endemism. In Neotropical Savannas and Seasonally Dry Forests: Plant Diversity, Biogeography, and Conservation; Pennington, R.T., Lewis, G.P., Ratter, J.A., Eds.; Crc Press-Taylor & Francis Group: Boca Raton, FL, USA, 2006; pp. 433–447. [Google Scholar]
- Cruz-Neto, O.; Machado, I.; Duarte, J., Jr.; Lopes, A. Synchronous phenology of hawkmoths (Sphingidae) and Inga species (Fabaceae-Mimosoideae): Implications for the restoration of the Atlantic forest of northeastern Brazil. Biodivers. Conserv. 2011, 20, 751–765. [Google Scholar] [CrossRef]
- Figueiredo, M.; Bruno, R.; Barros e Silva, A.; Nascimento, S.; Oliveira, I.; Felix, L. Intraspecific and interspecific polyploidy of Brazilian species of the genus Inga (Leguminosae: Mimosoideae). Genet. Mol. Res. 2014, 13, 3395–3403. [Google Scholar] [CrossRef]
- Dawson, I.; Hollingsworth, P.; Doyle, J.; Kresovich, S.; Weber, J.; Sotelo Montes, C.; Pennington, T.; Pennington, R. Origins and genetic conservation of tropical trees in agroforestry systems: A case study from the Peruvian Amazon. Conserv. Genet. 2008, 9, 361–372. [Google Scholar] [CrossRef]
- Pennington, T.D. Inga management. In The Genus Inga: Utilization; Pennington, T., Fernandes, E., Eds.; Royal Botanic Gardens, Kew: London, UK, 1998; pp. 159–167. [Google Scholar]
- Koptur, S. Outcrossing and pollinator limitation of fruit-set: Breeding systems of Neotropical Inga trees (Fabaceae: Mimosoideae). Evolution 1984, 38, 1130–1143. [Google Scholar] [CrossRef]
- Reynel, C.; Pennington, T.D. El Género Inga en el Perú: Morfología, Distribución y Usos; Royal Botanic Gardens, Kew: London, UK, 1997. [Google Scholar]
- León, J. Botánica de los Cultivos Tropicales; Servicio Editorial IICA: San José, Costa Rica, 1987; p. 445. [Google Scholar]
- Clement, C.R.; Denevan, W.M.; Heckenberger, M.J.; Junqueira, A.B.; Neves, E.G.; Teixeira, W.G.; Woods, W.I. The domestication of Amazonia before European conquest. Proc. R. Soc. Lond. B Biol. Sci. 2015, 282, 20150813. [Google Scholar] [CrossRef]
- Nichols, J.D.; Carpenter, F.L. Interplanting Inga edulis yields nitrogen benefits to Terminalia amazonia. For. Ecol. Manag. 2006, 233, 344–351. [Google Scholar] [CrossRef]
- Clement, C.R. 1492 and the loss of Amazonian crop genetic resources. I. The relation between domestication and human population decline. Econ. Bot. 1999, 53, 188. [Google Scholar] [CrossRef]
- Hollingsworth, P.M.; Dawson, I.K.; Goodall-Copestake, W.P.; Richardson, J.E.; Weber, J.C.; Sotelo Montes, C.; Pennington, R.T. Do farmers reduce genetic diversity when they domesticate tropical trees? A case study from Amazonia. Mol. Ecol. 2005, 14, 497–501. [Google Scholar] [CrossRef] [PubMed]
- Clement, C.; De Cristo-Araújo, M.; Coppens D’Eeckenbrugge, G.; Alves Pereira, A.; Picanço-Rodrigues, D. Origin and domestication of native Amazonian crops. Diversity 2010, 2, 72. [Google Scholar] [CrossRef]
- Cruz-Neto, O.; Aguiar, A.V.; Twyford, A.D.; Neaves, L.E.; Pennington, R.T.; Lopes, A.V. Genetic and ecological outcomes of Inga vera subsp affinis (Leguminosae) tree plantations in a fragmented tropical landscape. PLoS ONE 2014, 9. [Google Scholar] [CrossRef]
- Dawson, I.K.; Leakey, R.; Clement, C.R.; Weber, J.C.; Cornelius, J.P.; Roshetko, J.M.; Vinceti, B.; Kalinganire, A.; Tchoundjeu, Z.; Masters, E.; et al. The management of tree genetic resources and the livelihoods of rural communities in the tropics: Non-timber forest products, smallholder agroforestry practices and tree commodity crops. For. Ecol. Manag. 2014, 333, 9–21. [Google Scholar] [CrossRef]
- Rollo, A.; Lojka, B.; Honys, D.; Mandák, B.; Wong, J.A.C.; Santos, C.; Costa, R.; Quintela-Sabarís, C.; Ribeiro, M.M. Genetic diversity and hybridization in the two species Inga ingoides and Inga edulis: Potential applications for agroforestry in the Peruvian Amazon. Ann. For. Sci. 2016, 73, 425–435. [Google Scholar] [CrossRef]
- Dayanandan, S.; Bawa, K.; Kesseli, R. Conservation of microsatellites among tropical trees (Leguminosae). Am J Botany 1997, 84, 1658–1663. [Google Scholar] [CrossRef]
- Sokal, R.R.; Rohlf, F.J. Biometry, 2nd ed.; W.H. Freeman and Co.: San Francisco, CA, USA, 1981; p. 859. [Google Scholar]
- El Mousadik, A.; Petit, R.J. High level of genetic differentiation for allelic richness among populations of the argan tree [Argania spinosa (L.) Skeels] endemic to Morocco. Theor. Appl. Genet. 1996, 92, 832–839. [Google Scholar] [CrossRef]
- Nei, M. Molecular Evolutionary Genetics; Columbia University Press: New York, NY, USA, 1987; p. 333. [Google Scholar]
- Goudet, J. FSTAT (Version 1.2): A computer program to calculate F-statistics. J. Hered. 1995, 86, 485–486. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Rousset, F. Genepop’007: A complete re-implementation of the genepop software for Windows and Linux. Mol. Ecol. Resour. 2008, 8, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar]
- Hubisz, M.J.; Falush, D.; Stephens, M.; Pritchard, J.K. Inferring weak population structure with the assistance of sample group information. Mol. Ecol. Resour. 2009, 9, 1322–1332. [Google Scholar] [CrossRef]
- Wang, J. The computer program structure for assigning individuals to populations: Easy to use but easier to misuse. Mol. Ecol. Resour. 2017, 17, 981–990. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Earl, D.A.; vonHoldt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Jakobsson, M.; Rosenberg, N.A. CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 2007, 23, 1801–1806. [Google Scholar] [CrossRef]
- Ramasamy, R.K.; Ramasamy, S.; Bindroo, B.B.; Naik, V.G. STRUCTURE PLOT: A program for drawing elegant STRUCTURE bar plots in user friendly interface. Springerplus 2014, 3, 431. [Google Scholar] [CrossRef] [PubMed]
- Ormsby, T.; Napoleon, E.; Burke, R.; Groessl, C.; Bowden, L. Getting to Know ArcGIS Desktop; ESRI Press: Redlands, CA, USA, 2010; p. 592. [Google Scholar]
- León, J. Inga as shade for coffee, cacao, and tea: Historical aspects and present day utilization. In The Genus Inga: Utilization; Pennington, T., Fernandes, E., Eds.; Royal Botanic Gardens, Kew: London, UK, 1998; pp. 101–115. [Google Scholar]
- Miller, R.P.; Nair, P.K.R. Indigenous agroforestry systems in Amazonia: From Prehistory to Today. Agrofor. Syst. 2006, 66, 151–164. [Google Scholar] [CrossRef]
- Nybom, H. Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol. Ecol. 2004, 13, 1143–1155. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, M.M.; Plomion, C.; Petit, R.; Vendramin, G.G.; Szmidt, A.E. Variation in chloroplast single-sequence repeats in Portuguese maritime pine (Pinus pinaster Ait.). Theor. Appl. Genet. 2001, 102, 97–103. [Google Scholar] [CrossRef]
- Jones, T.H.; Steane, D.A.; Jones, R.C.; Pilbeam, D.; Vaillancourt, R.E.; Potts, B.M. Effects of domestication on genetic diversity in Eucalyptus globulus. For. Ecol. Manag. 2006, 234, 78–84. [Google Scholar] [CrossRef]
- Petit, R.J.; Duminil, J.; Fineschi, S.; Hampe, A.; Salvini, D.; Vendramin, G.G. Comparative organization of chloroplast, mitochondrial and nuclear diversity in plant populations. Mol. Ecol. 2005, 14, 689–701. [Google Scholar] [CrossRef]
- Eidt, R.C. Pioneer settlement in Eastern Peru. Ann. Assoc. Am. Geogr. 1962, 52, 255–278. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).