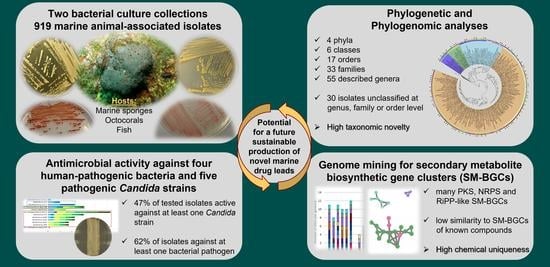
Marine Sponge and Octocoral-Associated Bacteria Show Versatile Secondary Metabolite Biosynthesis Potential and Antimicrobial Activities against Human Pathogens
Abstract
1. Introduction
2. Results
2.1. Taxonomic and Phylogenetic Diversity of the Two Marine Culture Collections
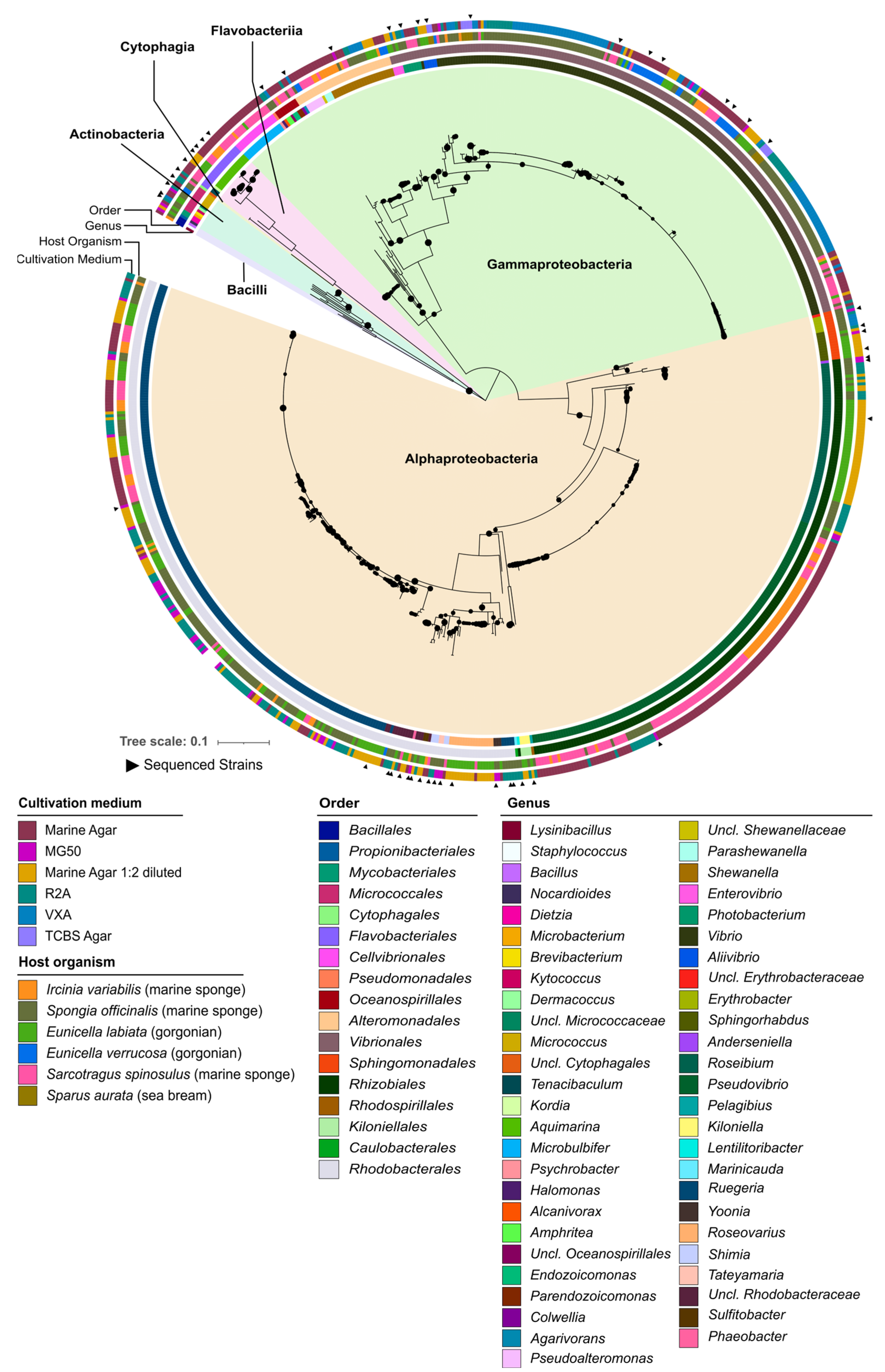
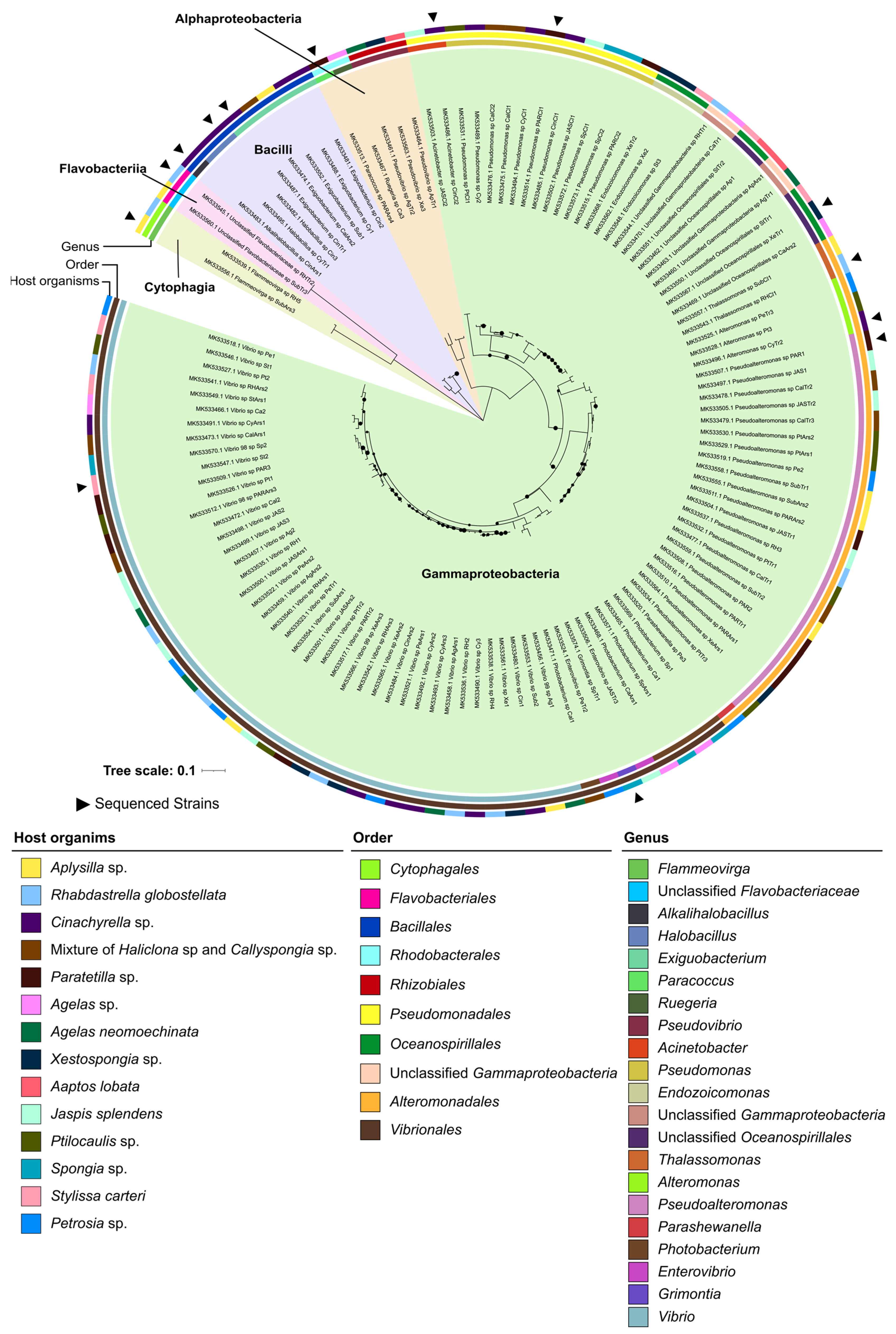
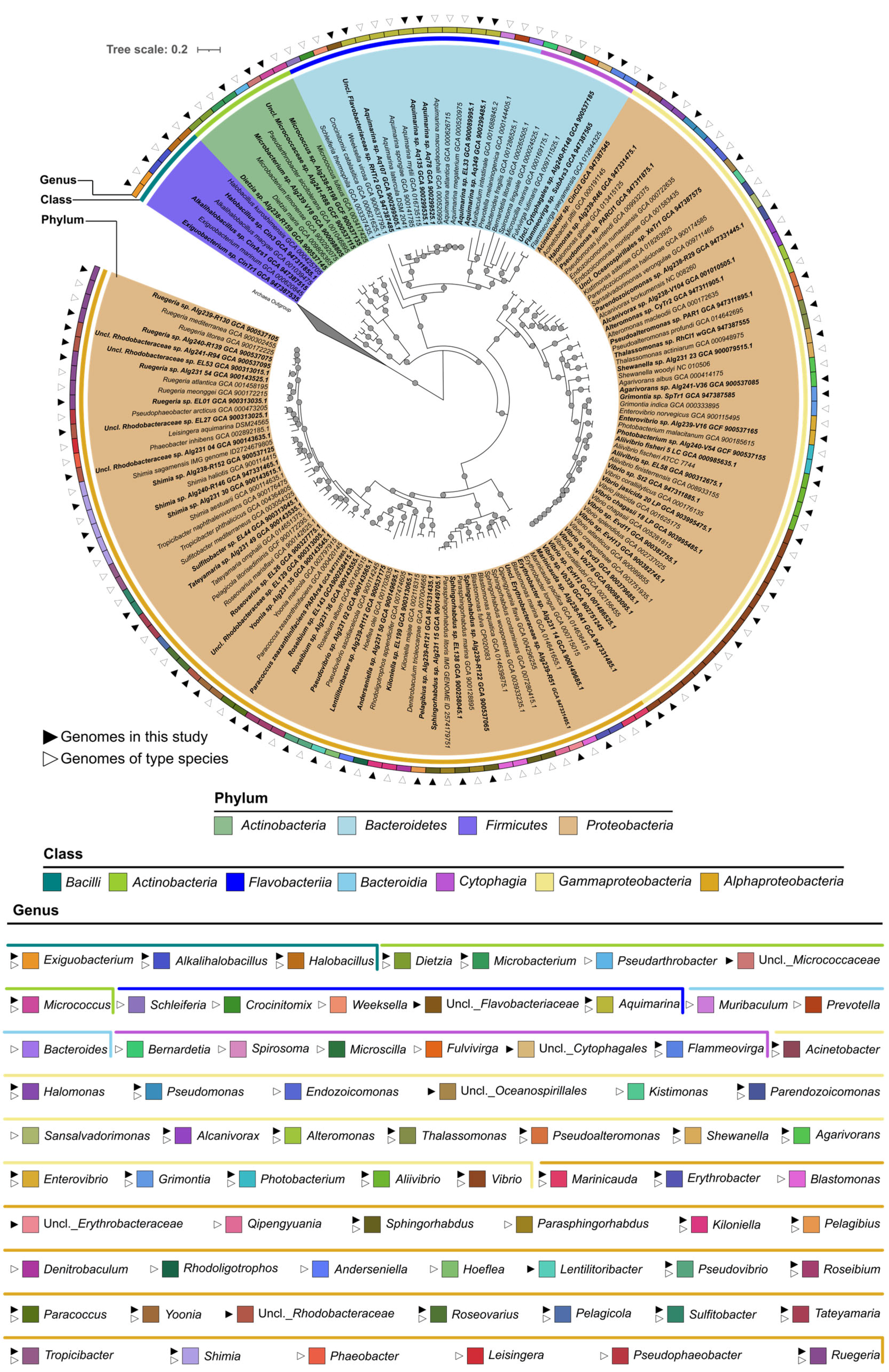
2.2. Phylogenomic Inference of Marine Host-Associated Bacteria
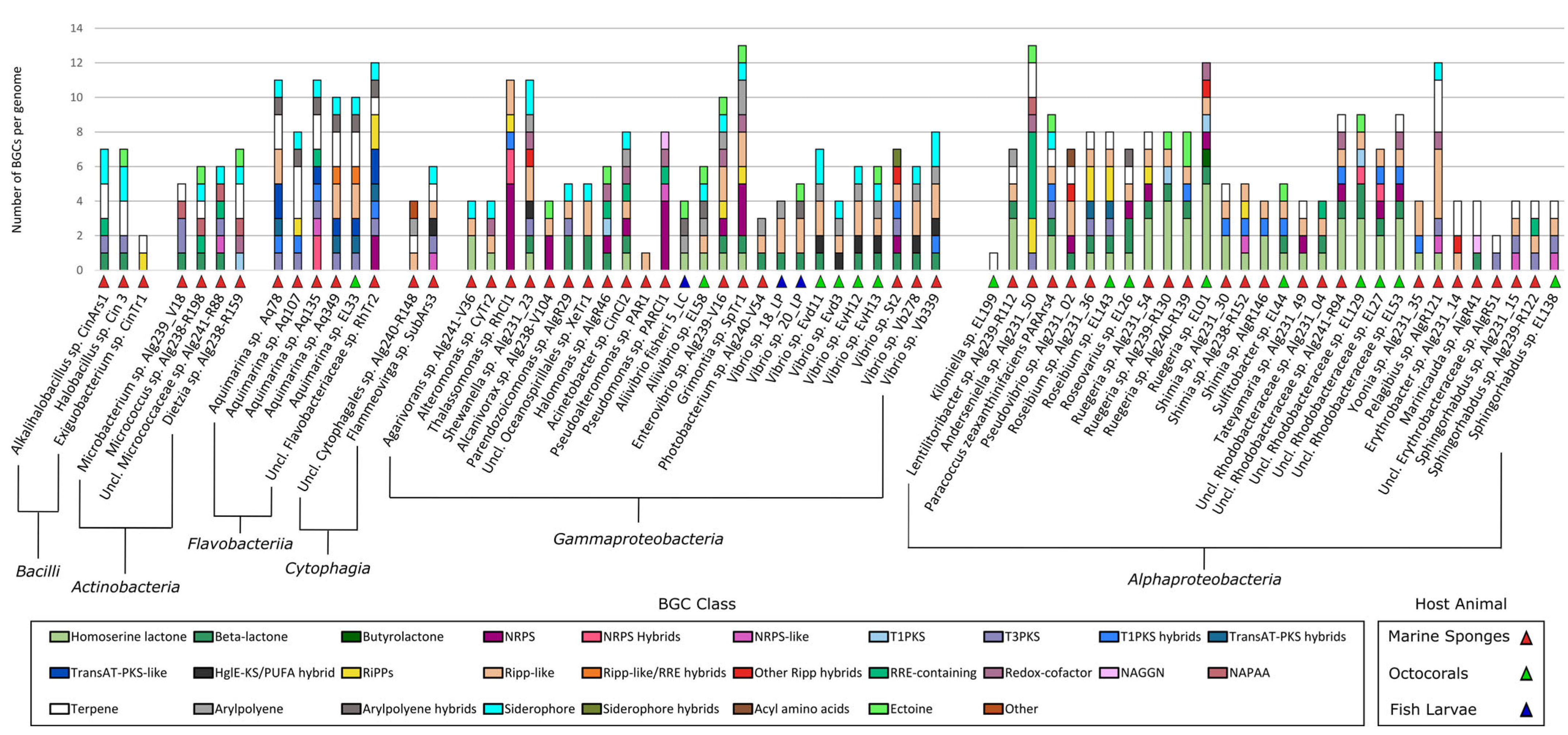
2.3. Identification of Secondary Metabolite Biosynthetic Gene Clusters (SM-BGCS) in Marine Bacterial Genomes
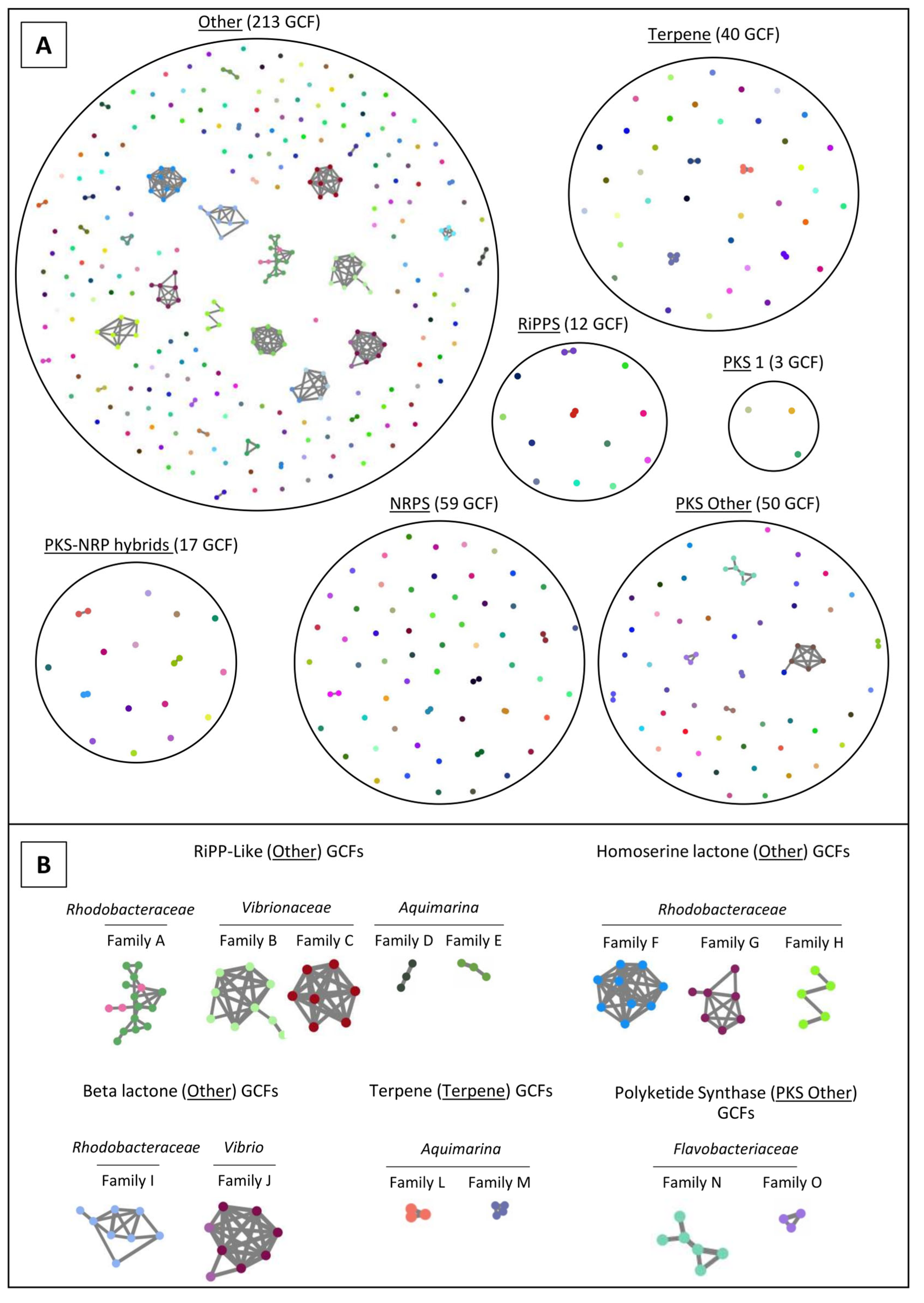
2.4. Network Analysis of SM-BGCs
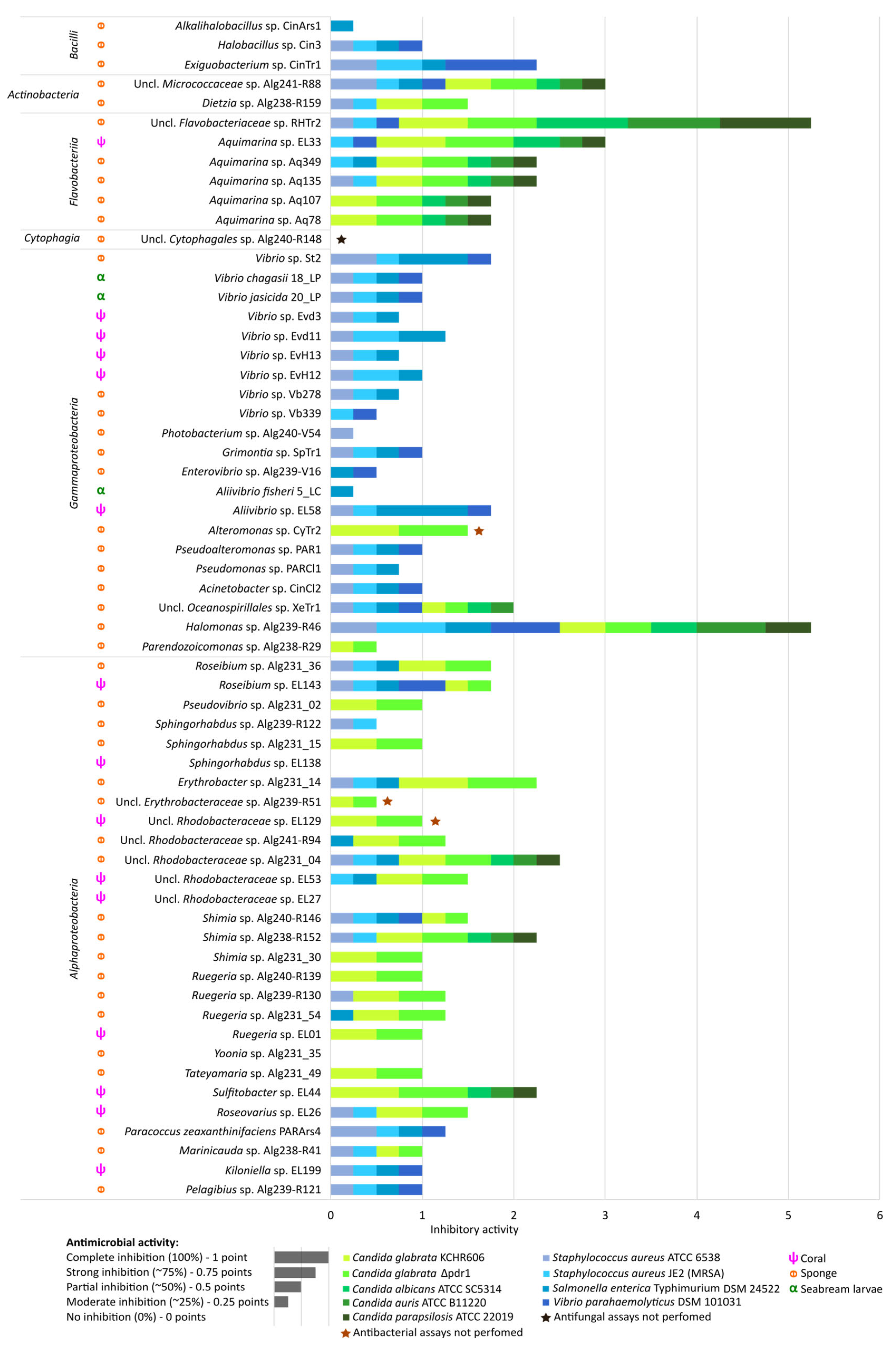
2.5. Antimicrobial Activities of Marine Bacterial Isolates
3. Discussion
3.1. The “MicroEcoEvo” and “EcoTech-SPONGE” Culture Collections Feature a Series of Underexplored and Taxonomically Novel Marine Bacteria
3.2. Both Culture Collections Comprise Taxa Typically Known for Their Symbiotic Lifestyles
3.3. Reduction of Carbon-Content and Prolonged Incubation Periods Promote an Increased Diversity of Culturable Marine Bacteria
3.4. The “MicroEcoEvo” and “EcoTech-SPONGE” Collections Are a Reservoir of Chemical Novelty with Most SM-BGCs Showing Little Homology to Those of Known Compounds
3.5. Taxon-Specific Differences in Antimicrobial Activity of Marine Bacteria Suggest Distinct Mechanisms Involved in Antagonism against Human-Pathogenic Bacteria versus Candida
4. Materials and Methods
4.1. The “MicroEcoEvo” and “EcoTech-SPONGE” Culture Collections
4.2. 16S rRNA Gene-Based Phylogenetic Analyses
4.3. Genome Sequencing and Assembly
4.4. Comparative Genomics and Phylogenomics Analysis
4.5. SM-BGC Identification and Network Analysis with antiSMASH and BiG-SCAPE
4.6. Marine Bacterial Isolates and Test Strains Used in Antimicrobial Assays
4.7. Cross-Streak Assays
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- O’Neil, J. Review on Antimicrobial Resistance. Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations; Amr-Review: London, UK, 2014. [Google Scholar]
- Algammal, A.M.; Hetta, H.F.; Elkelish, A.; Alkhalifah, D.H.H.; Hozzein, W.N.; Batiha, G.E.-S.; El Nahhas, N.; Mabrok, M.A. Methicillin-Resistant Staphylococcus aureus (MRSA): One Health Perspective Approach to the Bacterium Epidemiology, Virulence Factors, Antibiotic-Resistance, and Zoonotic Impact. Infect. Drug Resist. 2020, 13, 3255–3265. [Google Scholar] [CrossRef]
- Köck, R.; Becker, K.; Cookson, B.; van Gemert-Pijnen, J.E.; Harbarth, S.; Kluytmans, J.; Mielke, M.; Peters, G.; Skov, R.L.; Struelens, M.J.; et al. Methicillin-Resistant Staphylococcus aureus (MRSA): Burden of Disease and Control Challenges in Europe. Eurosurveillance 2010, 15, 19688. [Google Scholar] [CrossRef] [PubMed]
- Patra, S.D.; Mohakud, N.K.; Panda, R.K.; Sahu, B.R.; Suar, M. Prevalence and Multidrug Resistance in Salmonella enterica Typhimurium: An Overview in South East Asia. World J. Microbiol. Biotechnol. 2021, 37, 185. [Google Scholar] [CrossRef]
- Ghenem, L.; Elhadi, N.; Alzahrani, F.; Nishibuchi, M. Vibrio parahaemolyticus: A Review on Distribution, Pathogenesis, Virulence Determinants and Epidemiology. Saudi J. Med. Med. Sci. 2017, 5, 93–103. [Google Scholar] [CrossRef]
- Balasubramanian, R.; Im, J.; Lee, J.-S.; Jeon, H.J.; Mogeni, O.D.; Kim, J.H.; Rakotozandrindrainy, R.; Baker, S.; Marks, F. The Global Burden and Epidemiology of Invasive Non-Typhoidal Salmonella Infections. Hum. Vaccines Immunother. 2019, 15, 1421–1426. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Roy, S.; Behera, B.K.; Bossier, P.; Das, B.K. Acute Hepatopancreatic Necrosis Disease (AHPND): Virulence, Pathogenesis and Mitigation Strategies in Shrimp Aquaculture. Toxins 2021, 13, 524. [Google Scholar] [CrossRef]
- Nelapati, S.; Nelapati, K.; Chinnam, B.K. Vibrio parahaemolyticus—An Emerging Foodborne Pathogen—A Review. Vet. World 2011, 5, 48–62. [Google Scholar] [CrossRef]
- Turner, S.A.; Butler, G. The Candida Pathogenic Species Complex. Cold Spring Harb. Perspect. Med. 2014, 4, a019778. [Google Scholar] [CrossRef] [PubMed]
- Geddes-McAlister, J.; Shapiro, R.S. New Pathogens, New Tricks: Emerging, Drug-Resistant Fungal Pathogens and Future Prospects for Antifungal Therapeutics: Drug-Resistant Fungal Pathogens. Ann. N. Y. Acad. Sci. 2019, 1435, 57–78. [Google Scholar] [CrossRef]
- Lockhart, S.R. Candida auris and Multidrug Resistance: Defining the New Normal. Fungal Genet. Biol. 2019, 131, 103243. [Google Scholar] [CrossRef]
- Vivas, R.; Barbosa, A.A.T.; Dolabela, S.S.; Jain, S. Multidrug-Resistant Bacteria and Alternative Methods to Control Them: An Overview. Microb. Drug Resist. 2019, 25, 890–908. [Google Scholar] [CrossRef]
- Raimundo, I.; Silva, S.; Costa, R.; Keller-Costa, T. Bioactive Secondary Metabolites from Octocoral-Associated Microbes—New Chances for Blue Growth. Mar. Drugs 2018, 16, 485. [Google Scholar] [CrossRef] [PubMed]
- Anjum, K.; Abbas, S.Q.; Shah, S.A.A.; Akhter, N.; Batool, S.; Hassan, S.S. Marine Sponges as a Drug Treasure. Biomol. Ther. 2016, 24, 347–362. [Google Scholar] [CrossRef] [PubMed]
- Shetty, N.; Gupta, S. Eribulin Drug Review. South Asian J. Cancer 2014, 03, 057–059. [Google Scholar] [CrossRef] [PubMed]
- Cuevas, C.; Francesch, A. Development of Yondelis® (Trabectedin, ET-743). A Semisynthetic Process Solves the Supply Problem. Nat. Prod. Rep. 2009, 26, 322. [Google Scholar] [CrossRef]
- Pita, L.; Rix, L.; Slaby, B.M.; Franke, A.; Hentschel, U. The Sponge Holobiont in a Changing Ocean: From Microbes to Ecosystems. Microbiome 2018, 6, 46. [Google Scholar] [CrossRef]
- van de Water, J.A.J.M.; Allemand, D.; Ferrier-Pagès, C. Host-Microbe Interactions in Octocoral Holobionts—Recent Advances and Perspectives. Microbiome 2018, 6, 64. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. AntiSMASH 6.0: Improving Cluster Detection and Comparison Capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef]
- Karimi, E.; Ramos, M.; Gonçalves, J.M.S.; Xavier, J.R.; Reis, M.P.; Costa, R. Comparative Metagenomics Reveals the Distinctive Adaptive Features of the Spongia Officinalis Endosymbiotic Consortium. Front. Microbiol. 2017, 8, 2499. [Google Scholar] [CrossRef]
- Keller-Costa, T.; Lago-Lestón, A.; Saraiva, J.P.; Toscan, R.; Silva, S.G.; Gonçalves, J.; Cox, C.J.; Kyrpides, N.; Nunes da Rocha, U.; Costa, R. Metagenomic Insights into the Taxonomy, Function, and Dysbiosis of Prokaryotic Communities in Octocorals. Microbiome 2021, 9, 72. [Google Scholar] [CrossRef]
- Rodriguez Jimenez, A.; Dechamps, E.; Giaux, A.; Goetghebuer, L.; Bauwens, M.; Willenz, P.; Flahaut, S.; Laport, M.S.; George, I.F. The Sponges Hymeniacidon perlevis and Halichondria panicea Are Reservoirs of Antibiotic-producing Bacteria against Multi-drug Resistant Staphylococcus aureus. J. Appl. Microbiol. 2021, 131, 706–718. [Google Scholar] [CrossRef] [PubMed]
- Schultz, J.; Modolon, F.; Rosado, A.S.; Voolstra, C.R.; Sweet, M.; Peixoto, R.S. Methods and Strategies to Uncover Coral-Associated Microbial Dark Matter. mSystems 2022, 7, e00367-22. [Google Scholar] [CrossRef] [PubMed]
- Esteves, A.I.S.; Hardoim, C.C.P.; Xavier, J.R.; Gonçalves, J.M.S.; Costa, R. Molecular Richness and Biotechnological Potential of Bacteria Cultured from Irciniidae Sponges in the North-East Atlantic. FEMS Microbiol. Ecol. 2013, 85, 519–536. [Google Scholar] [CrossRef] [PubMed]
- Karimi, E.; Keller-Costa, T.; Slaby, B.M.; Cox, C.J.; da Rocha, U.N.; Hentschel, U.; Costa, R. Genomic Blueprints of Sponge-Prokaryote Symbiosis Are Shared by Low Abundant and Cultivatable Alphaproteobacteria. Sci. Rep. 2019, 9, 1999. [Google Scholar] [CrossRef]
- Karimi, E.; Costa, R. Isolation and Genome Sequencing of 14 Spongia sp. Bacterial Associates Expands the Taxonomic and Functional Breadth of the Cultivatable Marine Sponge Microbiome. BioRxiv 2020. [Google Scholar] [CrossRef]
- Keller-Costa, T.; Eriksson, D.; Gonçalves, J.M.S.; Gomes, N.C.M.; Lago-Lestón, A.; Costa, R. The Gorgonian Coral Eunicella labiata Hosts a Distinct Prokaryotic Consortium Amenable to Cultivation. FEMS Microbiol. Ecol. 2017, 93. [Google Scholar] [CrossRef] [PubMed]
- Sanches-Fernandes, G.M.M.; Califano, G.; Keller-Costa, T.; Castanho, S.; Soares, F.; Ribeiro, L.; Pousão-Ferreira, P.; Mata, L.; Costa, R. Draft Genome Sequence of Vibrio chagasii 18LP, Isolated from Gilthead Seabream (Sparus aurata) Larvae Reared in Aquaculture. Microbiol. Resour. Announc. 2021, 10, e00658-21. [Google Scholar] [CrossRef]
- Sanches-Fernandes, G.M.M.; Califano, G.; Keller-Costa, T.; Castanho, S.; Soares, F.; Ribeiro, L.; Pousão-Ferreira, P.; Mata, L.; Costa, R. Draft Genome Sequence of Vibrio jasicida 20LP, an Opportunistic Bacterium Isolated from Fish Larvae. Microbiol. Resour. Announc. 2021, 10, e00813-21. [Google Scholar] [CrossRef]
- Califano, G.; Franco, T.; Gonçalves, A.C.S.; Castanho, S.; Soares, F.; Ribeiro, L.; Mata, L.; Costa, R. Draft Genome Sequence of Aliivibrio fischeri Strain 5LC, a Bacterium Retrieved from Gilthead Sea Bream (Sparus aurata) Larvae Reared in Aquaculture. Genome Announc. 2015, 3, e00593-15. [Google Scholar] [CrossRef]
- Oliveira, V.; Polónia, A.R.M.; Cleary, D.F.R.; Huang, Y.M.; Voogd, N.J.; Rocha, U.N.; Gomes, N.C.M. Characterization of Putative Circular Plasmids in Sponge-associated Bacterial Communities Using a Selective Multiply-primed Rolling Circle Amplification. Mol. Ecol. Resour. 2021, 21, 110–121. [Google Scholar] [CrossRef]
- Oliveira, V.; Polónia, A.R.M.; Cleary, D.F.R.; Huang, Y.M.; de Voogd, N.J.; Keller-Costa, T.; Costa, R.; Gomes, N.C.M. Assessing the Genomic Composition, Putative Ecological Relevance and Biotechnological Potential of Plasmids from Sponge Bacterial Symbionts. Microbiol. Res. 2022, 265, 127183. [Google Scholar] [CrossRef] [PubMed]
- Cole, J.R.; Wang, Q.; Fish, J.A.; Chai, B.; McGarrell, D.M.; Sun, Y.; Brown, C.T.; Porras-Alfaro, A.; Kuske, C.R.; Tiedje, J.M. Ribosomal Database Project: Data and Tools for High Throughput RRNA Analysis. Nucleic Acids Res. 2014, 42, D633–D642. [Google Scholar] [CrossRef] [PubMed]
- Reasoner, D.J.; Geldreich, E.E. A New Medium for the Enumeration and Subculture of Bacteria from Potable Water. Appl. Env. Microbiol. 1985, 49, 1–7. [Google Scholar] [CrossRef]
- Sait, M.; Hugenholtz, P.; Janssen, P.H. Cultivation of Globally Distributed Soil Bacteria from Phylogenetic Lineages Previously Only Detected in Cultivation-Independent Surveys: Cultivation of Soil Bacteria. Environ. Microbiol. 2002, 4, 654–666. [Google Scholar] [CrossRef] [PubMed]
- Cúcio, A.C. Molecular Exploration of Bacterial Communities Associated with Azooxanthellate Gorgonians in the Coast of Algarve, South Portugal. Master’s Thesis, UALG, Faro, Portugal, 2011. [Google Scholar]
- Rodriguez-R, L.M.; Gunturu, S.; Harvey, W.T.; Rosselló-Mora, R.; Tiedje, J.M.; Cole, J.R.; Konstantinidis, K.T. The Microbial Genomes Atlas (MiGA) Webserver: Taxonomic and Gene Diversity Analysis of Archaea and Bacteria at the Whole Genome Level. Nucleic Acids Res. 2018, 46, W282–W288. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Muñoz, J.C.; Selem-Mojica, N.; Mullowney, M.W.; Kautsar, S.A.; Tryon, J.H.; Parkinson, E.I.; De Los Santos, E.L.C.; Yeong, M.; Cruz-Morales, P.; Abubucker, S.; et al. A Computational Framework to Explore Large-Scale Biosynthetic Diversity. Nat. Chem. Biol. 2020, 16, 60–68. [Google Scholar] [CrossRef]
- Ueno, K.; Uno, J.; Nakayama, H.; Sasamoto, K.; Mikami, Y.; Chibana, H. Development of a Highly Efficient Gene Targeting System Induced by Transient Repression of YKU80 Expression in Candida glabrata. Eukaryot. Cell 2007, 6, 1239–1247. [Google Scholar] [CrossRef][Green Version]
- Galkina, K.V.; Okamoto, M.; Chibana, H.; Knorre, D.A.; Kajiwara, S. Deletion of CDR1 Reveals Redox Regulation of Pleiotropic Drug Resistance in Candida glabrata. Biochimie 2020, 170, 49–56. [Google Scholar] [CrossRef]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in Vitro Evaluating Antimicrobial Activity: A Review. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef]
- Silva, S.G.; Blom, J.; Keller-Costa, T.; Costa, R. Comparative Genomics Reveals Complex Natural Product Biosynthesis Capacities and Carbon Metabolism across Host-associated and Free-living Aquimarina (Bacteroidetes, Flavobacteriaceae) Species. Env. Microbiol. 2019, 21, 4002–4019. [Google Scholar] [CrossRef]
- Silva, S.G.; Paula, P.; da Silva, J.P.; Mil-Homens, D.; Teixeira, M.C.; Fialho, A.M.; Costa, R.; Keller-Costa, T. Insights into the Antimicrobial Activities and Metabolomes of Aquimarina (Flavobacteriaceae, Bacteroidetes) Species from the Rare Marine Biosphere. Mar. Drugs 2022, 20, 423. [Google Scholar] [CrossRef]
- Helfrich, E.J.N.; Ueoka, R.; Dolev, A.; Rust, M.; Meoded, R.A.; Bhushan, A.; Califano, G.; Costa, R.; Gugger, M.; Steinbeck, C.; et al. Automated Structure Prediction of Trans-Acyltransferase Polyketide Synthase Products. Nat. Chem. Biol. 2019, 15, 813–821. [Google Scholar] [CrossRef] [PubMed]
- Dieterich, C.L.; Probst, S.I.; Ueoka, R.; Sandu, I.; Schäfle, D.; Dal Molin, M.; Minas, H.A.; Costa, R.; Oxenius, A.; Sander, P.; et al. Aquimarins, Peptide Antibiotics with Amino-Modified C-Termini from a Sponge-Derived Aquimarina sp. Bacterium. Angew. Chem. Int. Ed. Engl. 2021, 61, e202115802. [Google Scholar] [CrossRef]
- Steinert, G.; Whitfield, S.; Taylor, M.W.; Thoms, C.; Schupp, P.J. Application of Diffusion Growth Chambers for the Cultivation of Marine Sponge-Associated Bacteria. Mar. Biotechnol. 2014, 16, 594–603. [Google Scholar] [CrossRef] [PubMed]
- Munaganti, R.K.; Muvva, V.; Konda, S.; Naragani, K.; Mangamuri, U.K.; Dorigondla, K.R.; Akkewar, D. Antimicrobial Profile of Arthrobacter kerguelensis VL-RK_09 Isolated from Mango Orchards. Braz. J. Microbiol. 2016, 47, 1030–1038. [Google Scholar] [CrossRef]
- Alvarado, P.; Huang, Y.; Wang, J.; Garrido, I.; Leiva, S. Phylogeny and Bioactivity of Epiphytic Gram-Positive Bacteria Isolated from Three Co-Occurring Antarctic Macroalgae. Antonie Van Leeuwenhoek 2018, 111, 1543–1555. [Google Scholar] [CrossRef]
- Hewitt, O.H.; Díez-Vives, C.; Taboada, S. Microbial Insights from Antarctic and Mediterranean Shallow-Water Bone-Eating Worms. Polar Biol. 2020, 43, 1605–1621. [Google Scholar] [CrossRef]
- Baelum, J.; Borglin, S.; Chakraborty, R.; Fortney, J.L.; Lamendella, R.; Mason, O.U.; Auer, M.; Zemla, M.; Bill, M.; Conrad, M.E.; et al. Deep-Sea Bacteria Enriched by Oil and Dispersant from the Deepwater Horizon Spill: Enrichment of Oil Degraders from Gulf of Mexico. Environ. Microbiol. 2012, 14, 2405–2416. [Google Scholar] [CrossRef] [PubMed]
- Robinson, S.L.; Christenson, J.K.; Wackett, L.P. Biosynthesis and Chemical Diversity of β-Lactone Natural Products. Nat. Prod. Rep. 2019, 36, 458–475. [Google Scholar] [CrossRef]
- Bartz, J.-O.; Blom, J.; Busse, H.-J.; Mvie, J.B.; Hardt, M.; Schubert, P.; Wilke, T.; Goessmann, A.; Wilharm, G.; Bender, J.; et al. Parendozoicomonas haliclonae Gen. Nov. sp. Nov. Isolated from a Marine Sponge of the Genus Haliclona and Description of the Family Endozoicomonadaceae Fam. Nov. Comprising the Genera Endozoicomonas, Parendozoicomonas, and Kistimonas. Syst. Appl. Microbiol. 2018, 41, 73–84. [Google Scholar] [CrossRef]
- Neave, M.J.; Michell, C.T.; Apprill, A.; Voolstra, C.R. Endozoicomonas Genomes Reveal Functional Adaptation and Plasticity in Bacterial Strains Symbiotically Associated with Diverse Marine Hosts. Sci. Rep. 2017, 7, 40579. [Google Scholar] [CrossRef] [PubMed]
- Keller-Costa, T.; Kozma, L.; Silva, S.G.; Toscan, R.; Gonçalves, J.; Lago-Lestón, A.; Kyrpides, N.C.; Nunes da Rocha, U.; Costa, R. Metagenomics-Resolved Genomics Provides Novel Insights into Chitin Turnover, Metabolic Specialization, and Niche Partitioning in the Octocoral Microbiome. Microbiome 2022, 10, 151. [Google Scholar] [CrossRef] [PubMed]
- Raimundo, I.; Silva, R.; Meunier, L.; Valente, S.M.; Lago-Lestón, A.; Keller-Costa, T.; Costa, R. Functional Metagenomics Reveals Differential Chitin Degradation and Utilization Features across Free-Living and Host-Associated Marine Microbiomes. Microbiome 2021, 9, 43. [Google Scholar] [CrossRef] [PubMed]
- Sweet, M.; Villela, H.; Keller-Costa, T.; Costa, R.; Romano, S.; Bourne, D.G.; Cárdenas, A.; Huggett, M.J.; Kerwin, A.H.; Kuek, F.; et al. Insights into the Cultured Bacterial Fraction of Corals. mSystems 2021, 6, e01249-20. [Google Scholar] [CrossRef] [PubMed]
- Suantika, G. The Use of Indigenous Probiotic Halomonas aquamarina and Shewanella algae for White Shrimp (Litopenaeus vannamei Boone) Hatchery Productivity in Zero Water Discharge System. J. Aquac. Res. Dev. 2013, 04, 1000194. [Google Scholar] [CrossRef]
- Velmurugan, S.; Raman, K.; Thanga Viji, V.; Donio, M.B.S.; Adlin Jenifer, J.; Babu, M.M.; Citarasu, T. Screening and Characterization of Antimicrobial Secondary Metabolites from Halomonas salifodinae MPM-TC and Its in Vivo Antiviral Influence on Indian White Shrimp Fenneropenaeus indicus against WSSV Challenge. J. King Saud Univ. Sci. 2013, 25, 181–190. [Google Scholar] [CrossRef]
- Rosado, P.M.; Leite, D.C.A.; Duarte, G.A.S.; Chaloub, R.M.; Jospin, G.; Nunes da Rocha, U.; Saraiva, J.P.; Dini-Andreote, F.; Eisen, J.A.; Bourne, D.G.; et al. Marine Probiotics: Increasing Coral Resistance to Bleaching through Microbiome Manipulation. ISME J. 2019, 13, 921–936. [Google Scholar] [CrossRef]
- Dat, T.T.H.; Steinert, G.; Cuc, N.T.K.; Smidt, H.; Sipkema, D. Bacteria Cultivated From Sponges and Bacteria Not Yet Cultivated From Sponges—A Review. Front. Microbiol. 2021, 12, 737925. [Google Scholar] [CrossRef] [PubMed]
- Offret, C.; Desriac, F.; Le Chevalier, P.; Mounier, J.; Jégou, C.; Fleury, Y. Spotlight on Antimicrobial Metabolites from the Marine Bacteria Pseudoalteromonas: Chemodiversity and Ecological Significance. Mar. Drugs 2016, 14, 129. [Google Scholar] [CrossRef] [PubMed]
- Chau, R.; Pearson, L.A.; Cain, J.; Kalaitzis, J.A.; Neilan, B.A. A Pseudoalteromonas Clade with Remarkable Biosynthetic Potential. Appl. Env. Microbiol. 2021, 87, e02604-20. [Google Scholar] [CrossRef]
- Rodrigues, G.N.; Lago-Lestón, A.; Costa, R.; Keller-Costa, T. Draft Genome Sequence of Labrenzia sp. Strain EL143, a Coral-Associated Alphaproteobacterium with Versatile Symbiotic Living Capability and Strong Halogen Degradation Potential. Genome Announc. 2018, 6, e00132-18. [Google Scholar] [CrossRef] [PubMed]
- Pujalte, M.J.; Carmen Macián, M.; Arahal, D.R.; Garay, E. Stappia alba sp. Nov., Isolated from Mediterranean Oysters. Syst. Appl. Microbiol. 2005, 28, 672–678. [Google Scholar] [CrossRef] [PubMed]
- Raj Sharma, A.; Zhou, T.; Harunari, E.; Oku, N.; Trianto, A.; Igarashi, Y. Labrenzbactin from a Coral-Associated Bacterium Labrenzia sp. J. Antibiot. 2019, 72, 634–639. [Google Scholar] [CrossRef]
- Kačar, D.; Schleissner, C.; Cañedo, L.M.; Rodríguez, P.; de la Calle, F.; Galán, B.; García, J.L. Genome of Labrenzia sp. PHM005 Reveals a Complete and Active Trans-AT PKS Gene Cluster for the Biosynthesis of Labrenzin. Front. Microbiol. 2019, 10, 2561. [Google Scholar] [CrossRef]
- Couceiro, J.F.; Keller-Costa, T.; Marques, M.; Kyrpides, N.C.; Woyke, T.; Whitman, W.B.; Costa, R. The Roseibium album (Labrenzia alba) Genome Possesses Multiple Symbiosis Factors Possibly Underpinning Host-Microbe Relationships in the Marine Benthos. Microbiol. Resour. Announc. 2021, 10, e00320-21. [Google Scholar] [CrossRef] [PubMed]
- Parte, A.C.; Sardà Carbasse, J.; Meier-Kolthoff, J.P.; Reimer, L.C.; Göker, M. List of Prokaryotic Names with Standing in Nomenclature (LPSN) Moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 2020, 70, 5607–5612. [Google Scholar] [CrossRef] [PubMed]
- Bethesda (MD); National Library of Medicine (US); National Center for Biotechnology Information National Center for Biotechnology Information (NCBI)[Internet]. Available online: https://www.ncbi.nlm.nih.gov/ (accessed on 28 August 2022).
- Peixoto, R.S.; Rosado, P.M.; de Leite, D.C.A.; Rosado, A.S.; Bourne, D.G. Beneficial Microorganisms for Corals (BMC): Proposed Mechanisms for Coral Health and Resilience. Front. Microbiol. 2017, 8, 341. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed, A.K.; Hothersall, J.; Cooper, S.M.; Stephens, E.; Simpson, T.J.; Thomas, C.M. Characterization of the Mupirocin Biosynthesis Gene Cluster from Pseudomonas Fluorescens NCIMB 10586. Chem. Biol. 2003, 10, 419–430. [Google Scholar] [CrossRef] [PubMed]
- Sudek, S.; Lopanik, N.B.; Waggoner, L.E.; Hildebrand, M.; Anderson, C.; Liu, H.; Patel, A.; Sherman, D.H.; Haygood, M.G. Identification of the Putative Bryostatin Polyketide Synthase Gene Cluster from “Candidatus Endobugula Sertula”, the Uncultivated Microbial Symbiont of the Marine Bryozoan Bugula neritina. J. Nat. Prod. 2007, 70, 67–74. [Google Scholar] [CrossRef]
- Chakraborty, K.; Kizhakkekalam, V.K.; Joy, M. Chemical Mining of Heterotrophic Shewanella algae Reveals Anti-Infective Potential of Macrocyclic Polyketides against Multidrug-Resistant Pathogens. Bioorgan. Chem. 2021, 108, 104533. [Google Scholar] [CrossRef] [PubMed]
- Lemaire, O.N.; Méjean, V.; Iobbi-Nivol, C. The Shewanella Genus: Ubiquitous Organisms Sustaining and Preserving Aquatic Ecosystems. FEMS Microbiol. Rev. 2020, 44, 155–170. [Google Scholar] [CrossRef] [PubMed]
- Mitra, S.; Gachhui, R.; Mukherjee, J. Enhanced Biofilm Formation and Melanin Synthesis by the Oyster Settlement-Promoting Shewanella colwelliana Is Related to Hydrophobic Surface and Simulated Intertidal Environment. Biofouling 2015, 31, 283–296. [Google Scholar] [CrossRef] [PubMed]
- Vijayan, V.; Jasmin, C.; Anas, A.; Parakkaparambil Kuttan, S.; Vinothkumar, S.; Perunninakulath Subrayan, P.; Nair, S. Sponge-Associated Bacteria Produce Non-Cytotoxic Melanin Which Protects Animal Cells from Photo-Toxicity. Appl. Biochem. Biotechnol. 2017, 183, 396–411. [Google Scholar] [CrossRef]
- Cámara-Ruiz, M.; Balebona, M.C.; Moriñigo, M.Á.; Esteban, M.Á. Probiotic Shewanella putrefaciens (SpPdp11) as a Fish Health Modulator: A Review. Microorganisms 2020, 8, 1990. [Google Scholar] [CrossRef]
- Interaminense, J.A.; Vogeley, J.L.; Gouveia, C.K.; Portela, R.W.S.; Oliveira, J.P.; Andrade, H.A.; Peixoto, S.M.; Soares, R.B.; Buarque, D.S.; Bezerra, R.S. In Vitro and in Vivo Potential Probiotic Activity of Bacillus subtilis and Shewanella algae for Use in Litopenaeus vannamei Rearing. Aquaculture 2018, 488, 114–122. [Google Scholar] [CrossRef]
- Thetsana, C.; Ijichi, S.; Kaweewan, I.; Nakagawa, H.; Kodani, S. Heterologous Expression of a Cryptic Gene Cluster from a Marine Proteobacterium Thalassomonas actiniarum Affords New Lanthipeptides Thalassomonasins A and B. J. Appl. Microbiol. 2022, 132, 3629–3639. [Google Scholar] [CrossRef]
- Joerger, R. Alternatives to Antibiotics: Bacteriocins, Antimicrobial Peptides and Bacteriophages. Poult. Sci. 2003, 82, 640–647. [Google Scholar] [CrossRef]
- Li, Y.; Rebuffat, S. The Manifold Roles of Microbial Ribosomal Peptide–Based Natural Products in Physiology and Ecology. J. Biol. Chem. 2020, 295, 34–54. [Google Scholar] [CrossRef] [PubMed]
- Martens, T.; Gram, L.; Grossart, H.-P.; Kessler, D.; Müller, R.; Simon, M.; Wenzel, S.C.; Brinkhoff, T. Bacteria of the Roseobacter Clade Show Potential for Secondary Metabolite Production. Microb. Ecol. 2007, 54, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Buchan, A.; Mitchell, A.; Cude, W.N.; Campagna, S. Acyl-Homoserine Lactone-Based Quorum Sensing in Members of the Marine Bacterial Roseobacter Clade: Complex Cell-to-Cell Communication Controls Multiple Physiologies. In Stress and Environmental Regulation of Gene Expression and Adaptation in Bacteria; de Bruijn, F.J., Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2016; pp. 225–233. ISBN 978-1-119-00481-3. [Google Scholar]
- Pais, P.; Galocha, M.; Viana, R.; Cavalheiro, M.; Pereira, D.; Teixeira, M.C. Microevolution of the Pathogenic Yeasts Candida albicans and Candida glabrata during Antifungal Therapy and Host Infection. Microb. Cell 2019, 6, 142–159. [Google Scholar] [CrossRef] [PubMed]
- Pristov, K.E.; Ghannoum, M.A. Resistance of Candida to Azoles and Echinocandins Worldwide. Clin. Microbiol. Infect. 2019, 25, 792–798. [Google Scholar] [CrossRef] [PubMed]
- Shanthakumar, S.P.; Duraisamy, P.; Vishwanath, G.; Selvanesan, B.C.; Ramaraj, V.; Vasantharaj David, B. Broad Spectrum Antimicrobial Compounds from the Bacterium Exiguobacterium mexicanum MSSRFS9. Microbiol. Res. 2015, 178, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Romano, S.; Jackson, S.; Patry, S.; Dobson, A. Extending the “One Strain Many Compounds” (OSMAC) Principle to Marine Microorganisms. Mar. Drugs 2018, 16, 244. [Google Scholar] [CrossRef] [PubMed]
- Califano, G.; Castanho, S.; Soares, F.; Ribeiro, L.; Cox, C.J.; Mata, L.; Costa, R. Molecular Taxonomic Profiling of Bacterial Communities in a Gilthead Seabream (Sparus aurata) Hatchery. Front. Microbiol. 2017, 8, 204. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (ITOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Inkscape Project Inkscape. Available online: https://inkscape.org (accessed on 26 August 2021).
- Keller-Costa, T.; Silva, R.; Lago-Lestón, A.; Costa, R. Genomic Insights into Aquimarina sp. Strain EL33, a Bacterial Symbiont of the Gorgonian Coral Eunicella labiata. Genome Announc. 2016, 4, e00855-16. [Google Scholar] [CrossRef] [PubMed]
- Franco, T.; Califano, G.; Gonçalves, A.C.S.; Cúcio, C.; Costa, R. Draft Genome Sequence of Vibrio sp. Strain Evh12, a Bacterium Retrieved from the Gorgonian Coral Eunicella verrucosa. Genome Announc. 2016, 4, e01729-15. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Chen, I.-M.A.; Chu, K.; Palaniappan, K.; Ratner, A.; Huang, J.; Huntemann, M.; Hajek, P.; Ritter, S.; Varghese, N.; Seshadri, R.; et al. The IMG/M Data Management and Analysis System v.6.0: New Tools and Advanced Capabilities. Nucleic Acids Res. 2021, 49, D751–D763. [Google Scholar] [CrossRef]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2012, 41, D36–D42. [Google Scholar] [CrossRef] [PubMed]
- Arkin, A.P.; Cottingham, R.W.; Henry, C.S.; Harris, N.L.; Stevens, R.L.; Maslov, S.; Dehal, P.; Ware, D.; Perez, F.; Canon, S.; et al. KBase: The United States Department of Energy Systems Biology Knowledgebase. Nat. Biotechnol. 2018, 36, 566–569. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Kautsar, S.A.; Blin, K.; Shaw, S.; Navarro-Muñoz, J.C.; Terlouw, B.R.; van der Hooft, J.J.J.; van Santen, J.A.; Tracanna, V.; Suarez Duran, H.G.; Pascal Andreu, V.; et al. MIBiG 2.0: A Repository for Biosynthetic Gene Clusters of Known Function. Nucleic Acids Res. 2019, 48, D454–D458. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Almeida, J.F.; Marques, M.; Oliveira, V.; Egas, C.; Mil-Homens, D.; Viana, R.; Cleary, D.F.R.; Huang, Y.M.; Fialho, A.M.; Teixeira, M.C.; et al. Marine Sponge and Octocoral-Associated Bacteria Show Versatile Secondary Metabolite Biosynthesis Potential and Antimicrobial Activities against Human Pathogens. Mar. Drugs 2023, 21, 34. https://doi.org/10.3390/md21010034
Almeida JF, Marques M, Oliveira V, Egas C, Mil-Homens D, Viana R, Cleary DFR, Huang YM, Fialho AM, Teixeira MC, et al. Marine Sponge and Octocoral-Associated Bacteria Show Versatile Secondary Metabolite Biosynthesis Potential and Antimicrobial Activities against Human Pathogens. Marine Drugs. 2023; 21(1):34. https://doi.org/10.3390/md21010034
Chicago/Turabian StyleAlmeida, João F., Matilde Marques, Vanessa Oliveira, Conceição Egas, Dalila Mil-Homens, Romeu Viana, Daniel F. R. Cleary, Yusheng M. Huang, Arsénio M. Fialho, Miguel C. Teixeira, and et al. 2023. "Marine Sponge and Octocoral-Associated Bacteria Show Versatile Secondary Metabolite Biosynthesis Potential and Antimicrobial Activities against Human Pathogens" Marine Drugs 21, no. 1: 34. https://doi.org/10.3390/md21010034
APA StyleAlmeida, J. F., Marques, M., Oliveira, V., Egas, C., Mil-Homens, D., Viana, R., Cleary, D. F. R., Huang, Y. M., Fialho, A. M., Teixeira, M. C., Gomes, N. C. M., Costa, R., & Keller-Costa, T. (2023). Marine Sponge and Octocoral-Associated Bacteria Show Versatile Secondary Metabolite Biosynthesis Potential and Antimicrobial Activities against Human Pathogens. Marine Drugs, 21(1), 34. https://doi.org/10.3390/md21010034