Abstract
Plastid proteins are one of the main components in red algae. In order to clarify the angiotensin I converting enzyme (ACE) inhibitory peptides from red alga Palmaria sp. (Japan), we determined the plastid genome sequence. The genome possesses 205 protein coding genes, which were classified as genetic systems, ribosomal proteins, photosystems, adenosine triphosphate (ATP) synthesis, metabolism, transport, or unknown. After comparing ACE inhibitory peptides between protein sequences and a database, photosystems (177 ACE inhibitory peptides) were found to be the major source of ACE inhibitory peptides (total of 751). Photosystems consist of phycobilisomes, photosystem I, photosystem II, cytochrome complex, and a redox system. Among them, photosystem I (53) and II (51) were the major source of ACE inhibitory peptides. We found that the amino acid sequence of apcE (14) in phycobilisomes, psaA (18) and psaB (13) in photosystem I, and psbB (11) and psbC (10) in photosystem II covered a majority of bioactive peptide sequences. These results are useful for evaluating the bioactive peptides from red algae.
1. Introduction
Marine algae contain proteins, lipids, carbohydrates, vitamins, and minerals as nutrition. The amount of these elements vary depending on season and the area of production [,]. Seaweed can be used as a source of polysaccharides, such as alginate, carrageenan, and agar [,]. Asia has a long tradition of consuming seaweed and seaweed has recently become considered a health food worldwide [].
Among seaweeds, red algae contain a high amount of protein compared to green and brown algae [,]. The amount of protein varies according to environmental conditions and ranges from 7% to 30% [,]. The main components of protein in red algae are phycobiliproteins and ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco). Phycobiliproteins form the complex structure of phycobilisomes, with phycobiliproteins and chromophores that capture light energy for photosynthesis []. The chromophores are used as the antioxidant materials in this process [,]. The proteinase hydrolysate of the rod-shaped protein of phycobiliproteins and Rubisco has different bioactivities, such as inhibition of both angiotensin I converting enzyme (ACE) and dipeptidyl peptidase IV (DPP IV) [,,,,,,,,,,,]. Bioactive peptides have been reported in various protein sources [,]. The typical strategy for the identification of peptides includes a series of steps: peptide production using proteinases, preparation, inhibitory activity measurement, identification of peptide sequences, and confirmation of the activity using a synthesized peptide [,,,]. Some studies have confirmed this peptide inhibitory activity in animal experiments []. This method is useful for the identification of novel and major peptide sequences in samples. However, it is difficult to identify a small amount of peptide that has strong activity in a sample, as the peptide contributes its activity to the whole hydrolysate sample. The data for peptide sequences and inhibitory concentration (IC50) can be found in a database (http://www.uwm.edu.pl/biochemia/index.php/pl/biopep). These data were obtained from various protein sources. It has been speculated that the same value of biological activity would be expressed by peptides obtained from different sources. Therefore, it was hypothesized that finding the peptide sequences in the protein sequences from genomes would unveil functional peptides from natural sources.
In this study, we determined the complete plastid genome sequence of Palmaria sp. (Japan) and annotated protein coding genes (PCGs), which are the main source of proteins in red algae. To discover functional peptides, the relationship between protein sequences in the plastid and the database was evaluated.
2. Results and Discussion
2.1. General Features of Palmaria sp. (Japan) Plastid Genomes
The complete plastid genomes of Palmaria sp. (Japan) were determined using next-generation sequencing (NGS) methods. The contigs coding plastid were assembled using BLASTn before we obtained the draft circular plastid genome. The genes in the plastid were annotated manually and the gap or deletion in PCGs were confirmed using PCR amplification followed by Sanger sequencing using specific primers (Table S1). As a result, a total of 192,409 nt of the plastid genome was sequenced (Figure 1). The average coverage for the plastid genomes was 630×. The genome contained 205 PCGs (Table 1). The plastid sequence was deposited in DNA Data Bank of Japan (DDBJ) as AB807662.
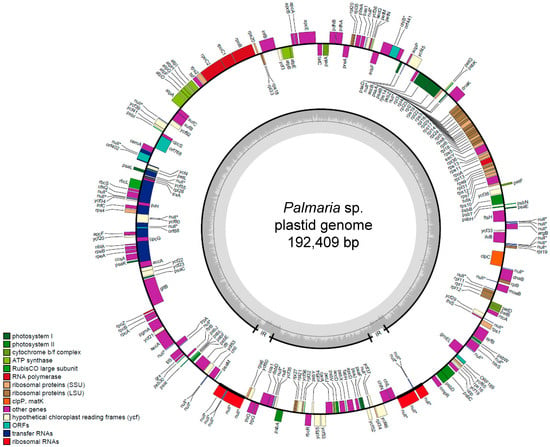
Figure 1.
The plastid genome map of Palmaria sp. (Japan).

Table 1.
Protein coding genes (PCGs) in Palmaria sp. (Japan).
When comparing the architecture of plastid genomes between Palmaria sp. (Japan) and the related species, the plastid genome was most similar to that of Palmaria palmata. This similarity was namely in terms of two introns, 205 PCGs, 33 tRNAs, and two copies of the ribosomal RNA operon (Table 2). Although the genes were completely conserved, Palmaria sp. (Japan) had a small total number of nt (192,409) and high GC content (34.6%) compared to P. palmata, which had 192,960 nt and 33.9% GC content.

Table 2.
Comparison of general plastid structure in red algae similar to Palmaria sp. (Japan).
2.2. Comparison of Amino Acid (AA) Composition between Palmaria sp. (Japan) Plastid Proteins and Proximate AA in P. palmata
The AA compositions of marine algae have been studied for a long period of time []. The AA compositions, which are an important source of protein, differ between algae species. This suggests that the differences may reflect the composition of the final product. Therefore, the AA composition of plastid proteins and the real composition in P. palmata were compared (Table 3). The AA composition was quite similar, except for aspartic acid and glycine in real protein, and isoleucine and leucine in real AA and protein. Mai et al. reported on the AA composition in various types of seaweed, and showed that the amount of aspartic acid and glycine was mostly stable in seaweed [,]. Therefore, we focused on the amounts of isoleucine and leucine. The amounts of isoleucine (9.0%) and leucine (10.1%) in plastid proteins was higher than the true AA and protein (isoleucine 5.3% and 3.7%; leucine 7.8% and 7.1%). The proportions found in plastid proteins showed that the proteins were equally expressed. Focusing on the classification of protein function, the amount of isoleucine and leucine in ribosomal protein (8.7% and 8.7%) and isoleucine in phycobilisomes (7.5%) was low. Therefore, considering the fact that ribosomal protein and phycobilisomes proteins are the main red algae proteins, the percentage of AA in the real seaweed would be close to the composition of plastid proteins. Although there is currently no information on nuclear and mitochondrial genomes, it would be expected that the proteins from these genomes would contain low amounts of isoleucine and leucine.

Table 3.
Composition of AAs in Palmaria sp. (Japan) plastid protein and AAs in P. palmata.
2.3. ACE Inhibitory Peptides in Plastid
ACE inhibitory peptides have been found in red algae proteins, which are namely the rod-like proteins of phycobilisomes and Rubisco, because these are the major components of soluble red algae proteins []. The increase in accessibility to the protein was previously studied [,]. However, 205 PCGs exist in the Palmaria sp. (Japan) plastid genome, which indicates that the insoluble or membrane proteins have potential as a source of bioactive peptides. Therefore, we screened the plastid proteins to confirm the possibility of using them as bioactive peptides. ACE inhibitory tripeptides with IC50 less than 20 μM were extracted from the biopep-uwm database, and a total of 89 peptides were selected. Although di-, tetra-, or longer peptides with ACE inhibitory activity were deposited in the database, we employed the tripeptide database to reduce overestimation. A large proportion of these peptides consisted of proline (34 peptides) or tyrosine (20 peptides) at the C-terminus. After comparing the plastid proteins and the peptide sequences, a total of 751 ACE inhibitory peptides were found (Table 4). When the peptide sources were classified according to protein function, photosystems contained the highest number with 177 peptides, followed by metabolism (176) and ribosomal proteins (128). The smallest number of peptides were involved in ATP synthesis (28), according to functional classification. This was due to a small proportion of total AAs involved in ATP synthesis.

Table 4.
Angiotensin I converting enzyme (ACE) inhibitory peptides from Palmaria sp. (Japan) plastid.
2.4. ACE Inhibitory Peptides in Photosystems
It has been reported that the proteins from photosystems are the major components of soluble proteins, with these proteins containing various types of bioactive peptides []. Photosystems contain a large number of ACE inhibitory peptides (Table 4). Therefore, we investigated the peptides in photosystems. The function of photosystem proteins was classified into phycobilisomes, photosystem I, photosystem II, cytochrome complex, and redox system. Among them, photosystem I had the highest number with 53 peptides, followed by photosystem II (51), and phycobilisomes (42) (Table 5). The ratio of the number of peptides to the total AA (peptide/AA (%)) was high in photosystem I (2.00%) and photosystem II (1.98%) compared with photosystems (1.59%) and plastid (1.49%). After this, we focused on the number of ACE inhibitory peptides in proteins. We found that the proteins of apcE, psaA, psaB, psbA, psbB, and psbC possessed a large number of the peptides (Table 6). The photosystem proteinspsaA, psaB, psbA, psbB, and psbC are the components of the integral membrane proteins in photosystem I and II, which are not easily obtained through water extraction as soluble proteins. Most ACE inhibitory peptides from red algae were from soluble proteins, that is, from the rod-like proteins of phycobilisomes and Rubisco. These data are useful for finding novel bioactive peptides from red algae proteins.

Table 5.
ACE inhibitory peptides from photosystems.

Table 6.
ACE inhibitory peptide in photosystem proteins.
2.5. Comparison of ACE Inhibitory Peptides in Palmaria sp. (Japan) and P. palmata
The plastid genomes of Palmaria sp. (Japan) and P. palmata were similar, and the number of PCGs was the same (205). To clarify the differences in ACE inhibitory peptides between Palmaria sp. (Japan) and P. palmata, the ACE inhibitory peptides were compared (Table 7). A total of 742 peptides were found in P. palmata, which was less than that found in Palmaria sp. (751). The difference was due to an unknown protein that had 80 peptides in Palmaria sp. and 72 peptides in P. palmata. Although the number of peptides among the other protein functional groups was almost the same, the peptide sequences differed between these groups (Table 4; Table 7). These data are useful for selecting peptide producing proteinases.

Table 7.
ACE inhibitory peptides from P. palmata plastid.
3. Materials and Methods
3.1. Plastid Genome Construction
Palmaria sp. was collected from Usujiri, Japan in February 2012. Genomic DNA was extracted using the hexadecyltrimethylammonium bromide (CTAB) method []. The genome sequence data were generated using the GS Junior Titanium Series system (Roche). After this, the DNA library was subjected to emulsion PCR (emPCR) using the emPCR Reagents kit (Lib-A) (Roche) according to the manufacturer’s protocol. After emPCR, DNA beads were enriched and placed on a picotiter plate (Roche) before we ran generation sequencing on this DNA using the GS Junior equipment (Roche). The contigs coding plastids were assembled with BLASTn using the red algal P. palmata plastid genes as a reference (NC_031147.1). After the reassembly, a circular plastid genome was obtained. The genes coding proteins were manually annotated using RNAmmer v1.2 server (http://www.cbs.dtu.dk/services/RNAmmer/), tRNAscan-SE 2.0 (http://lowelab.ucsc.edu/tRNAscan-SE/), ORF finder (https://www.ncbi.nlm.nih.gov/orffinder/). A gap in genes was confirmed by PCR amplification and Sanger sequencing (Table S1). The annotated plastid genomes were visualized using OrganellarGenomeDraw v1.2 [].
3.2. Collection of ACE Inhibitory Peptides and Comparison with Plastid Proteins
ACE inhibitory peptides were obtained from the biopep-uwm database (http://www.uwm.edu.pl/biochemia/index.php/pl/biopep) on 28 January 2019. From the database, tripeptides with IC50 less than 20 μM were selected. The peptide sequences in plastid proteins were manually annotated.
4. Conclusions
We determined the complete plastid genome sequence of the red alga Palmaria sp. (Japan) and annotated 205 PCGs. Comparing the plastid protein sequences and ACE inhibitory peptide sequences to a database, a large part of the peptide sequences was classified into photosystems (177) and metabolism (176). Among the photosystems, the proteins from apcE, psaA, psaB, psbA, psbB, and psbC possessed a large number of the peptides. Comparing protein sequences between Palmaria sp. (Japan) and P. palmata, the number of ACE inhibitory peptides was similar, although they had a different composition of peptides. We previously prepared ACE inhibitory peptides from water-extracted dulse protein as thermolysin hydrolysate []. The peptide sequences identified were mainly from phycobiliproteins. We therefore could not identify peptides from membrane proteins such as photosystem I and II. In silico analysis showed both the potential of membrane proteins for ACE inhibitory peptides and the characteristic C-terminal structure of ACE inhibitory peptides. Digestive enzymes such as pepsin (Aps, Glu, Leu, Phe, Trp, and Tyr), chymotrypsin (Phe, Trp, and Tyr), elastase (Ala, Gly, Ile, Leu, Ser, and Val), and prolyl endopeptidase (Pro) hydrolyzed the C-terminus of proteins, and would produce ACE inhibitory peptides. We expected that peptides from membrane proteins, which were not identified in in vitro experiments, would play a role in the inhibition of high blood pressure. In addition to ACE inhibitory activity, DPP IV inhibitory peptides were also identified in red algae protein hydrolysates, and in silico analysis would apply for finding novel bioactive peptides from red algae proteins.
Supplementary Materials
The following are available online at https://www.mdpi.com/1660-3397/17/3/190/s1, Table S1: Primers for Palmaria sp. (Japan) plastid sequence, Table S2: ACE inhibitory peptide from Palmaria sp. (Japan) plastid, Table S3: ACE inhibitory peptide from Palmaria sp. (Japan) photosystems, Table S4: ACE inhibitory peptide from Palmaria palmata plastid.
Author Contributions
H.K. and H.Y. conceived and designed the research; H.K., Y.M., and H.Y. contributed to sample collection; Y.M., T.T., K.A., and Y.K. performed the experiments and analyzed the data. Y.K. and H.K. contributed to writing and editing the manuscript.
Funding
This study was partially supported by the “Regional Innovation Cluster Program (Global Type), Ministry of Education, Culture, Sports, Science and Technology, Japan”.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Holdt, S.L.; Kraan, S. Bioactive compounds in seaweed: Functional food applications and legislation. J. Appl. Phycol. 2011, 23, 543–597. [Google Scholar] [CrossRef]
- Michalak, I.; Chojnacka, K. Algae as production systems of bioactive compounds. Eng. Life Sci. 2015, 15, 160–176. [Google Scholar] [CrossRef]
- Renn, D. Biotechnology and the red seaweed polysaccharide industry: Status, needs and prospects. Trends Biotechnol. 1997, 15, 9–14. [Google Scholar] [CrossRef]
- Wijesinghe, W.A.J.P.; Jeon, Y.J. Biological activities and potential industrial applications of fucose rich sulfated polysaccharides and fucoidans isolated from brown seaweeds: A review. Carbohydr. Polym. 2012, 88, 13–20. [Google Scholar] [CrossRef]
- Maehre, H.K.; Edvinsen, G.K.; Eilertsen, K.E.; Elvevoll, E.O. Heat treatment increases the protein bioaccessibility in the red seaweed dulse (Palmaria Palmata), but not in the brown seaweed winged kelp (Alaria Esculenta). J. Appl. Phycol. 2016, 28, 581–590. [Google Scholar] [CrossRef]
- Mai, K.; Mercer, J.P.; Donlon, J. Comparative studies on the nutrition of two species of abalone, Haliotis tuberculata L. and Haliotis discus hannai Ino: II. Amino acid composition of abalone and six species of macroalgae with an assessment of their nutritional value. Aquaculture 1994, 128, 115–130. [Google Scholar] [CrossRef]
- Grote, B. Recent Developments in aquaculture of Palmaria palmata (Linnaeus) (Weber & Mohr 1805): Cultivation and uses. Rev. Aquac. 2019, 11, 25–41. [Google Scholar] [CrossRef]
- Gantt, E. Phycobilisomes. Annu. Rev. Plant Physiol. 1981, 32, 327–347. [Google Scholar] [CrossRef]
- Sato, N.; Furuta, T.; Takeda, T.; Miyabe, Y.; Ura, K.; Takagi, Y.; Yasui, H.; Kumagai, Y.; Kishimura, H. Antioxidant activity of proteins extracted from red alga dulse harvested in Japan. J. Food Biochem. 2018, 43, e12709. [Google Scholar] [CrossRef]
- Sonani, R.R.; Singh, N.K.; Kumar, J.; Thakar, D.; Madamwar, D. Concurrent purification and antioxidant activity of phycobiliproteins from Lyngbya Sp. A09DM: An antioxidant and anti-aging potential of phycoerythrin in Caenorhabditis Elegans. Process Biochem. 2014, 49, 1757–1766. [Google Scholar] [CrossRef]
- Beaulieu, L.; Sirois, M.; Tamigneaux, É. Evaluation of the in vitro biological activity of protein hydrolysates of the edible red alga, Palmaria palmata (dulse) harvested from the Gaspe coast and cultivated in tanks. J. Appl. Phycol. 2016, 28, 3101–3115. [Google Scholar] [CrossRef]
- Cao, D.; Lv, X.; Xu, X.; Yu, H.; Sun, X.; Xu, N. Purification and identification of a novel ACE inhibitory peptide from marine alga Gracilariopsis lemaneiformis protein hydrolysate. Eur. Food Res. Technol. 2017, 243, 1829–1837. [Google Scholar] [CrossRef]
- Suetsuna, K.; Nakano, T. Identification of an antihypertensive peptide from peptic digest of wakame (Undaria pinnatifida). J. Nutr. Biochem. 2000, 11, 450–454. [Google Scholar] [CrossRef]
- Fitzgerald, C.; Mora-Soler, L.; Gallagher, E.; O’Connor, P.; Prieto, J.; Soler-Vila, A.; Hayes, M. Isolation and characterization of bioactive pro-peptides with in vitro renin inhibitory activities from the macroalga Palmaria palmata. J. Agric. Food Chem. 2012, 60, 7421–7427. [Google Scholar] [CrossRef]
- Furuta, T.; Miyabe, Y.; Yasui, H.; Kinoshita, Y.; Kishimura, H. Angiotensin I converting enzyme inhibitory peptides derived from phycobiliproteins of dulse Palmaria palmata. Mar. Drugs 2016, 14, 32. [Google Scholar] [CrossRef]
- Harnedy, P.A.; FitzGerald, R.J. In vitro assessment of the cardioprotective, anti-diabetic and antioxidant potential of Palmaria palmata protein hydrolysates. J. Appl. Phycol. 2013, 25, 1793–1803. [Google Scholar] [CrossRef]
- Harnedy, P.A.; Soler-Vila, A.; Edwards, M.D.; FitzGerald, R.J. The effect of time and origin of harvest on the in vitro biological activity of Palmaria palmata protein hydrolysates. Food Res. Int. 2014, 62, 746–752. [Google Scholar] [CrossRef]
- He, H.L.; Chen, X.L.; Wu, H.; Sun, C.Y.; Zhang, Y.Z.; Zhou, B.C. High throughput and rapid screening of marine protein hydrolysates enriched in peptides with angiotensin-I-converting enzyme inhibitory activity by capillary electrophoresis. Bioresour. Technol. 2007, 98, 3499–3505. [Google Scholar] [CrossRef] [PubMed]
- Kitade, Y.; Miyabe, Y.; Yamamoto, Y.; Takeda, H.; Shimizu, T.; Yasui, H.; Kishimura, H. Structural characteristics of phycobiliproteins from red alga Mazzaella japonica. J. Food Biochem. 2018, 42, 1–10. [Google Scholar] [CrossRef]
- Miyabe, Y.; Furuta, T.; Takeda, T.; Kanno, G.; Shimizu, T.; Tanaka, Y.; Gai, Z.; Yasui, H.; Kishimura, H. Structural properties of phycoerythrin from dulse Palmaria palmata. J. Food Biochem. 2017, 41, e12301. [Google Scholar] [CrossRef]
- Cian, R.E.; Alaiz, M.; Vioque, J.; Drago, S.R. Enzyme proteolysis enhanced extraction of ACE inhibitory and antioxidant compounds (peptides and polyphenols) from Porphyra columbina residual cake. J. Appl. Phycol. 2013, 25, 1197–1206. [Google Scholar] [CrossRef]
- Cha, S.H.; Lee, K.W.; Jeon, Y.J. Screening of extracts from red algae in Jeju for potentials marine angiotensin-I converting enzyme (ACE) inhibitory activity. Algae 2006, 21, 343–348. [Google Scholar] [CrossRef]
- Korhonen, H.; Pihlanto, A. Bioactive peptides: Production and functionality. Int. Dairy J. 2006, 16, 945–960. [Google Scholar] [CrossRef]
- Sato, M.; Hosokawa, T.; Yamaguchi, T.; Nakano, T.; Muramoto, K.; Kahara, T.; Funayama, K.; Kobayashi, A.; Nakano, T. Angiotensin I-converting enzyme inhibitory peptides derived from wakame (Undaria pinnatifida) and their antihypertensive effect in spontaneously hypertensive rats. J. Agric. Food Chem. 2002, 50, 6245–6252. [Google Scholar] [CrossRef]
- Cho, C.H.; Choi, J.W.; Lam, D.W.; Kim, K.M.; Yoon, H.S. Plastid genome analysis of three nemaliophycidae red algal species suggests environmental adaptation for iron limited habitats. PLoS ONE 2018, 13, e0196995. [Google Scholar] [CrossRef] [PubMed]
- Janouskovec, J.; Liu, S.L.; Martone, P.T.; Carre, W.; Leblanc, C.; Collen, J.; Keeling, P.J. Evolution of red algal plastid genomes: Ancient architectures, introns, horizontal gene transfer, and taxonomic utility of plastid markers. PLoS ONE 2013, 8, e59001. [Google Scholar] [CrossRef] [PubMed]
- Salomaki, E.D.; Nickles, K.R.; Lane, C.E. The Ghost Plastid of Choreocolax Polysiphoniae. J. Phycol. 2015, 51, 217–221. [Google Scholar] [CrossRef]
- Lee, J.M.; Cho, C.H.; Park, S.I.; Choi, J.W.; Song, H.S.; West, J.A.; Bhattacharya, D.; Yoon, H.S. Parallel evolution of highly conserved plastid genome architecture in red seaweeds and seed plants. BMC Biol. 2016, 14, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Kim, K.M.; Yang, E.C.; Miller, K.A.; Boo, S.M.; Bhattacharya, D.; Yoon, H.S. Reconstructing the complex evolutionary history of mobile plasmids in red algal genomes. Sci. Rep. 2016, 6, 23744. [Google Scholar] [CrossRef] [PubMed]
- Murata, M.; Nakozoe, J. Production and use of marine algae in Japan. Jpn. Agric. Res. Q. 2001, 35, 281–290. [Google Scholar] [CrossRef]
- Galland-Irmouli, A.V.; Fleurence, J.; Lamghari, R.; Luçon, M.; Rouxel, C.; Barbaroux, O.; Bronowicki, J.P.; Villaume, C.; Guéant, J.L. Nutritional value of proteins from edible seaweed Palmaria palmata (dulse). J. Nutr. Biochem. 1999, 10, 353–359. [Google Scholar] [CrossRef]
- Sfriso, A.A.; Gallo, M.; Baldi, F. Phycoerythrin productivity and diversity from five red macroalgae. J. Appl. Phycol. 2018, 30, 2523–2531. [Google Scholar] [CrossRef]
- Fleurence, J.; Massiani, L.; Guyader, O.; Mabeau, S. Use of enzymatic cell wall degradation for improvement of protein extraction from Chondrus crispus, Gracilaria verrucosa and Palmaria palmata. J. Appl. Phycol. 1995, 7, 393–397. [Google Scholar] [CrossRef]
- Cota-Sánchez, J.H.; Remarchuk, K.; Ubayasena, K. Ready-to-use DNA extracted with a CTAB method adapted for herbarium specimens and mucilaginous plant tissue. Plant Mol. Biol. Rep. 2006, 24, 161–167. [Google Scholar] [CrossRef]
- Lohse, M.; Drechsel, O.; Kahlau, S.; Bock, R. OrganellarGenomeDRAW—A suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 2013, 41, 575–581. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).