Abstract
Alzheimer’s disease (AD) is a progressive, non-curable neurodegenerative disorder that poses persistent challenges for early diagnosis due to its gradual onset and the difficulty in distinguishing pathological changes from normal aging. Neuroimaging, particularly MRI and PET, plays a key role in detection; however, limitations in data availability and the complexity of early structural biomarkers constrain traditional diagnostic approaches. This review investigates the use of generative models, specifically Generative Adversarial Networks (GANs) and Diffusion Models, as emerging tools to address these challenges. These models are capable of generating high-fidelity synthetic brain images, augmenting datasets, and enhancing machine learning performance in classification tasks. The review synthesizes findings across multiple studies, revealing that GAN-based models achieved diagnostic accuracies up to 99.70%, with image quality metrics such as SSIM reaching 0.943 and PSNR up to 33.35 dB. Diffusion Models, though relatively new, demonstrated strong performance with up to 92.3% accuracy and FID scores as low as 11.43. Integrating generative models with convolutional neural networks (CNNs) and multimodal inputs further improved diagnostic reliability. Despite these advancements, challenges remain, including high computational demands, limited interpretability, and ethical concerns regarding synthetic data. This review offers a comprehensive perspective to inform future AI-driven research in early AD detection.
1. Introduction
Recent advancements in medical sciences and healthcare technologies have led to remarkable progress in improved disease detection, early diagnosis, better therapeutic interventions, and enhanced patient outcomes. These innovations have significantly contributed to increased global life expectancy and better quality of life. As a result, the world is experiencing a significant demographic shift, with the proportion of elderly individuals in the global population steadily rising. According to the World Health Organization (WHO), the number of individuals aged 60 years and older is expected to rise from 1 billion in 2020 to 2.1 billion by 2050 []. This surge in the elderly population is accompanied by an increased prevalence of age-related illnesses, particularly AD, which has emerged as one of the most prevalent and debilitating neurodegenerative disorders worldwide.
Alzheimer’s disease is the most common form of dementia worldwide, accounting for approximately 60–80% of cases []. It is also recognized as a leading cause of disability and dependence among older adults. It is estimated that more than 55 million people globally are living with dementia, a number expected to rise to 78 million by 2030 and 139 million by 2050, largely driven by population aging []. As of 2025, Alzheimer’s disease affects nearly 7.2 million adults aged 65 and older in the United States with projections indicating it will nearly double to 13 million by 2050. Beyond its clinical progression, the disease imposes a heavy emotional burden on families and caregivers, while generating substantial healthcare and societal costs. Among older adults, Alzheimer’s is the fifth leading cause of death preceded only by heart disease, cancer, stroke, and chronic lower respiratory conditions underscoring its significance as a major public health concern []. It is a progressive, irreversible brain disorder characterized by memory loss, cognitive dysfunction, reasoning, language, problem-solving abilities, and behavioral changes, which gradually impair a person’s ability to perform everyday tasks. As it advances, patients often require full-time care and medical support, placing an enormous emotional and economic burden on families and healthcare systems.
The exact cause of AD still remains unknown. Factors like growing older, family history, level of education, and daily habits are known to play a role. In addition, several brain-related conditions can contribute to the development of dementia symptoms []. The disease often begins with subtle symptoms such as difficulty recalling recent events and gradually advances to severe memory loss, disorientation, and loss of speech and motor skills. Pathologically, AD is linked to the accumulation of amyloid-beta plaques and tau neurofibrillary tangles in the brain. These protein aggregates are believed to interfere with neuron-to-neuron communication and trigger inflammation and cell death []. It initially begins in the entorhinal cortex and hippocampus, critical regions for memory and learning, and subsequently spreads to the cerebral cortex, affecting language, judgment, social behavior, and motor functions []. These changes lead to a gradual decline in cognitive function and behavioral disturbances.
Diagnosing Alzheimer’s in its early stages remains a significant challenge in clinical practice. In its initial stages, symptoms include mild memory loss, difficulty in recalling recent events, confusion, challenges in completing familiar tasks, language disturbances, and mood or personality changes. These symptoms can be subtle and are often mistaken for normal aging or other forms of dementia, resulting in many cases remaining undiagnosed until the disease has significantly progressed, and irreversible brain damage has already occurred. As the disease progresses, these symptoms worsen, severely impairing daily functioning and quality of life. Thus, early and accurate diagnosis is essential not only to initiate timely treatments that may slow the progression, but also to give patients and families the opportunity to prepare, seek support, and improve quality of life.
Early detection of Alzheimer’s disease is critical for improving patient outcomes and guiding timely interventions. Identifying AD at the preclinical stage allows for lifestyle changes, pharmaceutical trials, and supportive care to be initiated when they are most effective. Moreover, early diagnosis can alleviate uncertainty for patients and caregivers, aid in planning, and reduce healthcare costs associated with late-stage care. Recent research has emphasized the importance of biomarkers for the diagnosis and monitoring of Alzheimer’s disease. Several biomarkers have been identified as indicators of AD, such as changes in cerebrospinal fluid (CSF), blood-based biomarkers, and particularly neuroimaging biomarkers. Brain imaging has emerged as a critical tool in Alzheimer’s research and clinical diagnosis. Modalities like Magnetic Resonance Imaging (MRI), Positron Emission Tomography (PET), and functional MRI (fMRI), and Diffusion Tensor Imaging (DTI) provide critical insights into structural and functional brain changes. Making these widely usable to detect structural and functional abnormalities associated with AD. These imaging techniques provide valuable insights into brain atrophy, amyloid deposition, and neuronal activity patterns, serving as non-invasive tools for early diagnosis []. Collectively, these imaging modalities enable early-stage identification of pathological changes, often before overt clinical symptoms emerge.
Despite their diagnostic value, neuroimaging data presents significant challenges. High-dimensionality, inter-subject variability, limited availability of labeled data, and the cost of image acquisition limit their broader use in population-scale screening. These challenges underscore the need for intelligent systems capable of extracting complex patterns from high-dimensional imaging data. In recent years, generative artificial intelligence (AI) has shown tremendous promise in tackling these limitations. Unlike traditional discriminative models, generative models are capable of learning the underlying data distribution to generate new, synthetic samples that are statistically similar to the original data. Among the most, Generative Adversarial Networks (GANs) and Diffusion Models have emerged as leading frameworks in medical image analysis. These models are capable of generating high-quality synthetic medical images, which can be used to augment small datasets, improve image resolution, and even translate one imaging modality to another (e.g., MRI to PET).
GANs introduced by Goodfellow et al. in 2014 [], with their adversarial architecture, have been applied to a wide range of AD tasks including neuroimaging data augmentation, generating missing imaging modalities, enhancing image resolution, and modelling disease progression. These applications help to overcome the common issue of data imbalance, enhance resolution, and simulate realistic brain pathologies to improve model generalization [,,].
Most recently, Diffusion Models, a newer class of generative models based on a stepwise denoising process that reconstructs clean high-fidelity synthetic image data from random noise []. These models offer better training stability and high-quality images which makes them suitable for Alzheimer’s disease imaging.
The integration of GANs and Diffusion Models into AD diagnostic pipelines is transforming the landscape of early detection. By producing high-quality synthetic neuroimages, enhancing data diversity, and improving model performance, GANs and diffusion models enable more accurate and scalable diagnostic systems. Their ability to learn intricate patterns in imaging data makes them a highly suitable diagnostic framework for Alzheimer’s disease research and real-world clinical care.
This review paper aims to comprehensively analyze recent research applying GANs and Diffusion Models to AD detection using neuroimaging. We summarize current methodologies, model architectures, applications, assess their performance metrics, strengths, limitations, and clinical potential. Through these insights from recent studies, we aim to guide researchers and practitioners on ongoing efforts at the intersection of AI innovation and neurodegenerative disease diagnosis research. The rest of this review paper is structured as follows. To introduce the essential background on AD, Section 2 and Section 3 define the stages of AD and exploring the most important neuroimaging modalities that are used for AD diagnosis. Section 4 presents a comparative review of the available literature that apply these generative models to AD diagnosis. It reviews different types of methods applied in those reviewed papers, identified their key research challenges and gaps. It also includes an evaluation of various performance metrics, with particular emphasis on experimental results and diagnostic accuracy reported in the reviewed articles. Section 5 discusses the most common datasets used in AD research. Section 6 and Section 7 are devoted to describing different types of GANs and Diffusion models, respectively. Finally, Section 8 summarizes and discusses the study’s key findings, results, challenges, and potential future directions.
2. Alzheimer’s Disease Stages
Alzheimer’s disease tends to progress slowly over many years, gradually affecting different regions of the human brain, causing memory loss, behavioral changes and hampering the process of thinking. It starts with mild cognitive impairment (MCI), where individuals experience noticeable memory issues that do not yet disrupt daily activities. Later on, it may lead to AD, while not everyone with MCI develops AD. As the disease progresses, the emotional and psychological impact both patients and caregivers, underscoring the importance of strong mental health support throughout the journey []. Understanding the stages of AD is essential for providing the required support. Alzheimer’s disease typically progresses through five general stages []. It often begins with a Preclinical Alzheimer’s disease stage. The second stage is called MCI. Which is divided into two types: progressive MCI (pMCI) where a person with MCI will develop AD and stable MCI (sMCI) indicate that it will not progress towards AD. The disease moves to a third stage called Mild dementia (MD) in which patients develop memory issues that interfere with their daily activities. The fourth stage is Moderate dementia (MoD). Finally, the fifth stage is called Severe dementia due to Alzheimer’s disease. In this stage, the disease becomes severe and full-time care becomes essential. Figure 1 shows the AD progression stages.
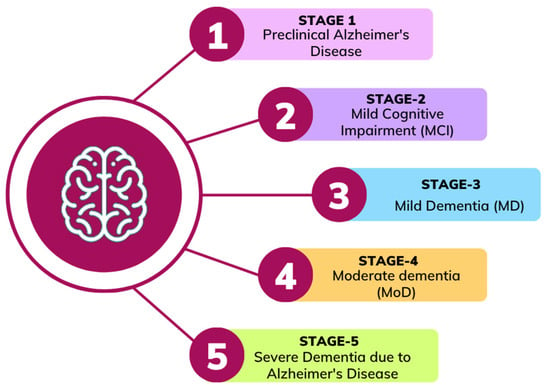
Figure 1.
Alzheimer’s disease progression stages.
3. Neuroimaging Modalities
Neuroimaging is an essential tool to understand, diagnose, and monitor AD in its early stages. Data collected using different modalities varies in format, quality, and the type of information it reveals each offering distinct insights into the brain’s anatomy and functional activity. Neuroimaging can extract measurable indicators, or quantitative biomarkers, that can identify specific neurological conditions and predict dementia []. With the integration of artificial intelligence, these modalities become a vital diagnostic tool to detect and predict the progress of AD. Application of different neuroimaging modalities to support the diagnosis of Alzheimer’s disease is shown in Figure 2. In this section, different neuroimaging modalities and their approaches towards AD diagnosis are discussed.
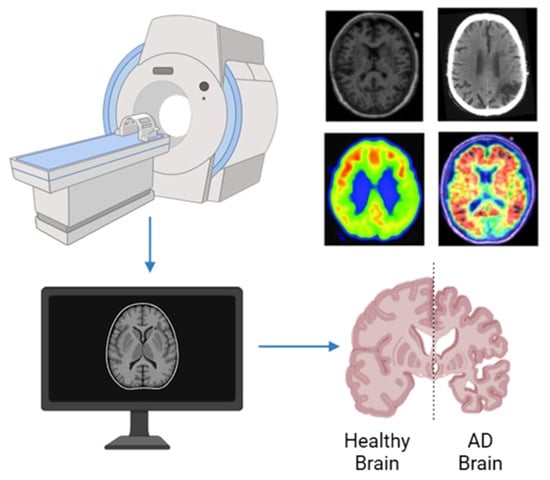
Figure 2.
Applications of Neuroimaging modalities in AD diagnosis.
3.1. Magnetic Resonance Imaging (MRI)
MRI is widely used regarded as one of the most reliable neuroimaging techniques in Alzheimer’s research and clinical assessment due to its ability to reveal detailed brain structures without invasive procedures. Its ability to produce high-resolution images makes it especially useful in identifying brain atrophy particularly in areas like the hippocampus and entorhinal cortex, which often show signs of shrinkage in the early stages of the disease []. These structural insights help to differentiate Alzheimer’s disease from other neurodegenerative conditions and to assess disease progression. Figure 3 [] illustrates MRI scans comparing healthy individuals and patients with AD, showcasing the visible differences used in diagnosis.
Figure 3.
MRI brain image of healthy and AD patients.
3.2. Functional Magnetic Resonance Imaging (fMRI)
Functional MRI (fMRI) is a non-invasive scan that shows the connectivity between different areas of the brain by tracking changes in blood flow. In Alzheimer’s disease, it often reveals early disruptions in brain networks like the Default Mode Network (DMN), which is linked to memory and self-reflection. These functional changes can appear before structural damage is visible, making fMRI a helpful tool for early detection []. Figure 4 [] shows fMRI scans comparing healthy and AD-affected brains. It also helps researchers understand how Alzheimer’s affects thinking and memory during mental tasks.
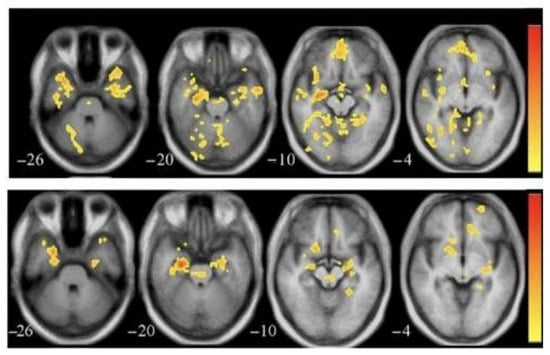
Figure 4.
fMRI brain image of healthy (top) and AD patients (bottom).
3.3. Positron Emission Tomography (PET)
Positron Emission Tomography (PET) is an imaging technique that uses small amounts of radioactive tracers to create 2D or 3D images of the brain’s chemical activity. It is especially useful for tracking things like blood flow, oxygen use, and glucose metabolism, giving a clearer insight into how different brain regions function. PET scans can detect early metabolic changes before major structural damage occurs, helping to identify the AD at a progressive stage and monitor its advancement through measurable brain activity patterns []. Figure 5 [] shows PET imaging of an AD patient.
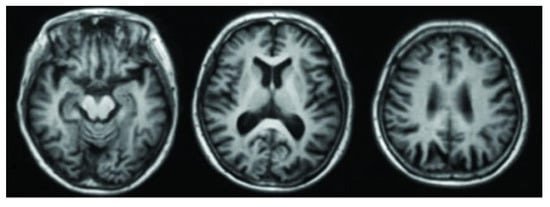
Figure 5.
PET scan of an AD patient.
3.4. Fluorodeoxyglucose Positron Emission Tomography (FDG-PET)
FDG-PET monitors and identify reduced glucose metabolism in different brain regions. By highlighting brains dysfunctionality, it can distinguish AD from other types of dementia. Figure 6 [] shows scanning patterns of dementia using FDG-PET imaging of an AD patient.
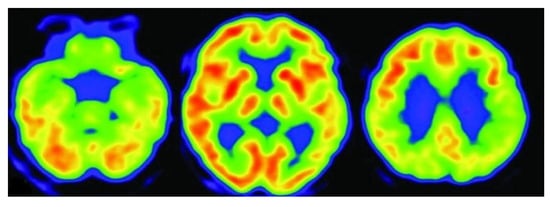
Figure 6.
FDG-PET scan of an AD patient.
3.5. Computed Tomography (CT)
CT scans are fast and widely available tools that help create 3D images of the brain, making it helpful in the early evaluation of neurological conditions, including suspected dementia. Though less detailed than MRI, it is still really helpful for checking memory issues and ruling out other conditions like strokes or tumors that can mimic Alzheimer’s symptoms.
3.6. Diffusion Tensor Imaging (DTI)
It is a specialized form of MRI that captures the movement of water molecules within brain tissue to reveal early structural changes related to Alzheimer’s disease []. It generates three-dimensional tensor fields that reveal the direction and strength of water diffusion. DTI makes it possible to reconstruct white matter tracts and evaluate the brain’s structural connectivity and integrity. Key metrics like fractional anisotropy (FA) and mean diffusivity (MD) are used to detect early signs of white matter damage []. Table 1 highlights different neuroimaging modalities used in Alzheimer’s diagnosis.

Table 1.
Neuroimaging modalities in Alzheimer’s disease diagnosis.
4. Background Study
4.1. Paper Selection Strategy
For this study the PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) framework selection was followed. Figure 7 shows the PRISMA flow chart. An extensive search was carried out using trusted academic databases, including IEEE Xplore, PubMed, ScienceDirect, SpringerLink, Nature, and Google Scholar. The search strategy combined keywords such as Alzheimer’s disease, early detection, Generative Adversarial Networks, Diffusion Models, neuroimaging, and synthetic medical imaging, among others. This helped capture a diverse range of studies relevant to the intersection of artificial intelligence and Alzheimer’s diagnosis.
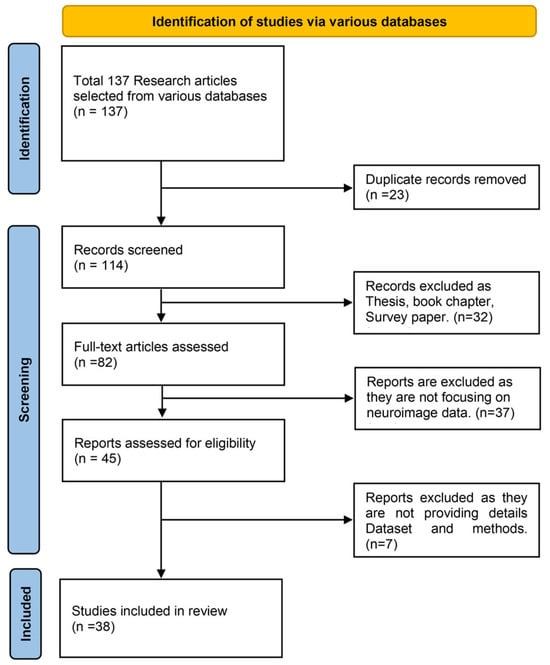
Figure 7.
PRISMA flow chart for study selection process.
4.2. Literature Review
This section provides a comprehensive synthesis of notable research articles from 2013 to 2025 highlighting generative models, particularly GANs and diffusion models, in enhancing the early diagnosis of AD using various neuroimaging modalities. Table 2 highlights the medical image dataset being used in the research articles included in this. It also provides the modality and total number of participants used in their study. Table 3 provides an explanation of the different methods used by the researchers and their performance metrics according to the model used. Table 4 presents the models used in the reviewed literature highlighting their key challenges and limitations.
Raj et al. [] developed a network diffusion model to simulate the prion-like spread of dementia across brain networks. By analyzing brain scans from healthy individuals, they predicted where atrophy would appear in Alzheimer’s and frontotemporal dementia patients and their predictions closely matched real MRI data. The model performed impressively, with diagnostic accuracy exceeding an AUC of 0.90, even outperforming traditional PCA methods. However, it relied on a static brain map and a small sample, which may limit its flexibility.
Lee et al. [] used an SVM-based method to detect early-stage Alzheimer’s by analyzing DTI brain scans, focusing on white matter integrity and fiber pathways especially those linked to the thalamus. Their model achieved 100% accuracy, 100% sensitivity, and 100% specificity in both 10-fold cross-validation and independent testing. This highlights its strong potential for distinguishing MCI from healthy aging. However, its dependence on pre-defined seed regions and differences in imaging sources may limit generalizability. Future improvements could include standardizing imaging protocols and incorporating other biomarkers for broader clinical use.
Pan et al. [] introduced a two-stage deep learning framework to improve Alzheimer’s diagnosis using MRI and missing PET data. First, they used a 3D CycleGAN to synthesize PET scans from corresponding MRIs. Then, they applied a multi-modal learning model (LM3IL) to classify AD and predict MCI conversion. Tested on ADNI datasets, their method achieved strong performance 92.5% accuracy, 89.94% sensitivity, and 94.53% specificity for AD vs. HC classification. The generated PET scans were visually realistic and had a PSNR of 24.49. While effective, the method depends on careful image alignment and patch extraction. Future work could explore more flexible, end-to-end models for better generalization.
Han et al. [] proposed an unsupervised GAN-based method to detect Alzheimer’s disease by reconstructing MRI slice sequences using WGAN-GP with L1 loss. Trained on healthy brain scans from the OASIS-3 dataset, the model identifies anomalies based on reconstruction errors. It achieved AUCs of 0.780 (CDR 0.5), 0.833 (CDR 1), and 0.917 (CDR 2), effectively detecting early to advanced AD stages. The approach requires no labeled pathological data, making it scalable, but it may overlook abnormalities outside selected brain regions. Future work could explore full-brain coverage and multimodal data for broader detection.
Shin et al. [] proposed GANDALF, a GAN-based model that combines MRI-to-PET synthesis with AD classification using discriminator-adaptive loss fine-tuning. Trained end-to-end on the ADNI dataset, it achieved 85.2% accuracy for AD/CN, 78.7% for AD/MCI/CN (Precision: 0.83, Recall: 0.66), and 37.0% for AD/LMCI/EMCI/CN classification. While it outperformed baseline methods in multi-class tasks, binary performance was similar to CNN-only models. More advanced architecture and tuning could further enhance results.
Islam and Zhang [] designed a GAN-based approach to generate synthetic brain PET images for different stages of Alzheimer’s disease Normal Control (NC), Mild Cognitive Impairment (MCI), and Alzheimer’s Disease (AD). Using a Deep Convolutional GAN (DCGAN), they trained the model on real PET scans from the ADNI dataset to produce high-quality synthetic images. The generated images showed strong visual and statistical similarity to real ones, achieving a PSNR of 32.83 and SSIM of 77.48. When used to train a CNN classifier, these synthetic images improved classification accuracy by 10%, reaching 71.45%. Although promising, the method’s limitations include the need to train separate models for each class and the reliance on 2D image slices.
Hu et al. [] introduced a Bidirectional GAN to generate realistic brain PET images from MR scans, aiming to preserve individual brain structure differences. Their model uses a ResU-Net generator and ResNet-based encoder, combining adversarial, pixel-wise, and perceptual losses for better image quality. Tested on 680 subjects from the ADNI dataset, it achieved PSNR of 27.36, SSIM of 0.88, and improved AD vs. CN classification accuracy to 87.82%. While effective, the method could benefit from improved handling of latent vector injection for finer image details.
To address the issue of class imbalance in Alzheimer’s datasets, Hu et al. [] developed a DCGAN-based approach that generates synthetic PET images for underrepresented AD cases. These generated images were added to the training set to balance the data, improving the performance of a DenseNet-based classifier. As a result, classification accuracy increased from 67% to 74%. The synthetic images were evaluated using MMD (1.78) and SSIM (0.53), showing good diversity and realism. While effective, the method uses 2D generation and manual filtering. Future work could involve 3D GANs and smarter sample selection for better scalability.
Zhao et al. [] presented a 3D Multi-Information GAN (mi-GAN) framework that predicts Alzheimer’s Disease progression by generating future 3D brain MRI scans conditioned on baseline scans and patient metadata (age, gender, education, and APOE status). The model uses a 3D U-Net generator and DenseNet-based multi-class classifier optimized with focal loss. Evaluated on the ADNI dataset, mi-GAN achieved a high SSIM of 0.943, and the classifier reached 76.67% accuracy, with a pMCI vs. sMCI accuracy of 78.45%, outperforming previous cGAN and cross-entropy-based models. Limitations include less accuracy in gray matter prediction and short-term progression. Future work could benefit from improved gray matter modeling and broader clinical feature integration.
A novel unsupervised anomaly detection framework MADGAN was proposed by Han et al. (2021) [], that reconstructs multiple adjacent MRI slices to detect subtle brain anomalies like Alzheimer’s disease (AD) and brain metastases. The model uses a self-attention GAN architecture trained solely on healthy MRI slices to predict the next 3 slices from the previous 3, comparing reconstruction loss to detect anomalies. The method achieved promising AUCs for AD detection: 0.727 for early-stage (MCI) and 0.894 for late-stage AD, and 0.921 for brain metastases, highlighting its effectiveness across different disease types and stages. The key advantage of MADGAN lies in leveraging healthy data only, mimicking a physician’s diagnostic intuition. However, the reconstruction is less stable in texture consistency, especially for contrast-enhanced (T1c) images, and detection performance can vary with SA module configurations. Future work could involve enhancing attention mechanisms and exploring additional loss functions to improve generalizability and lesion localization.

Table 2.
Overview of medical imaging datasets and their modalities used in studies.
Table 2.
Overview of medical imaging datasets and their modalities used in studies.
| Reference | Dataset | Modality | Total Number of Participants |
|---|---|---|---|
| [] | ADNI-like MRI data (T1w), Diffusion MRI of healthy subjects | Structural MRI | 18 AD, 18 bvFTD, 19 control: 14 healthy for connectome |
| [] | ADNI-1 and ADNI-2 | MRI, PET | ADNI-1: 821; ADNI-2: 636 |
| [] | OASIS-3 | MRI | Training: 408 subjects; Test: 113 healthy, 99 (CDR 0.5), 61 (CDR 1), 4 (CDR 2) |
| [] | ADNI | MRI, PET | 1033 (722 train, 104 val, 207 test) |
| [] | ADNI | PET | 411 PET scans (98 AD, 105 NC, 208 MCI) |
| [] | ADNI | MRI, PET | 680 subjects |
| [] | ADNI-1, ADNI-2 | MRI, PET | ADNI-1 (PET AD only), ADNI-2 (100 NC, 20 AD, 80 AD MRI-only) |
| [] | ADNI-GO, ADNI-2, OASIS | 3D MRI | 210 (mi-GAN), 603 (classifier), 48 (validation) |
| [] | OASIS-3, Internal dataset | T1, T1c MRI | 408 (T1), 135 (T1c healthy) |
| [] | ADNI, AIBL, NACC | MRI (1.5-T and 3-T) | ADNI: 151 (training), AIBL: 107, NACC: 565 |
| [] | ADNI-1, ADNI-2 | MRI + PET | ADNI-1: 821; ADNI-2: 534 |
| [] | ADNI-1 | T1 MRI | 833 (221 AD, 297 MCI, 315 NC) |
| [] | ADNI (268 subjects) | rs-fMRI + DTI | 268 |
| [] | ADNI (13,500 3D MRI images after augmentation) | 3D Structural MRI | 138 (original), 13,500 (augmented scans) |
| [] | ADNI (1732 scan-pairs, 873 subjects) | MRI → Synthesized PET | 873 |
| [] | ADNI | T1-weighted MRI | 632 participants |
| [] | ADNI2 | T1-weighted MRI | 169 participants, 27,600 image pairs |
| [] | Custom MRI dataset (Kaggle) | T1-weighted Brain MRI | 6400 images (approx.) |
| [] | ADNI2, NIFD (in-domain), NACC (external) | T1-weighted MRI | 3319 MRI scans |
| [] | ADNI | MRI and PET (multimodal) | ~2400 (14,800 imaging sessions) |
| [] | ADNI (Discovery), SMC (Practice) | T1-weighted MRI, Demographics, Cognitive scores | 538 (ADNI) + 343 (SMC) |
| [] | ADNI | T1-weighted MRI | 362 (CN: 87, MCI: 211, AD: 64) |
| [] | ADNI1, ADNI3, AIBL | 1.5 T & 3 T MRI | ~168 for SR cohort, ~1517 for classification |
| [] | ADNI | T1-weighted MRI | 6400 images across 4 stages (Non-Demented, Very Mild, Mild, Moderate Demented) |
| [] | ADNI | Cognitive Features | 819 participants (5013 records) |
| [] | ADNI, OASIS-3, Centiloid | Low-res PET + MRI → High-res PET | ADNI: 334; OASIS-3: 113; Centiloid: 46 |
| [] | ADNI | MRI T1WI, FDG PET (Synth.) | 332 subjects, 1035 paired scans |
| [] | ADNI, OASIS, UK Biobank | 3D T1-weighted MRI | ADNI: 1188, OASIS: 600, UKB: 38,703 |
| [] | Alzheimer MRI (6400 images) | T1-weighted MRI | Alzheimer: 6400 images |
| [] | OASIS-3 | T1-weighted MRI | 300 (100 AD, 100 MCI, 100 NC) |
| [] | ADNI-3, In-house | Siemens ASL MRI (T1, M0, CBF) | ADNI Siemens: 122; GE: 52; In-house: 58 |
| [] | ADNI | MRI + Biospecimen (Aβ, t-tau, p-tau) | 50 subjects |
| [] | OASIS | MRI | 300 subjects (100 AD, 100 MCI, 100 NC) |
| [] | ADNI | MRI | 311 (AD: 65, MCI: 67, NC: 102, cMCI: 77) |
| [] | ADNI | Structural MRI → Aβ-PET, Tau-PET (synthetic) | 1274 |
| [] | Kaggle | MRI | 6400 images (4 AD classes) |
| [] | ADNI | sMRI, DTI, fMRI (multimodal) | 5 AD stages (NC, SMC, EMCI, LMCI, AD) |
Zhou et al. [] explored GANs to enhance 1.5 T MRI scans into 3 T-like images (3-T*) for Alzheimer’s classification using an FCN. These enhanced images not only looked better based on quality scores (BRISQUE, NIQE), but also helped a deep learning model improve its AD prediction accuracy boosting AUC from 0.907 to 0.932. However, the study was limited by a small sample size and did not include MCI cases. Expanding the dataset and refining the models could make this approach even more effective in future work.
A hybrid deep learning model combining TPA-GAN and PT-DCN to generate missing PET scans from MRI and classify Alzheimer’s disease is developed by Gao et al. (2022) []. Tested on ADNI-1/2, it achieved 90.7% accuracy (AUC 0.95) for AD vs. CN and 85.2% accuracy (AUC 0.89) for pMCI vs. sMCI. Despite strong results, its reliance on PET-MRI pairs and dataset-specific training limits generalizability. Future work could address this with domain adaptation.
Yu et al. [] proposed THS-GAN, a semi-supervised generative adversarial network that introduces tensor-train decomposition and high-order pooling (GSP block) for Alzheimer’s disease (AD) and mild cognitive impairment (MCI) classification from T1-weighted MRI scans. Their framework improves over traditional GANs by using a three-player cooperative game—generator, discriminator, and classifier—while tensor-train layers reduce parameters and preserve brain structural information. Global second-order pooling enhances discriminative feature representation. Trained on 833 ADNI MRI scans, the model achieved an AUC of 95.92% for AD vs. NC, 88.72% for MCI vs. NC, and 85.35% for AD vs. MCI, outperforming SS-GAN and triple-GAN baselines by a notable margin. THS-GAN also proved more data-efficient, delivering strong results with fewer labeled samples. Limitations include sensitivity to TT-rank tuning and GSP block positioning.
Pan and Wang [] developed CT-GAN, an innovative deep learning model that blends brain structure and function by combining DTI and rs-fMRI scans to improve Alzheimer’s diagnosis. By using attention-based transformers and GANs, the model effectively captured complex brain patterns and outperformed existing methods with accuracies of 94.44% (AD vs. NC), 93.55% (LMCI vs. NC), and 92.68% (EMCI vs. NC). It also highlighted key brain regions like the hippocampus and precuneus. However, the study was limited by a small dataset and reliance on predefined brain regions.
Thota and Vasumathi [] introduced WGANGP-DTL, a classification framework combining Wasserstein GAN with Gradient Penalty and Deep Transfer Learning for Alzheimer’s detection using 3D MRI scans. It uses WGANGP for data augmentation, 3DS-FCM for segmentation, Inception v3 for feature extraction, and a Deep Belief Network for classification. Tested on 13,500 augmented images, the model achieved 99.70% accuracy, 99.09% sensitivity, and 99.82% specificity, outperforming other deep learning models. While highly effective, the method depends on extensive preprocessing and fine-tuning.
Zhang et al. [] introduced BPGAN, a 3D BicycleGAN-based model that synthesizes PET scans from MRI to tackle missing modality issues in AD diagnosis. Trained on 1732 MRI-PET pairs, it outperformed existing methods with strong image quality (e.g., SSIM 0.7294) and boosted AD classification accuracy by 1–4%, reaching 85.03% on Dataset-B. While effective, the model’s reliance on complex preprocessing and limited diagnostic improvement suggest future work should explore adaptive ROI localization and broader clinical use.
Yuan et al. [] developed ReMiND, a diffusion model (DDPM)-based method to generate missing 3D MRI scans in longitudinal Alzheimer’s studies. Using past or past-and-future scans, it outperformed autoencoders and simple methods in preserving brain structure, achieving SSIM of 0.895 and PSNR of 28.96 dB. However, it currently relies on fixed scan intervals and only nearby timepoints. Future work could include multiple timepoints and multimodal data for improved tracking of disease progression.
Huang et al. [] proposed a wavelet-guided diffusion model to enhance low-resolution MRI scans, aiming to improve Alzheimer’s diagnosis. Using a Wavelet U-Net with DDPM, the model achieved SSIM of 0.8201, PSNR of 27.15, and FID of 13.15, also improving classification accuracy. However, it is computationally intensive and limited to T1-weighted MRIs. Future work could expand to multi-modal and longitudinal data for broader clinical use.
Boyapati et al. [] introduced a deep learning approach that combines CNNs with GANs to boost Alzheimer’s detection from MRI scans. By generating synthetic images and using filters to enhance image quality, their method achieved a strong 96% accuracy outperforming standard CNN models. Still, the study was limited by a small number of real moderate AD cases and the lack of other imaging types like PET or fMRI.
Nguyen et al. [] developed a deep learning method to distinguish AD, FTD, and healthy individuals using structural MRI. By combining brain atrophy data with grading maps from an ensemble of 3D U-Nets, their model achieved 86% accuracy and offered interpretable results. While it generalized well to other datasets, it is resource-intensive and limited to baseline MRI. Future work could include multi-modal and longitudinal data to better track disease progression.
Uday Sekhar et al. [] combined MRI and PET scans with GAN-generated synthetic data to improve early Alzheimer’s detection. By training an ensemble of models, including CNNs and LSTMs, they boosted diagnostic performance reaching an F1-score of 0.82 and an AUC of 0.93. The study shows how combining real and synthetic data can enhance accuracy and generalization. However, it was limited by a small number of real Alzheimer’s cases and lacked testing on outside datasets. Future work could benefit from more diverse and long-term data to better capture disease progression.

Table 3.
Performance metrics of GAN-based methods and Diffusion Models used in the reviewed literature.
Table 3.
Performance metrics of GAN-based methods and Diffusion Models used in the reviewed literature.
| Reference | Technique/Method | Model | Results |
|---|---|---|---|
| [] | Network Diffusion Model | Network eigenmode diffusion model | Strong correlation between predicted and actual atrophy maps; eigenmodes accurately classified AD/bvFTD; ROC AUC higher than PCA |
| [] | 3D CycleGAN + LM3IL | Two-stage: PET synthesis (3D-cGAN) + classification (LM3IL) | AD vs. HC—Accuracy: 92.5%, Sensitivity: 89.94%, Specificity: 94.53%; PSNR: 24.49 ± 3.46 |
| [] | WGAN-GP + L1 loss (MRI slice reconstruction) | WGAN-GP-based unsupervised reconstruction + anomaly detection using L2 loss | AUC: 0.780 (CDR 0.5), 0.833 (CDR 1), 0.917 (CDR 2) |
| [] | GANDALF: GAN with discriminator-adaptive loss for MRI-to-PET synthesis and AD classification | GAN + Classifier | Binary (AD/CN): 85.2% Acc3-class: 78.7% Acc, F2: 0.69, Prec: 0.83, Rec: 0.664-class: 37.0% Acc |
| [] | DCGAN to generate PET images for NC, MCI, and AD | DCGAN | PSNR: 32.83, SSIM: 77.48, CNN classification accuracy improved to 71.45% with synthetic data |
| [] | Bidirectional GAN with ResU-Net generator, ResNet-34 encoder, PatchGAN discriminator | Bidirectional GAN | PSNR: 27.36, SSIM: 0.88; AD vs. CN classification accuracy: 87.82% with synthetic PET |
| [] | DCGAN for PET synthesis from noise; DenseNet classifier for AD vs. NC | DCGAN + DenseNet | Accuracy improved from 67% to 74%; MMD: 1.78, SSIM: 0.53 |
| [] | 3D patch-based mi-GAN with baseline MRI + metadata; 3D DenseNet with focal loss for classification | mi-GAN + DenseNet | SSIM: 0.943, Multi-class Accuracy: 76.67%, pMCI vs. sMCI Accuracy: 78.45% |
| [] | MADGAN: GAN with multiple adjacent slice reconstruction using WGAN-GP + ℓ1 loss and self-attention | 7-SA MADGAN | AUC for AD: 0.727 (MCI), 0.894 (late AD); AUC for brain metastases: 0.921 |
| [] | Generative Adversarial Network (GAN), Fully Convolutional Network (FCN) | GAN + FCN | Improved AD classification with accuracy increases up to 5.5%. SNR, BRISQUE, and NIQE metrics showed significant image quality improvements. |
| [] | TPA-GAN for PET imputation, PT-DCN for classification | TPA-GAN + PT-DCN | AD vs. CN: ACC 90.7%, SEN 91.2%, SPE 90.3%, F1 90.9%, AUC 0.95; pMCI vs. sMCI: ACC 85.2%, AUC 0.89 |
| [] | THS-GAN: Tensor-train semi-supervised GAN with high-order pooling and 3D-DenseNet | THS-GAN | AD vs. NC: AUC 95.92%, Acc 95.92%; MCI vs. NC: AUC 88.72%, Acc 89.29%; AD vs. MCI: AUC 85.35%, Acc 85.71% |
| [] | CT-GAN with Cross-Modal Transformer and Bi-Attention | GAN + Transformer with Bi-Attention | AD vs. NC: Acc = 94.44%, Sen = 93.33%, Spe = 95.24%LMCI vs. NC: Acc = 93.55%, Sen = 90.0%, Spe = 95.24%EMCI vs. NC: Acc = 92.68%, Sen = 90.48%, Spe = 95.0% |
| [] | WGANGP-DTL (Wasserstein GAN with Gradient Penalty + Deep Transfer Learning using Inception v3 and DBN) | WGANGP + Inception v3 + DBN | Accuracy: 99.70% Sensitivity: 99.09% Specificity: 99.82% F1-score: >99% |
| [] | BPGAN (3D BicycleGAN with Multiple Convolution U-Net, Hybrid Loss) | 3D BicycleGAN (BPGAN) with MCU Generator | Dataset-A: MAE = 0.0318, PSNR = 26.92, SSIM = 0.7294Dataset-B: MAE = 0.0396, PSNR = 25.08, SSIM = 0.6646 Diagnosis Acc = 85.03% (multi-class, MRI + Synth. PET) |
| [] | ReMiND (Diffusion-based MRI Imputation) | Denoising Diffusion Probabilistic Model (DDPM) with modified U-Net | SSIM: 0.895, PSNR: 28.96; no classification metrics reported |
| [] | Wavelet-guided Denoising Diffusion Probabilistic Model (Wavelet Diffusion) | Wavelet Diffusion with Wavelet U-Net | SSIM: 0.8201, PSNR: 27.15, FID: 13.15 (×4 scale); Recall ~90% (AD vs. NC); improved classification performance overall |
| [] | CNN + GAN (DCGAN to augment data; CNN for classification) | CNN + DCGAN (data augmentation) | Accuracy: 96% (with GAN), 69% (without GAN); classification across 4 AD stages |
| [] | Deep Grading + Multi-layer Perceptron + SVM Ensemble (Structure Grading + Atrophy) | 125 3D U-Nets + Ensemble (MLP + SVM) | In-domain (3-class): Accuracy: 86.0%, BACC: 84.7%, AUC: 93.8%, Sensitivity (CN/AD/FTD): 89.6/83.2/81.3; Out-of-domain: Accuracy: 87.1%, BACC: 81.6%, AUC: 91.6%, Sensitivity (CN/AD/FTD): 89.6/76.9/78.4 |
| [] | GAN for synthetic MRI generation + Ensemble deep learning classifiers | GAN + CNN, LSTM, Ensemble Networks | GAN results: Precision: 0.84, Recall: 0.76, F1-score: 0.80, AUC-ROC: 0.91, Proposed Ensemble: Precision: 0.85, Recall: 0.79, F1-score: 0.82, AUC-ROC: 0.93 |
| [] | Modified HexaGAN (Deep Generative Framework) | Modified HexaGAN (GAN + Semi-supervised + Imputation) | ADNI: AUROC 0.8609, Accuracy 0.8244, F1-score 0.7596, Sensitivity 0.8415, Specificity 0.8178; SMC: AUROC 0.9143, Accuracy 0.8528, Sensitivity 0.9667, Specificity 0.8286. |
| [] | Conditional Diffusion Model for Data Augmentation | Conditional DDPM + U-Net | Best result (Combine 900): Accuracy: 74.73%, Precision: 77.28%, Recall (Sensitivity): 66.52%, F1-score: 0.6968, AUC: 0.8590; Specificity: not reported |
| [] | Latent Diffusion Model (d3T*) for MRI super-resolution + DenseNet Siamese Network for AD/MCI/NC classification | Latent Diffusion-based SR + Siamese DenseNet | AD classification: Accuracy 92.3%, AUROC 93.1%, F1-score 91.9%; Significant improvement over 1.5 T and c3T*; Comparable to real 3 T MRI |
| [] | GAN-based data augmentation + hybrid CNN-InceptionV3 model for multiclass AD classification | GAN + Transfer Learning (CNN + InceptionV3) | Accuracy: 90.91%; metrics like precision, recall, and F1-score also reported high performance |
| [] | DeepCGAN (GAN + BiGRU with Wasserstein Loss) | Encoder–Decoder GAN with BiGRU layers | Accuracy: 97.32%, Recall (Sensitivity): 95.43%, Precision: 95.31%, F1-Score: 95.61%, AUC: 99.51% |
| [] | Latent Diffusion Model for Resolution Recovery (LDM-RR) | Latent Diffusion Model (LDM-RR) | Recovery coefficient: 0.96; Longitudinal p-value: 1.3 × 10−10; Cross-tracer correlation: r = 0.9411; Harmonization p = 0.0421 |
| [] | Diffusion-based multi-view learning (one-way and two-way synthesis) | U-NET-based Diffusion Model with MLP Classifier | Accuracy: 82.19%, SSIM: 0.9380, PSNR: 26.47, Sensitivity: 95.19%, Specificity: 92.98%, Recall: 82.19% |
| [] | Conditional DDPM and LDM with counterfactual generation and DenseNet121 classifier | LDM + 3D DenseNet121 CNN | AUC: 0.870, F1-score: 0.760, Sensitivity: 0.889, Specificity: 0.837 (ADNI test set after fine-tuning) |
| [] | GANs, VAEs, Diffusion (DDIM) models for MRI generation + DenseNet/ResNet classifiers | DDIM (Diffusion Model) + DenseNet | Accuracy: 80.84%, Precision: 86.06%, Recall: 78.14%, F1-Score: 80.98% (Alzheimer’s, DenseNet + DDIM) |
| [] | GAN-based data generation + EfficientNet for multistage classification | GAN for data augmentation + EfficientNet CNN | Accuracy: 88.67% (1:0), 87.17% (9:1), 82.50% (8:2), 80.17% (7:3); Recall/Sensitivity/Specificity not separately reported |
| [] | Conditional Latent Diffusion Model (LDM) for M0 image synthesis from Siemens PASL | Conditional LDM + ML classifier | SSIM: 0.924, PSNR: 33.35, CBF error: 1.07 ± 2.12 mL/100 g/min; AUC: 0.75 (Siemens), 0.90 (GE) in AD vs. CN classification |
| [] | Multi-modal conditional diffusion model for image-to-image translation (prognosis prediction) | Conditional Diffusion Model + U-Net | PSNR: 31.99 dB, SSIM: 0.75, FID: 11.43 |
| [] | GAN for synthetic MRI image generation + EfficientNet for multi-stage classification | GAN + EfficientNet | Validation accuracy improved from 78.48% to 85.11%, training accuracy from 90.16% to 98.68% with GAN data |
| [] | Dual GAN + Pyramid Attention + CNN | Dual GAN with Pyramid Attention and CNN | Accuracy: 98.87%, Recall/Sensitivity: 95.67%, Specificity: 98.78%, Precision: 99.78%, F1-score: 99.67% |
| [] | Prior-information-guided residual diffusion model with CLIP module and intra-domain difference loss | Residual Diffusion Model with CLIP guidance | SSIM: 92.49% (Aβ), 91.44% (Tau); PSNR: 26.38 dB (Aβ), 27.78 dB (Tau); AUC: 90.74%, F1: 82.74% (Aβ); AUC: 90.02%, F1: 76.67% (Tau) |
| [] | Hybrid of Deep Super-Resolution GAN (DSR-GAN) for image enhancement + CNN for classification | DSR-GAN + CNN | Accuracy: 99.22%, Precision: 99.01%, Recall: 99.01%, F1-score: 99.01%, AUC: 100%, PSNR: 29.30 dB, SSIM: 0.847, MS-SSIM: 96.39% |
| [] | Bidirectional Graph GAN (BG-GAN) + Inner Graph Convolution Network + Balancer for stable multimodal connectivity generation | BG-GAN + InnerGCN | Accuracy > 96%, Precision/Recall/F1 ≈ 0.98–1.00, synthetic data outperformed real in classification |
Hwang et al. (2023) [] introduced a modified HexaGAN model to predict amyloid positivity in cognitively normal individuals using MRI, cognitive scores, and demographic data. Built to handle missing data and imbalanced classes, the model achieved strong performance with AUROCs of 0.86 (ADNI) and 0.91 (clinical dataset). While promising, its complexity and need for fine-tuning limit scalability. Future work could focus on adding multi-modal and longitudinal data to boost clinical usefulness.
Yao et al. (2023) [] introduced a conditional diffusion model to generate synthetic MRI slices for Alzheimer’s diagnosis, helping to balance limited and uneven datasets. Using diagnostic labels to guide image creation, the model improved classification performance achieving 74.73% accuracy and an AUC of 0.8590, outperforming GAN-based methods. However, it used a small, static dataset and lacked specificity reporting.
Yoon et al. (2024) [] introduced a diffusion-based MRI super-resolution model (d3T) to enhance Alzheimer’s and MCI diagnosis by upgrading 1.5 T scans to 3 T quality. The improved images boosted diagnostic accuracy to 92.3% and helped predict MCI-to-AD conversion more reliably. While the results are promising, the model was tested only on ADNI data and lacks validation on higher-resolution scans or multi-center datasets. Future work could explore 3 T-to-7 T enhancement and integrate multi-modal imaging for broader clinical use.
Tufail et al. (2024) [] combined InceptionV3 with GANs to improve Alzheimer’s diagnosis from MRI scans, especially in imbalanced datasets. By generating synthetic images to boost underrepresented classes, their model achieved an AUC of 87% and showed better accuracy for early-stage AD. However, it was limited to one dataset and lacked multi-modal input. Future work should be tested on diverse datasets and include other imaging types like PET for more reliable diagnosis.
Ali et al. (2024) [] developed DeepCGAN, a deep learning model that uses cognitive test data over time rather than brain scans to detect Alzheimer’s early. With a BiGRU-enhanced GAN design and advanced loss functions, it achieved impressive results: 97.32% accuracy and a near-perfect AUC of 99.51%. While powerful, the model is complex and resource-heavy. Future improvements could involve combining it with imaging data or simplifying the architecture for easier clinical use.
Shah et al. (2024) [] developed a latent diffusion model (LDM-RR) to boost the clarity and accuracy of amyloid PET scans by generating high-resolution images using paired low-res PET and MRI data. Trained in synthetic examples and tested across major datasets like ADNI and OASIS-3, the model delivered strong results in a recovery coefficient of 0.96, better detection of amyloid buildup over time (p = 1.3 × 10−10), and improved consistency across different PET tracers (r = 0.9411, p = 0.0421). Though standard metrics like accuracy and sensitivity were not detailed, the model clearly outperformed older methods. Its main drawbacks are high computational demands and reliance on synthetic data. The authors suggest future improvements like faster, self-supervised training and broader real-world validation.
Chen et al. (2024) [] introduced a diffusion-based method to generate FDG PET scans from MRI T1 images for Alzheimer’s diagnosis. Their two-way diffusion approach transforming MRI to PET via a diffusion and reconstruction process outperformed the simpler one-way method, achieving 82.19% accuracy and high sensitivity (95.19%) and specificity (92.98%). While effective, the method is computationally intensive. The authors suggest that combining diffusion with other generative models like GANs could enhance performance and efficiency.
Dhinagar et al. (2024) [] developed an interpretable diffusion-based method to generate 3D brain MRIs for Alzheimer’s diagnosis. Using DDPMs and Latent Diffusion Models, they created realistic and even counterfactual scans imagining how an AD patient’s brain might look if healthy. These synthetic images helped pre-train a 3D CNN, which achieved strong results on the ADNI dataset (AUC 0.87, sensitivity 88.9%, specificity 83.7%) and generalized well to OASIS. Despite the high performance, the approach is computationally heavy and may benefit from efficiency improvements and added clinical data in the future.
Gajjar et al. (2024) [] compared GANs, VAEs, and diffusion models (DDIMs) to generate MRI scans for diagnosing Alzheimer’s and Parkinson’s diseases. Diffusion models delivered the best results of 80.84% accuracy for Alzheimer’s and 92.42% for Parkinson’s though they were slow to train. VAEs produced lower-quality images but still supported decent classification. The study highlights the potential of generative models to boost diagnostic accuracy with limited data, though future work should focus on making diffusion models faster and more efficient for clinical use.
Wong et al. (2024) [] tackled the challenge of limited MRI data in Alzheimer’s research by using GANs to generate synthetic images. These were combined with real scans to train an EfficientNet model for classifying AD, MCI, and NC stages. Even with less real data, the model maintained strong accuracy up to 88.67% with full data and 80.17% with just 70% real data. While GAN training had some stability issues and too much synthetic data slightly hurt performance, the approach proved that GANs can effectively boost diagnosis when data is scarce.
Shou et al. (2024) [] addressed the issue of missing M0 calibration images in Siemens ASL MRI scans, which was key for measuring brain blood flow in Alzheimer’s research by using a conditional latent diffusion model to generate them. Their model produced high-quality images (SSIM 0.924, PSNR ~33.35) and accurate CBF values, aligning well with known disease patterns. It also showed solid classification performance, especially with GE data (AUC up to 0.90). While the method helps bridge gaps across MRI vendors, it is limited by data imbalances and the absence of ground truth in some datasets. Future work could focus on the standardization and demographic adjustments to improve robustness.
Hwang et al. (2024) [] created a multi-modal diffusion model to predict Alzheimer’s progression by generating future MRI scans using early imaging and clinical biomarkers like Aβ, t-tau, and p-tau. Their model outperformed GANs and image-only diffusion methods, producing clearer, more detailed scans (PSNR 31.99, SSIM 0.75, FID 11.43). Adding clinical data notably improved results. However, the study used only 50 subjects and did not report diagnostic metrics like accuracy, suggesting the need for larger datasets and broader clinical inputs to boost real-world impact.
Wong et al. (2025) [] addressed the challenge of limited and imbalanced MRI data for Alzheimer’s diagnosis by using GANs to generate synthetic brain images. Trained on OASIS data, these images boosted the performance of an EfficientNet model classifying AD, MCI, and NC stages. Accuracy rose from 78.48% to 85.11% when using entirely synthetic data, though GAN training faced stability issues and some overfitting. The study shows promise for GAN-based augmentation, with future work focusing on improving training stability and exploring more robust GAN models.
Zhang and Wang (2024) [] developed a dual GAN framework with pyramid attention and a CNN classifier to boost Alzheimer’s detection from MRI scans. Trained on ADNI data, the model achieved outstanding results in 98.87% accuracy and a 99.67% F1-score far outperforming standard CNNs. While powerful, its reliance on a single dataset and lack of clinical interpretability limit real-world use. The authors recommend adding data like cognitive scores or genetics and enhancing explainability to improve clinical relevance.
Ou et al. (2024) [] introduced a diffusion model to generate Aβ and tau PET images from MRI scans, aiming to reduce the cost and radiation of PET imaging in Alzheimer’s diagnosis. Their method uses residual learning, prior info like age and gender, and a novel loss function to boost image quality and modality distinction. Tested on 1274 ADNI subjects, it outperformed other models in image quality (SSIM~92%) and achieved strong biomarker classification results (AUC~90%). Though promising, it relies on accurate prior data and high computing power. Future work will aim to improve efficiency and adapt the model to diverse populations.

Table 4.
Overview of generative models used in literature to diagnose Alzheimer’s disease research highlighting their key challenges and limitations.
Table 4.
Overview of generative models used in literature to diagnose Alzheimer’s disease research highlighting their key challenges and limitations.
| Reference | Dataset | Model | Challenges and Limitations |
|---|---|---|---|
| [] | ADNI-like MRI data (T1w), Diffusion MRI of healthy subjects | Network eigenmode diffusion model | Small sample size; no conventional ML metrics (accuracy, F1); assumes static connectivity; limited resolution in tractography; noise in MRI volumetrics. |
| [] | ADNI-1 & ADNI-2 | Two-stage: PET synthesis (3D-cGAN) + classification (LM3IL) | Requires accurate MRI–PET alignment; patch-based learning may limit generalization. |
| [] | OASIS-3 | WGAN-GP-based unsupervised reconstruction + anomaly detection using L2 loss | Region-limited detection (hippocampus/amygdala); may miss anomalies outside selected areas. |
| [] | ADNI | GAN + Classifier | Binary classification was not better than CNN-only models; requires more tuning and architectural exploration. |
| [] | ADNI | DCGAN | Trained separate GANs per class; used 2D slices only; lacks unified 3D modeling approach. |
| [] | ADNI | Bidirectional GAN | Limited fine detail in some outputs; latent vector injection mechanism could be improved for better synthesis. |
| [] | ADNI-1, ADNI-2 | DCGAN + DenseNet | Used 2D image generation; manual filtering of outputs; lacks 3D modeling and automation. |
| [] | ADNI-GO, ADNI-2, OASIS | mi-GAN + DenseNet | Lower performance on gray matter prediction; limited short-term progression prediction; improvement possible with better feature modeling. |
| [] | OASIS-3, Internal dataset | 7-SA MADGAN | Reconstruction instability on T1c scans; limited generalization; fewer healthy T1c scans; needs optimized attention modules. |
| [] | ADNI, AIBL, NACC | GAN + FCN | Small sample size for GAN training (151 participants). Limited to AD vs. normal cognition (no MCI). |
| [] | ADNI-1, ADNI-2 | TPA-GAN + PT-DCN | Requires paired modalities; model trained/tested on ADNI-1/2 independently; limited generalization. |
| [] | ADNI-1 | THS-GAN | Requires careful TT-rank tuning; performance varies with GSP block position; validation limited to ADNI dataset. |
| [] | ADNI (268 subjects) | GAN + Transformer with Bi-Attention | Limited dataset size, dependency on predefined ROIs, potential overfitting; lacks validation on other neurodegenerative disorders. |
| [] | ADNI (13,500 3D MRI images after augmentation) | WGANGP + Inception v3 + DBN | Heavy reliance on data augmentation, complex pipeline requiring multiple preprocessing and tuning steps. |
| [] | ADNI (1732 scan-pairs, 873 subjects) | 3D BicycleGAN (BPGAN) with MCU Generator | High preprocessing complexity, marginal diagnostic gains, requires broader validation and adaptive ROI exploration. |
| [] | ADNI | Denoising Diffusion Probabilistic Model (DDPM) with modified U-Net | Uses only adjacent timepoints; assumes fixed intervals; no classification; no sensitivity/specificity; computationally intensive. |
| [] | ADNI2 | Wavelet Diffusion with Wavelet U-Net | High computational cost; limited to T1 MRI; does not incorporate multi-modal data or longitudinal timepoints. |
| [] | Custom MRI dataset (Kaggle) | CNN + DCGAN (data augmentation) | Risk of overfitting due to small original dataset; no reporting of sensitivity/specificity; limited to image data. |
| [] | ADNI2, NIFD (in-domain), NACC (external) | 125 3D U-Nets + Ensemble (MLP + SVM) | High computational cost (393 M parameters, 25.9 TFLOPs); inference time ~1.6 s; only baseline MRI used; limited by class imbalance and absence of multimodal or longitudinal data. |
| [] | ADNI | GAN + CNN, LSTM, Ensemble Networks | Limited real Alzheimer’s samples; reliance on synthetic augmentation; needs more external validation and data diversity. |
| [] | ADNI (Discovery), SMC (Practice) | Modified HexaGAN (GAN + Semi-supervised + Imputation) | High model complexity; requires fine-tuning across datasets; limited to MRI and tabular inputs. |
| [] | ADNI | Conditional DDPM + U-Net | Small dataset, imbalanced classes; specificity not reported; limited to static MRI slices; no multi-modal or longitudinal data. |
| [] | DNI1, ADNI3, AIBL | Latent Diffusion-based SR + Siamese DenseNet | High computational cost; needs advanced infrastructure for training; diffusion SR takes longer than CNN-based methods. |
| [] | ADNI | GAN + Transfer Learning (CNN + InceptionV3) | Class imbalance still impacts performance slightly; more detailed metrics (sensitivity/specificity) not reported. |
| [] | ADNI | Encoder–Decoder GAN with BiGRU layers | Computational complexity, GAN training instability, underutilization of multimodal data (e.g., neuroimaging). |
| [] | ADNI, OASIS-3, Centiloid | Latent Diffusion Model (LDM-RR) | High computational cost; trained on synthetic data; limited interpretability; real-time deployment needs optimization. |
| [] | ADNI | U-NET-based Diffusion Model with MLP Classifier | One-way synthesis introduces variability; computational intensity; requires improvement in generalization and speed. |
| [] | ADNI, OASIS, UK Biobank | LDM + 3D DenseNet121 CNN | High computational cost, requires careful fine-tuning, limited by resolution/memory constraints. |
| [] | Alzheimer MRI (6400 images) | DDIM (Diffusion Model) + DenseNet | Diffusion models are computationally intensive; VAE had low image quality; fine-tuning reduced accuracy in some cases; computational cost vs. performance tradeoff. |
| [] | OASIS-3 | GAN for data augmentation + EfficientNet CNN | GAN training instability; performance drop when synthetic data exceeds real data; no separate sensitivity/specificity metrics reported. |
| [] | ADNI-3, In-house | Conditional LDM + ML classifier | No ground truth M0 for Siemens data; SNR difference between PASL/pCASL; class imbalance; vendor variability. |
| [] | ADNI | Conditional Diffusion Model + U-Net | Small sample size; no classification metrics reported; requires broader validation with more diverse data. |
| [] | OASIS | GAN + EfficientNet | GAN training instability; overfitting in CNN; limited dataset size; scope to explore alternate GAN models for robustness. |
| [] | ADNI | Dual GAN with Pyramid Attention and CNN | Dependent on ADNI dataset quality; generalization affected by population diversity; limited interpretability; reliance on image features for AD detection. |
| [] | ADNI | Residual Diffusion Model with CLIP guidance | Dependent on accurate prior info (e.g., age, gender); high computational cost; needs optimization for broader demographic generalization. |
| [] | Kaggle | DSR-GAN + CNN | High computational complexity; SR trained on only 1700 images; generalizability and real-time scalability remain open challenges. |
| [] | ADNI | BG-GAN + InnerGCN | Difficulty in precise structure-function mapping due to fMRI variability; biological coordination model can be improved. |
A hybrid model combining Deep Super-Resolution GAN (DSR-GAN) with a CNN to classify Alzheimer’s into four stages using MRI scans is presented by Oraby et al. (2025) []. The GAN enhanced image clarity, helping the CNN achieve high accuracy of 99.22% with nearly perfect precision, recall, F1-score (all ~99%), and an AUC of 100%. The super-resolution also improved image quality (PSNR 29.30, SSIM 0.847). Despite its strong results, the model requires heavy computation and has limited scalability. Future work should expand datasets, including other imaging types like PET, and add explainability to support clinical use.
Zhou et al. (2025) [] developed BG-GAN, a generative model that captures relationships between brain structure (sMRI/DTI) and function (fMRI) for Alzheimer’s diagnosis. The model combines a Bidirectional Graph GAN, Inner Graph Convolutional Network (InnerGCN), and a Balancer module to stabilize training and improve data generation. Trained on the ADNI dataset across five subject categories, Normal Control (NC), Subjective Memory Complaint (SMC), Early Mild Cognitive Impairment (EMCI), Late MCI (LMCI), and Alzheimer’s Disease (AD). Results showed that BG-GAN outperformed baseline GCN, GAE, and GAT models, achieving classification accuracy above 96%, with precision, recall, and F1-scores nearing 0.98–1.00 on multi-modal inputs. Interestingly, synthetic data generated by BG-GAN even yielded better classification performance than real empirical data, indicating the effectiveness of the generative modeling approach. However, challenges remain in fully capturing structural-functional brain mappings due to fMRI variability. The authors recommend integrating more biologically grounded coordination models and testing applicability across other brain disorders.
5. Dataset
Alzheimer’s Disease Neuroimaging Initiative (ADNI): It is one of the most trusted and widely used datasets in Alzheimer’s research. Started in 2003 across 50 sites in the U.S. and Canada and led by Dr. Michael W. Weiner. Over the years, the study has expanded through phases ADNI-1, ADNI-2, and ADNI-3 and includes adults aged 55 to 90 []. Each dataset offers a wide range of imaging modalities including sMRI, fMRI, PET and FDG PET. ADNI-1 consists of 95 patients with Alzheimer’s disease (AD), 206 with mild cognitive impairment (MCI), and 102 cognitively normal (NC) controls. Meanwhile, the ADNI-3 cohort expands with 122 AD patients, 387 individuals with MCI, and 605 healthy controls [].
Open Access Series of Imaging Studies (OASIS): Most widely used, open-access neuroimaging dataset designed to support dementia and aging research. It includes three versions: OASIS-1 features 434 cross-sectional MRI scans from 416 individuals; OASIS-2 provides 373 longitudinal MRIs from 150 older adults; and OASIS-3, the largest, offers over 2000 MRI and PET scans from 1098 participants aged 42 to 96 []. The dataset is ideal for studying brain aging, disease progression, and cognitive decline.
Kaggle: It is a popular online platform that hosts a publicly available MRI dataset for dementia classification, widely used in AI research. This dataset includes a total of 6400 MRI scans, labeled across four classes: 3200 Non-Demented (ND), 2240 Very Mild Demented (VMD), 896 Mild Demented (MD), and 64 Moderate Demented (MoD) cases [].
Minimal Interval Resonance Imaging in Alzheimer’s Disease (MIRIAD): The dataset was designed to study brain changes in Alzheimer’s disease over short periods. It includes 708 T1-weighted MRI scans collected from 69 older adults 46 with mild-to-moderate Alzheimer’s and 23 healthy controls. All scans were performed using the same machine and technician, helping to ensure consistency in the data []. Participants were grouped based on their MMSE scores, making it easier to track how brain structure differs between healthy aging and Alzheimer’s.
Australian Imaging, Biomarkers and Lifestyle Study of Aging (AIBL): AIBL is a longitudinal study based in Australia, involving over 1100 participants aged 60 and above, recruited from cities such as Melbourne and Perth. It includes healthy controls, MCI, and AD patients. It provides MRI, PET, blood biomarkers, genetic data, and cognitive assessments. The dataset is especially useful for studying early detection and how lifestyle and biological factors contribute to Alzheimer’s progression [].
6. Generative Adversarial Networks (GANs)
Originally introduced by Goodfellow et al. [], they have become increasingly popular in medical imaging research, especially for diseases like Alzheimer’s where data can be limited. A typical GAN framework involves two neural networks: a generator that tries to produce realistic synthetic images from random noise, and a discriminator that tries to tell the difference between real and fake images. These two networks train together in a competitive setup, which can be expressed with the following objective function:
In Equation (1) is the generator network that maps noise z to synthetic data, D is discriminator network, x is real image from the true data distribution , z represents latent noise vector drawn from prior . discriminator’s probability estimate that x is real, generator’s synthetic output.
Over time, this basic idea has evolved into several powerful variants, each designed to solve specific challenges in medical imaging. In this section, we highlighted some of the most relevant GAN models used in Alzheimer’s disease research. A standard GAN architecture has been illustrated in Figure 8.
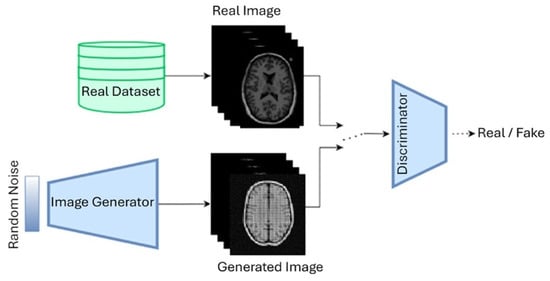
Figure 8.
GAN architecture.
6.1. Deep Convolutional Gan (DCGAN)
DCGAN [] replaces traditional fully connected layers with convolutional ones, allowing it to better capture spatial features in images. This model is especially useful for generating realistic-looking synthetic MRI scans and addressing the instability of the basic GAN. In Alzheimer’s studies, researchers have used DCGAN to generate more training data, which helps improve the performance of diagnostic models, especially when real data is scarce.
Here, output of the generator given noise input weight matrices for each transposed convolutional layer. is the activation function and hyperbolic tangent activation function.
6.2. Conditional Gan (CGAN)
Unlike standard GANs, CGAN [] allows you to guide the image generation process using extra information such as disease stage, diagnosis label, or other metadata. This is incredibly helpful when you want to generate images specific to the early, mild, or advanced stages of Alzheimer’s. The training objective is adjusted accordingly:
This label conditioning makes it possible to create highly relevant training data for classification models. In Equation (3) is the discriminator output and is the generator output conditioned on .
6.3. CycleGAN
CycleGAN [] is a great option when you have two different imaging modalities (like MRI and PET scans) but no matched pairs. It learns to translate images from one domain to another and back again using a cycle-consistency loss. This ensures that an MRI converted into a PET image and then back again still resembles the original MRI:
This technique is especially useful in Alzheimer’s research to enrich data when multi-modal data alignment is not possible. is cycle-consistency loss used to enforce bijective mappings between two domains. Here generator maps data from X to Y domain. reconstructed image in domain X after translating to Y via , then back via F. reconstructed image in domain Y after translating to X via F, then back via . and are real sample from source domain X and domain Y.
6.4. StyleGAN
StyleGAN [] introduces a different approach by adding “style” control at different layers of the generator. This allows researchers to manipulate specific features in the generated images like shape, contrast, or structure more independently. Although not as widely adopted yet in Alzheimer’s work, StyleGAN’s ability to generate detailed, high-resolution images opens up exciting possibilities for creating realistic brain scans that capture subtle disease characteristics.
where is the mapping network that refines the input noise, and is the synthesis network modulated by styles .
6.5. Wasserstein GAN (WGAN)
One of the common issues with traditional GANs is instability during training, which often results in mode collapse or non-converging models. To address these issues, [] introduced the Wasserstein GAN (WGAN), which replaces the JS divergence with the Earth Mover (EM) or Wasserstein-1 distance. This change provides more meaningful gradients, improving both convergence and training stability. WGAN is also relatively simple to implement and offers better control over training dynamics, helps generate more reliable and realistic brain scans. The drawback of WGAN is its slow optimization compared to traditional GANs.
Figure 9 Illustrates the similarities and relationships between various GANs (DCGAN, CGAN, CycleGAN, StyleGAN, WGAN) and Diffusion Models (conditional DDPMs, CDM, LDM, SGMS).
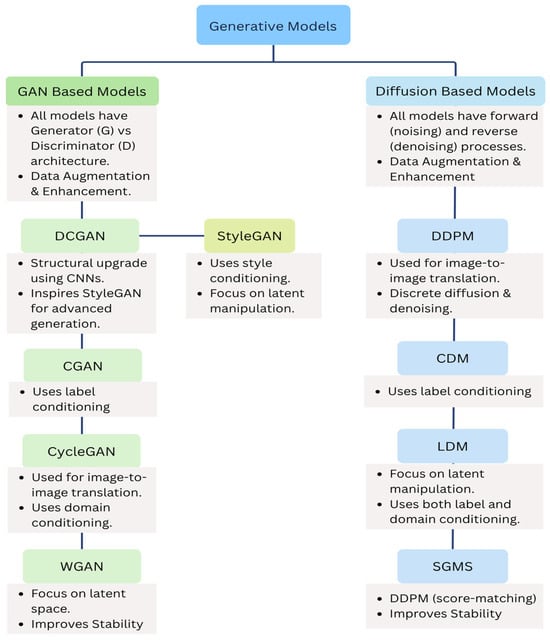
Figure 9.
Similarities among various GANs and Diffusion model architectures.
7. Diffusion Models
Recent advancements in generative modeling have introduced Diffusion models as a powerful generative framework capable of learning complex data distributions, especially for their application in neurodegenerative disease diagnosis such as Alzheimer’s Disease (AD), where limited labeled data often hinders robust model development. These models show great promise in generating realistic synthetic neuroimages, enhancing diagnostic prediction, and simulating disease progression over time. Diffusion models work by gradually adding Gaussian noise to clean data over some time steps through a forward diffusion process and then learning to reverse this process to recover or generate clean data. Figure 10 illustrates the typical architecture of the diffusion model.
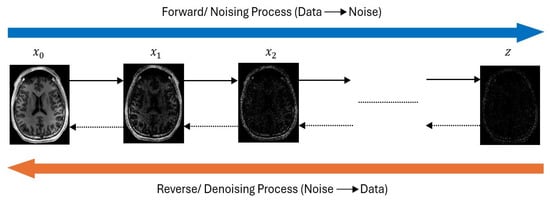
Figure 10.
Diffusion Model architecture.
7.1. Denoising Diffusion Probabilistic Models (DDPM)
DDPMs, introduced by Ho et al. (2020) [], are based on a Markovian forward process where Gaussian noise is incrementally added to input data over several time steps. The process is defined as:
where is the forward diffusion process that gradually adds noise. is gaussian distribution. , are data sample at time steps t − 1 and t, respectively. is predefined variance schedule at time step t and is identity matrix.
After that the model performs a reverse denoising process using a neural network trained to recover the original data.
where is the reverse denoising distribution parameterized by neural network , is noisy image at diffusion step t and denoised image one step backward. and represents predicted mean of the denoised sample and predicted variance at step t − 1, respectively.
7.2. Convolutional Diffusion Models (CDM)
CDM incorporates auxiliary variables such as cognitive test scores, genetic risk factors, or biomarker concentrations into the denoising process. The study by [] used this model to predict future MRI degeneration patterns using baseline imaging alongside cerebrospinal fluid metrics like Aβ, total tau, and phosphorylated tau.
7.3. Latentl Diffusion Models (LDM)
To reduce computational demands and handle large high-resolution data efficiently, Latent diffusion models compressed the image before applying the diffusion process. Instead of working directly on the high-resolution data, these models encode input images into a lower-dimensional representation and then decode the final result back into a full image. This makes efficient training and faster inference while retaining key structural details. Researchers have found these models particularly helpful when working with large-scale imaging studies [].
7.4. Score-Based Generative Models (SGMS)
Score-based diffusion models, also referred to as score-matching networks, adopt a different generative approach. Instead of learning to denoise step-by-step, they estimate the direction in which the data density increases, essentially learning the gradient of the data’s log-probability and using this information to reconstruct clean images from noisy ones. New samples are generated using Langevin dynamics []:
In Equation (8) is the noisy sample at time step t, is the updated sample at time step t − 1. is the step size or learning rate for Langevin dynamics. is data distribution at time t and is the score function. Z is the gaussian noise. Score-based methods have been explored for cleaning up noisy MRI scans or recovering missing parts of an image, especially in situations where image quality is affected by movement or poor scanner settings.
Figure 11 demonstrates the key differences among these models by comparing criteria such as input conditions, data domain, training dynamics, and output fidelity.
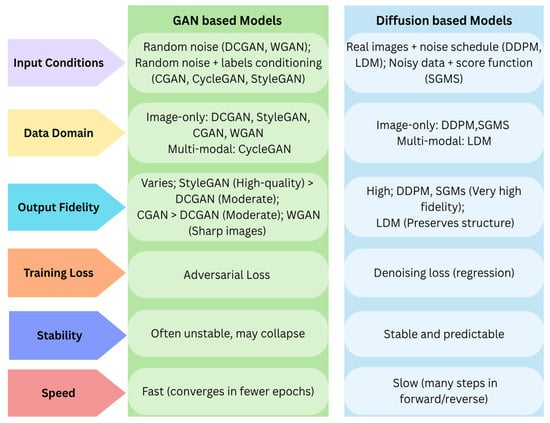
Figure 11.
Key differences between GAN and Diffusion Models.
8. Discussion and Conclusions
Alzheimer’s disease (AD) remains one of the most complex and difficult neurodegenerative disorders to diagnose, especially during its early stages when intervention can be most impactful. Accurate and timely detection is crucial, as early diagnosis allows clinicians to initiate therapeutic strategies that can slow disease progression, preserve cognitive function, and improve patients’ quality of life. This review underscores the emerging potential of generative artificial intelligence (AI), particularly generative models such as Generative Adversarial Networks (GANs) and Diffusion Models as a powerful complement to conventional neuroimaging analysis in the quest for early-stage detection. By synthesizing high-quality brain images, these models not only help overcome the challenge of limited datasets but also enhance feature extraction capabilities. This enables downstream classifiers to detect subtler neurodegenerative patterns that might be overlooked by traditional techniques.
One major challenge in Alzheimer’s diagnosis is the lack of high-quality, labeled neuroimaging data. Generative models help overcome this by producing synthetic MRI and PET scans that closely mimic real ones, enhancing data diversity and improving diagnostic accuracy. Some studies even reported up to 99.7% accuracy with GANs and high-quality metrics from Diffusion Models (e.g., SSIM > 0.92, PSNR > 30 dB). Unlike traditional reviews that just list ML models, this paper focuses on how generative approaches like DCGAN, CycleGAN, WGAN, and conditional DDPMs actively enhance early detection by augmenting data, capturing disease patterns, and modeling progression.
Generative models stand out for their ability to integrate diverse data types like MRI, PET scans, and CSF biomarkers, offering a fuller view of Alzheimer’s progression. When combined with CNNs, these models do not just aid diagnosis they elevate it. But there is a trade-off to consider. While these models often deliver excellent results, they can be hard to interpret due to a lack of transparency. In clinical settings, trust matters, and clinicians need to understand how a model reaches its decision. Unfortunately, most GANs and diffusion models still operate like black boxes. Coupled with their high computational demands, these limitations currently hinder their widespread adoption in clinical practice. Many DCGAN-based studies (e.g., [,]) generate 2D slices only, losing volumetric context and cross-slice biomarkers. Implementing 3D GAN architectures (e.g., 3D-CycleGAN [], 3D-BicycleGAN []) or spatio-temporal diffusion models are recommended to preserve full brain topology.
Even though the research landscape is promising, not all findings are consistent. Models trained on datasets like ADNI often struggle when tested on others like OASIS, raising concerns about generalizability. GAN classifiers fine-tuned on ADNI drop 10–15% accuracy on OASIS or AIBL, likely due to scanner/vendor differences. Domain adaptation methods, such as adversarial feature alignment or style-transfer approaches (e.g., Cycle GAN-based harmonization), along with fine-tuning using small subsets from target datasets, can help overcome these domain discrepancies. In some cases, adding synthetic images did not improve accuracy much, especially when the generated data lacked diversity or detail. DCGAN outputs often collapse to a few modes, leading to overfitting when used for augmentation [,]. Strategies such as using WGAN-GP or Spectral Normalization to stabilize training, enforce latent-space regularization, or switch to diffusion models which naturally sample diverse modes could mitigate these problems. Another challenge is validation. Metrics like SSIM and PSNR measure image quality, but they do not guarantee medical accuracy. Many studies still lack clinical validation through expert reviews or pathology comparisons, making it hard to assess their real-world utility. Fewer than 20% of reviewed papers report radiologist or neuropathologist agreement on synthetic images. Future research should incorporate double-blind reader studies where experts rate synthetic vs. real scans, and correlate GAN/diffusion outputs with biomarker levels or longitudinal outcomes.
To bridge the remaining gaps, future work should prioritize the development of more interpretable and adaptable generative architectures. Which could make generative models even more effective and easier to use. Transfer learning is one promising strategy. By fine-tuning models that were trained on large datasets, researchers can adapt them to new patient groups or imaging modalities with relatively little data. Integrating explainable AI (XAI) methodologies, including Grad-CAM, SHAP or visualization tools, to help clinicians understand and trust what the model is doing.
Emerging approaches like federated learning allow hospitals to train models collaboratively without sharing patient data, preserving privacy. Graph-based generative models (e.g., BG-GAN) also show promise in capturing the complex progression of Alzheimer’s. Together, these advances could make generative tools more accurate, scalable, and clinically trustworthy.
Through a comprehensive critical evaluation of the strengths and limitations of current generative methods, this review advances the state of the art by not merely summarizing existing work, but clearly outlining targeted recommendations and directions for future research. It offers a solid framework for developing next-generation diagnostic tools that are not only accurate and efficient but also interpretable, clinically practical, and ethically responsible. In doing so, it bridges technical innovation with the broader goal of enhancing early diagnosis and long-term care for individuals living with Alzheimer’s disease.
Author Contributions
Conceptualization, M.M.A. and S.L.; methodology, M.M.A.; software, M.M.A.; validation, M.M.A. and S.L.; formal analysis, M.M.A.; investigation M.M.A.; resources, M.M.A.; writing—original draft preparation, M.M.A.; writing—review and editing, M.M.A. and S.L.; visualization, M.M.A. and S.L.; supervision, S.L.; project administration, S.L.; funding acquisition, S.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
No data was created, and no specific dataset was used. The pertinent information is properly cited throughout the manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
| GAN | Generative Adversarial Network |
| MRI | Magnetic Resonance Imaging |
| MCI | Mild Cognitive Impairment |
| AD | Alzheimer’s Disease |
References
- World Health Organization. Ageing and Health; WHO (World Health Organization): Geneva, Switzerland, 2024. [Google Scholar]
- Bronzuoli, M.R.; Iacomino, A.; Steardo, L.; Scuderi, C. Targeting neuroinflammation in Alzheimer’s disease. J. Inflamm. Res. 2016, 9, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Alzheimer’s Disease International. Alzheimer’s Disease International; Alzheimer’s Disease International: London, UK, 2024. [Google Scholar]
- Alzheimer and Association. American Perspectives on Early Detection of Alzheimer’s Disease in the Era of Treatment; Alzheimer’s Association: Chicago, IL, USA, 2025. [Google Scholar]
- Uddin, J.; Shumi, S.S.; Khandoker, F.; Fariha, T. Factors Contributing to Alzheimer’s disease in Older Adult Populations: A Narrative Review. Int. J. Sci. Res. Multidiscip. Stud. 2024, 10, 76–82. [Google Scholar]
- Braak, H.; Braak, E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991, 82, 239–259. [Google Scholar] [CrossRef] [PubMed]
- Ávila-Villanueva, M.; Dolado, A.M.; Gómez-Ramírez, J.; Fernández-Blázquez, M. Brain Structural and Functional Changes in Cognitive Impairment Due to Alzheimer’s Disease. Front. Psychol. 2022, 13, 886619. [Google Scholar] [CrossRef] [PubMed]
- Jack, C.R., Jr.; Bennett, D.A.; Blennow, K.; Carrillo, M.C.; Dunn, B.; Haeberlein, S.B.; Holtzman, D.M.; Jagust, W.; Jessen, F.; Karlawish, J.; et al. NIA-AA Research Framework: Toward a biological definition of Alzheimer’s disease. Alzheimer’s Dement. 2018, 14, 535–562. [Google Scholar] [CrossRef] [PubMed]
- Goodfellow, I.J.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative Adversarial Networks. arXiv 2014, arXiv:1406.2661. [Google Scholar] [CrossRef]
- Bowles, C.; Chen, L.; Guerrero, R.; Bentley, P.; Gunn, R.; Hammers, A.; Dickie, D.A.; Hernández, M.V.; Wardlaw, J.; Rueckert, D. GAN Augmentation: Augmenting Training Data using Generative Adversarial Networks. arXiv 2018, arXiv:1810.10863. [Google Scholar] [CrossRef]
- Chartsias, A.; Joyce, T.; Giuffrida, M.V.; Tsaftaris, S.A. Multimodal MR Synthesis via Modality-Invariant Latent Representation. IEEE Trans. Med. Imaging 2018, 37, 803–814. [Google Scholar] [CrossRef] [PubMed]
- You, C.; Li, G.; Zhang, Y.; Zhang, X.; Shan, H.; Ju, S.; Zhao, Z.; Zhang, Z.; Cong, W.; Vannier, M.W.; et al. CT Super-Resolution GAN Constrained by the Identical, Residual, and Cycle Learning Ensemble (GAN-CIRCLE). IEEE Trans. Med. Imaging 2020, 39, 188–203. [Google Scholar] [CrossRef] [PubMed]
- Ho, J.; Jain, A.; Abbeel, P. Denoising Diffusion Probabilistic Models. arXiv 2020, arXiv:2006.11239. [Google Scholar] [CrossRef]
- Khandoker, F.; Uddin, J.; Fariha, T.; Shumi, S.S. Importance of Psychological Well-being after disasters in Bangladesh: A Narrative Review. Int. J. Sci. Res. Multidiscip. Stud. 2024, 10, 1–6. [Google Scholar]
- Clinic, M. Alzheimer’s Stages: How the Disease Progresses. Available online: https://www.mayoclinic.org/diseases-conditions/alzheimers-disease/in-depth/alzheimers-stages/art-20048448 (accessed on 4 July 2025).
- Ahmed, M.R.; Zhang, Y.; Feng, Z.; Lo, B.; Inan, O.T.; Liao, H. Neuroimaging and Machine Learning for Dementia Diagnosis: Recent Advancements and Future Prospects. IEEE Rev. Biomed. Eng. 2019, 12, 19–33. [Google Scholar] [CrossRef] [PubMed]
- Scheltens, P.; Fox, N.; Barkhof, F.; De Carli, C. Structural magnetic resonance imaging in the practical assessment of dementia: Beyond exclusion. Lancet Neurol. 2002, 1, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Jeong, M.; Stiles, W.R.; Choi, H.S. Neuroimaging Modalities in Alzheimer’s Disease: Diagnosis and Clinical Features. Int. J. Mol. Sci. 2022, 23, 6079. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Shan, Y.; Ding, J. A literature review of MRI techniques used to detect amyloid-beta plaques in Alzheimer’s disease patients. Ann. Palliat. Med. 2021, 10, 10062–10074. [Google Scholar] [CrossRef] [PubMed]
- Alves, G.S.; Knöchel, V.O.; Knöchel, C.; Carvalho, A.F.; Pantel, J.; Engelhardt, E.; Laks, J. Integrating Retrogenesis Theory to Alzheimer’s Disease Pathology: Insight from DTI-TBSS Investigation of the White Matter Microstructural Integrity. Biomed. Res. Int. 2015, 2015, 291658. [Google Scholar] [CrossRef] [PubMed]
- Raj, A.; Kuceyeski, A.; Weiner, M. A Network Diffusion Model of Disease Progression in Dementia. Neuron 2012, 73, 1204–1215. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.; Park, B.; Han, K. Classification of diffusion tensor images for the early detection of Alzheimer’s disease. Comput. Biol. Med. 2013, 43, 1313–1320. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Liu, M.; Lian, C.; Zhou, T.; Xia, Y.; Shen, D. Synthesizing missing PET from MRI with cycle-consistent generative adversarial networks for Alzheimer’s disease diagnosis. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2018, Proceedings of the 21st International Conference, Granada, Spain, 16–20 September 2018; Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer: Cham, Switzerland, 2018; pp. 455–463. [Google Scholar] [CrossRef]
- Han, C.; Rundo, L.; Murao, K.; Milacski, Z.Á.; Umemoto, K.; Sala, E.; Nakayama, H.; Satoh, S. GAN-based Multiple Adjacent Brain MRI Slice Reconstruction for Unsupervised Alzheimer’s Disease Diagnosis. arXiv 2019, arXiv:1906.06114. [Google Scholar]
- Shin, H.-C.; Ihsani, A.; Xu, Z.; Mandava, S.; Sreenivas, S.T.; Forster, C.; Cha, J.; Alzheimer’s Disease Neuroimaging Initiative. GANDALF: Generative Adversarial Networks with Discriminator-Adaptive Loss Fine-Tuning for Alzheimer’s Disease Diagnosis from MRI. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2020, Proceedings of the 23rd International Conference, Lima, Peru, 4–8 October 2020; Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer Science and Business Media Deutschland GmbH: Berlin, Germany, 2020; pp. 688–697. [Google Scholar] [CrossRef]
- Islam, J.; Zhang, Y. GAN-based synthetic brain PET image generation. Brain Inf. 2020, 7, 3. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Shen, Y.; Wang, S.; Lei, B. Brain MR to PET Synthesis via Bidirectional Generative Adversarial Network. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2020, Proceedings of the 23rd International Conference, Lima, Peru, 4–8 October 2020; Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer Science and Business Media Deutschland GmbH: Berlin, Germany, 2020; pp. 698–707. [Google Scholar] [CrossRef]
- Hu, S.; Yu, W.; Chen, Z.; Wang, S. Medical Image Reconstruction Using Generative Adversarial Network for Alzheimer Disease Assessment with Class-Imbalance Problem. In Proceedings of the 2020 IEEE 6th International Conference on Computer and Communications, ICCC 2020, Chengdu, China, 11–14 December 2020; pp. 1323–1327. [Google Scholar] [CrossRef]
- Zhao, Y.; Ma, B.; Jiang, P.; Zeng, D.; Wang, X.; Li, S. Prediction of Alzheimer’s Disease Progression with Multi-Information Generative Adversarial Network. IEEE J. Biomed. Health Inform. 2021, 25, 711–719. [Google Scholar] [CrossRef] [PubMed]
- Han, C.; Rundo, L.; Murao, K.; Noguchi, T.; Shimahara, Y.; Milacski, Z.A.; Koshino, S.; Sala, E.; Nakayama, H.; Satoh, S. MADGAN: Unsupervised medical anomaly detection GAN using multiple adjacent brain MRI slice reconstruction. BMC Bioinform. 2021, 22, 31. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Qiu, S.; Joshi, P.S.; Xue, C.; Killiany, R.J.; Mian, A.Z.; Chin, S.P.; Au, R. Enhancing magnetic resonance imaging-driven Alzheimer’s disease classification performance using generative adversarial learning. Alzheimer’s Res. Ther. 2021, 13, 60. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Shi, F.; Shen, D.; Liu, M.; Disease, T.A. Task-Induced Pyramid and Attention GAN for Multimodal Brain Image Imputation and Classification in Alzheimer’s Disease. IEEE J. Biomed. Health Inform. 2022, 26, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Lei, B.; Ng, M.K.; Cheung, A.C.; Shen, Y.; Wang, S. Tensorizing GAN with High-Order Pooling for Alzheimer’s Disease Assessment. arXiv 2020, arXiv:2008.00748. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Wang, S. Cross-Modal Transformer GAN: A Brain Structure-Function Deep Fusing Framework for Alzheimer’s Disease. arXiv 2022, arXiv:2206.13393. [Google Scholar]
- Thota, N.R.; Vasumathi, D. Wasserstein Gan-Gradient Penalty with Deep Transfer Learning Based Alzheimer Disease Classification on 3D Mri Scans. I-Manag. J. Image Process. 2022, 9, 9–20. [Google Scholar]
- Zhang, J.; He, X.; Qing, L.; Gao, F.; Wang, B. BPGAN: Brain PET synthesis from MRI using generative adversarial network for multi-modal Alzheimer’s disease diagnosis. Comput. Methods Programs Biomed. 2022, 217, 106676. [Google Scholar] [CrossRef] [PubMed]
- Yuan, C.; Duan, J.; Tustison, N.J.; Xu, K.; Hubbard, R.A.; Linn, K.A. ReMiND: Recovery of Missing Neuroimaging using Diffusion Models with Application to Alzheimer’s Disease. medRxiv 2023. [Google Scholar] [CrossRef]
- Huang, G.; Chen, X.; Shen, Y.; Wang, S. MR Image Super-Resolution Using Wavelet Diffusion for Predicting Alzheimer’s Disease. In Brain Informatics, Proceedings of the 16th International Conference, BI 2023, Hoboken, NJ, USA, 1–3 August 2023; Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer Science and Business Media Deutschland GmbH: Berlin, Germany, 2023; pp. 146–157. [Google Scholar] [CrossRef]
- Boyapati, N.; Tej, M.B.; Darshitha, M.; Shreya, P.; Naveen, S.N.; Gandhi, C.R.; Amrutha, V. Alzheimer’s Disease Prediction using Convolutional Neural Network (CNN) with Generative Adversarial Network (GAN). In Proceedings of the 2023 International Conference on Data Science, Agents and Artificial Intelligence, ICDSAAI 2023, Chennai, India, 21–23 December 2023. [Google Scholar] [CrossRef]
- Nguyen, H.D.; Clément, M.; Planche, V.; Mansencal, B.; Coupé, P. Deep grading for MRI-based differential diagnosis of Alzheimer’s disease and Frontotemporal dementia. Artif. Intell. Med. 2023, 144, 102636. [Google Scholar] [CrossRef] [PubMed]
- Sekhar, U.S.; Vyas, N.; Dutt, V.; Kumar, A. Multimodal Neuroimaging Data in Early Detection of Alzheimer’s Disease: Exploring the Role of Ensemble Models and GAN Algorithm. In Proceedings of the International Conference on Circuit Power and Computing Technologies, ICCPCT 2023, Kollam, India, 10–11 August 2023; pp. 1664–1669. [Google Scholar] [CrossRef]
- Hwang, U.; Kim, S.-W.; Jung, D.; Kim, S.; Seo, S.W.; Seong, J.-K.; Yoon, S.; Alzheimer’s Disease Neuroimaging Initiative. Real-world prediction of preclinical Alzheimer’s disease with a deep generative model. Artif. Intell. Med. 2023, 144, 102654. [Google Scholar] [CrossRef] [PubMed]
- Yao, W.; Shen, Y.; Nicolls, F.; Wang, S.Q. Conditional Diffusion Model-Based Data Augmentation for Alzheimer’s Prediction. In Communications in Computer and Information Science; Springer Science and Business Media Deutschland GmbH: Berlin, Germany, 2023; pp. 33–46. [Google Scholar] [CrossRef]
- Yoon, D.; Myong, Y.; Kim, Y.G.; Sim, Y.; Cho, M.; Oh, B.-M.; Kim, S. Latent diffusion model-based MRI superresolution enhances mild cognitive impairment prognostication and Alzheimer’s disease classification. Neuroimage 2024, 296, 120663. [Google Scholar] [CrossRef] [PubMed]
- Tufail, H.; Ahad, A.; Puspitasari, I.; Shayea, I.; Coelho, P.J.; Pires, I.M. Deep Learning in Smart Healthcare: A GAN-based Approach for Imbalanced Alzheimer’s Disease Classification. Procedia Comput. Sci. 2024, 241, 146–153. [Google Scholar] [CrossRef]
- Ali, I.; Saleem, N.; Alhussein, M.; Zohra, B.; Aurangzeb, K.; Haq, Q.M.U. DeepCGAN: Early Alzheimer’s detection with deep convolutional generative adversarial networks. Front. Med. 2024, 11, 1443151. [Google Scholar] [CrossRef] [PubMed]
- Shah, J.; Che, Y.; Sohankar, J.; Luo, J.; Li, B.; Su, Y.; Wu, T.; For the Alzheimer’s Disease Neuroimaging Initiative. Enhancing Amyloid PET Quantification: MRI-Guided Super-Resolution Using Latent Diffusion Models. Life 2024, 14, 1580. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Weng, Y.; Huang, Y.; Zhang, Y.; Dening, T.; Hosseini, A.A.; Xiao, W. A multi-view learning approach with diffusion model to synthesize FDG PET from MRI T1WI for diagnosis of Alzheimer’s disease. Alzheimer’s Dement. 2024, 21, e14421. [Google Scholar] [CrossRef] [PubMed]
- Dhinagar, N.J.; Thomopoulos, S.I.; Laltoo, E.; Thompson, P.M. Counterfactual MRI Generation with Denoising Diffusion Models for Interpretable Alzheimer’s Disease Effect Detection. In Proceedings of the 2024 46th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Orlando, FL, USA, 15–19 July 2024; pp. 1–6. [Google Scholar] [CrossRef]
- Gajjar, P.; Garg, M.; Desai, S.; Chhinkaniwala, H.; Sanghvi, H.A.; Patel, R.H.; Gupta, S.; Pandya, A.S. An Empirical Analysis of Diffusion, Autoencoders, and Adversarial Deep Learning Models for Predicting Dementia Using High-Fidelity MRI. IEEE Access 2024, 12, 131231–131243. [Google Scholar] [CrossRef]
- Wong, P.C.; Abdullah, S.S.; Shapiai, M.I. Exceptional performance with minimal data using a generative adversarial network for alzheimer’s disease classification. Sci. Rep. 2024, 14, 17037. [Google Scholar] [CrossRef] [PubMed]
- Shou, Q.; Cen, S.; Chen, N.-K.; Ringman, J.M.; Wen, J.; Kim, H.; Wang, D.J.; Alzheimer’s Disease Neuroimaging Initiative. Diffusion model enables quantitative CBF analysis of Alzheimer’s Disease. medRxiv 2024. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.; Shin, J. Prognosis Prediction of Alzheimer’s Disease Based on Multi-Modal Diffusion Model. In Proceedings of the 2024 18th International Conference on Ubiquitous Information Management and Communication, IMCOM 2024, Kuala Lumpur, Malaysia, 3–5 January 2024. [Google Scholar] [CrossRef]
- Ching, W.P.; Abdullah, S.S.; Shapiai, M.I.; Islam, A.K.M.M. Performance Enhancement of Alzheimer’s Disease Diagnosis Using Generative Adversarial Network. J. Adv. Res. Appl. Sci. Eng. Technol. 2025, 45, 191–201. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, L. Early Diagnosis of Alzheimer’s Disease Using Dual GAN Model with Pyramid Attention Networks; Taylor and Francis Ltd.: Oxfordshire, UK, 2024. [Google Scholar] [CrossRef]
- Ou, Z.; Jiang, C.; Pan, Y.; Zhang, Y.; Cui, Z.; Shen, D. A Prior-information-guided Residual Diffusion Model for Multi-modal PET Synthesis from MRI. In Proceedings of the Thirty-Third International Joint Conference on Artificial Intelligence Main Track, Jeju-si, Republic of Korea, 3–9 August 2024. [Google Scholar]
- Oraby, S.; Emran, A.; El-Saghir, B.; Mohsen, S. Hybrid of DSR-GAN and CNN for Alzheimer disease detection based on MRI images. Sci. Rep. 2025, 15, 12727. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Ding, C.; Jing, C.; Liu, F.; Hung, K.; Pham, H.; Mahmud, M.; Lyu, Z.; Qiao, S.; Wang, S.; et al. BG-GAN: Generative AI Enable Representing Brain Structure-Function Connections for Alzheimer’s Disease. IEEE Trans. Consum. Electron. 2025. [Google Scholar] [CrossRef]
- Gavidia-Bovadilla, G.; Kanaan-Izquierdo, S.; Mataroa-Serrat, M.; Perera-Lluna, A. Early prediction of Alzheimer’s disease using null longitudinal model-based classifiers. PLoS ONE 2017, 12, e0168011. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Zhang, J.; Adeli, E.; Shen, D. Landmark-based deep multi-instance learning for brain disease diagnosis. Med. Image Anal. 2018, 43, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Radford, A.; Metz, L.; Chintala, S. Unsupervised Representation Learning with Deep Convolutional Generative Adversarial Networks. arXiv 2015, arXiv:1511.06434. [Google Scholar]
- Mirza, M.; Osindero, S. Conditional Generative Adversarial Nets. arXiv 2014, arXiv:1411.1784. [Google Scholar] [CrossRef]
- Kazeminia, S.; Baur, C.; Kuijper, A.; Van Ginneken, B.; Navab, N.; Albarqouni, S.; Mukhopadhyay, A. GANs for medical image analysis. Artif. Intell. Med. 2020, 109, 101938. [Google Scholar] [CrossRef] [PubMed]
- Karras, T.; Laine, S.; Aila, T. A Style-Based Generator Architecture for Generative Adversarial Networks. In Proceedings of the 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–20 June 2019; pp. 4396–4405. [Google Scholar] [CrossRef]
- Arjovsky, M.; Chintala, S.; Bottou, L. Wasserstein Generative Adversarial Networks. In Proceedings of the 34th International Conference on Machine Learning, Sydney, Australia, 6–11 August 2017. [Google Scholar]
- Song, Y.; Ermon, S. Generative Modeling by Estimating Gradients of the Data Distribution. arXiv 2019, arXiv:1907.05600. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).